Abstract
Frequent convergent evolution in phylogenetically unrelated taxa points to the importance of ecological factors during evolution, whereas convergent evolution in closely related taxa indicates the importance of favourable pre-existing characters (pre-adaptations). We investigated the transitions to arboreal life in oribatid mites (Oribatida, Acari), a group of mostly soil-living arthropods. We evaluated which general force—ecological factors, historical constraints or chance—was dominant in the evolution of arboreal life in oribatid mites. A phylogenetic study of 51 oribatid mite species and four outgroup taxa, using the ribosomal 18S rDNA region, indicates that arboreal life evolved at least 15 times independently. Arboreal oribatid mite species are not randomly distributed in the phylogenetic tree, but are concentrated among strongly sclerotized, sexual and evolutionary younger taxa. They convergently evolved a capitate sensillus, an anemoreceptor that either precludes overstimulation in the exposed bark habitat or functions as a gravity receptor. Sexual reproduction and strong sclerotization were important pre-adaptations for colonizing the bark of trees that facilitated the exploitation of living resources (e.g. lichens) and served as predator defence, respectively. Overall, our results indicate that ecological factors are most important for the observed pattern of convergent evolution of arboreal life in oribatid mites, supporting an adaptationist view of evolution.
Keywords: convergent evolution, adaptation, ecological niche, pre-adaptation, oribatid mites, constraints
1. Introduction
Convergent evolution is the development of similar traits in different evolutionary lineages. Famous examples of convergence are the similar body forms and lifestyles of marsupial and eutherian mammals, camera eyes in vertebrates and cephalopods, and electrogeneration (and perception) in the platypus and in a number of fishes, but myriad evolutionary convergences have been discovered in molecules, physiological traits and complex morphological adaptations (Morris 2003). The haemoglobins in animals, plants, protists and prokaryotes probably have an independent evolutionary origin (Hardison 1996), echolocation call design evolved convergently in bats (Jones & Teeling 2006) and eusociality evolved convergently in insects, shrimps and mammals (O'Riain et al. 2000). However, despite the large number of observed cases of convergent evolution, its importance and implications are subjects of intense debate.
For many, convergence derives from frequent and independent adaptations and thereby points to the importance of ecological factors during evolution (Sinclair et al. 2003; Langerhans & DeWitt 2004; Zhang 2006; Marks 2007). In this view, convergent evolution indicates the limits of potential evolutionary pathways, such that different evolutionary trajectories resulted in similar solutions to the same ecological problem. For example, Morris (2003) used convergent evolution as evidence for directed evolution resulting in similar endpoints, although this teleological view has been criticized (Lenski 2003). Convergent evolution may also be a product of chance, as there can be more than one optimum for a trait (Gould & Lewontin 1979; Doolittle 1981; Gould 1989; Zhang & Kumar 1997; Marks 2007). An ecological challenge could have been solved in a similar way by two or more species through chance alone. Further, convergent evolution may result from historical contingencies of certain groups of organisms. Taxa may have certain pre-existing conditions, i.e. pre-adaptations, that result in fast radiation when environmental conditions change or when new habitats are colonized.
Oribatid mites may serve as model organisms to study the hypothesis of the relative importance of adaptation versus chance during evolution. They are an evolutionarily old group that probably has existed for at least 380 million years (Norton et al. 1988), and they slowly but continuously radiated to a large number of species; about 10 000 species are described but overall 100 000 may exist (Walter & Proctor 1999; Schatz 2002).
We investigated whether independent adaptations (caused by ecological factors), pre-adaptations or chance events were the important factors for the evolution of arboreal life in oribatid mites by examining how often, and in which taxonomic groups, arboreal life evolved. Oribatid mites are primarily soil-living organisms, but numerous taxa include species with an arboreal lifestyle. Oribatid mites on trees live in particular arboreal microhabitats such as bark or lichens, on both trunks and twigs (Proctor et al. 2002; Lindo & Winchester 2006; Behan-Pelletier et al. 2007; Erdmann et al. 2007). Because arboreal oribatid mite species permanently live on trees, they probably share morphological or behavioural traits (Walter & Behan-Pelletier 1999; Karasawa & Hijii 2004) including sexual reproduction (Behan-Pelletier & Winchester 1998; Erdmann et al. 2006). In contrast to oribatid mites on trees, of which in temperate forests 95 per cent of all individuals reproduce sexually (Erdmann et al. 2006), parthenogenetic reproduction dominates in soil-living taxa; in soils of temperate forests, about 80 per cent of the individuals are parthenogenetic (Maraun et al. 2003; Cianciolo & Norton 2006; Domes et al. 2007a). In soil, both oribatid mites with cuticles hardened by sclerotization or mineralization and soft-bodied species coexist, whereas on trees species with soft-bodied adults are virtually absent. Soil oribatid mites are characterized by a large and often ornamented sensillus, whereas in tree-living species a capitate sensillus predominates (Aoki 1973). Further, in contrast to soil-living oribatid mites, many tree-living oribatid mite species feed on lichens (Seyd & Seaward 1984; Erdmann et al. 2007).
Using a large collection of oribatid mite taxa representing most of the known tree-living taxa, we investigated how often oribatid mites independently colonized trees. A molecular phylogeny was constructed using the ribosomal 18S region (18S rDNA). We also tested whether tree-living in oribatid mites is correlated with the traits noted above, sexual reproduction, a capitate sensillus and strong sclerotization, using information from the literature (e.g. Seyd & Seaward 1984; Weigmann 2006; Erdmann et al. 2007; B. Fischer 2007, unpublished data).
2. Material and methods
(a). Species and gene selection
For covering all major lineages of oribatid mites, we investigated members of five out of six commonly recognized groups (table 1): Palaeosomata (three spp. included), Enarthronota (three spp.), Mixonomata (three spp.), Desmonomata (12 spp.) and Brachypylina (30 spp.) (Grandjean 1969; Weigmann 2006); the species-poor Parhyposomata were not sampled. The middle-derivative Desmonomata and the higher Brachypylina (=Circumdehiscentiae) were most heavily sampled. All specimens were collected from the field and determined to species level. Habitat (soil or bark), reproductive mode, feeding mode and type of sensillus were extracted from the literature (Seyd & Seaward 1984; Cianciolo & Norton 2006; Weigmann 2006; Erdmann et al. 2007) or determined by us (table 1). The degree of sclerotization was estimated from the darkness of the cuticle of mature adults. Outgroup taxa, necessary for the rooting of the tree, included members of Araneae, Ricinulei (an arachnid lineage often linked to Acari), Opilioacariformes and Ixodidae (Parasitiformes). Their sequences were obtained from GenBank (see table 1 for accession numbers).
Table 1.
Phylogenetic affiliation, full species name, fragment length, GenBank accession numbers, reproductive mode, type of sensillus and degree of sclerotization of oribatid mite taxa studied and outgroups (bark-living taxa in bold). Sequences other than those labelled ‘a’ were obtained from GenBank (http://www.ncbi.nlm.nih.gov/GenBank).
| taxa | fragment length (bp) | GenBank accession number | reproductive mode | type of sensillus | degree of sclerotization | |
|---|---|---|---|---|---|---|
| outgroups | ||||||
| Araneae | Liphistius bicoloripes (Ono 1988) | 1617 | AF007104 | sexual | — | — |
| Ricinulei | Pseudocellus pearsei (Chamberlin & Ivie 1938) | 1619 | PPU91489 | sexual | — | — |
| Ixodidae | Amblyomma sphenodonti (Dubleton 1943) | 1621 | DQ507238 | sexual | — | — |
| Opilioacaridae | Opilioacarus texanus (Chamberlin & Mulaik 1942) | 1619 | AF124935 | sexual | — | — |
| Enarthronota | ||||||
| Hypochthoniidae | Hypochthonius rufulus (C. L. Koch 1835) | 1664 | EF091427 | thelytokous | non-clavate | weak |
| Eniochthoniidae | Eniochthonius minutissimus (Berlese 1903) | 1643 | EF091428 | thelytokous | non-clavate | weak |
| Lohmanniidae | Lohmannia banksi (Norton et al. 1978) | 1676 | AF022036 | thelytokous | non-clavate | weak |
| Palaeosomata | ||||||
| Acaronychidae | Stomacarus ligamentifer (Hammer 1967) | 1620 | EU433992 | sexual | non-clavate | weak |
| Zachvatkinella sp. (Lange 1954) | 1619 | EF203776 | sexual | non-clavate | weak | |
| Palaeacaridae | Palaeacarus hystricinus (Trägardh 1932) | 1618 | EF204472 | thelytokous | non-clavate | weak |
| Mixonomata | ||||||
| Phthiracaridae | Steganacarus magnus (Nicolet 1855) | 1616 | AF022040 | sexual | non-clavate | strong |
| Atropacarus striculus (C. L. Koch 1835) | 1625 | EF091416 | thelytokous | non-clavate | strong | |
| Euphthiracaroidea | Rhysotritia duplicata (Grandjean 1953) | 1624 | EF091417 | thelytokous | non-clavate | strong |
| Desmonomata | ||||||
| Camisiidae | Camisia biurus(Koch 1839) | 1624 | EF081302 | thelytokous | clavate | strong |
| Camisia horrida(Hermann 1804)a | 1624 | EU432207 | thelytokous | clavate | strong | |
| Camisia invenusta (Michael 1888)a | 1624 | EU432208 | thelytokous | clavate | strong | |
| Camisia segnis(Hermann 1804)a | 1624 | EU432209 | thelytokous | clavate | strong | |
| Camisia spinifer(C. L. Koch 1835) | 1624 | EF091420 | thelytokous | clavate | strong | |
| Platynothrus peltifer (C. L. Koch 1839) | 1624 | EF091422 | thelytokous | non-clavate | strong | |
| Crotoniidae | Crotonia brachyrostrum(Hammer 1966) | 1624 | EF081303 | sexual | clavate | strong |
| Malaconothridae | Malaconothrus gracilis v.d. (Hammen 1952) | 1624 | EF091424 | thelytokous | no sensillus | weak |
| Trimalaconothrus sp. (Berlese 1916)a | 1624 | EU432210 | thelytokous | no sensillus | weak | |
| Nothridae | Nothrus silvestris (Nicolet 1855) | 1624 | EF091425 | thelytokous | non-clavate | strong |
| Trhypochthoniidae | Archegozetes longisetosus (Aoki 1965) | 1631 | AF022027 | thelytokous | non-clavate | intermediate |
| Trhypochthonius tectorum(Berlese 1896) | 1623 | AF022041 | thelytokous | clavate | intermediate | |
| Brachypylina (non-Poronota) | ||||||
| Carabodidae | Carabodes subarcticus (Trägardh 1902) | 1623 | EF091429 | sexual | clavate | strong |
| Odontocepheus elongatus(Michael 1879)a | 1625 | EU432200 | sexual | clavate | strong | |
| Ceratoppiidae | Ceratoppia bipilis (Hermann 1804)a | 1624 | EU432204 | sexual | clavate | intermediate |
| Cepheidae | Cepheus latus (Koch 1835)a | 1624 | EU432206 | sexual | clavate | strong |
| Cymbaeremaeidae | Cymbaeremaeus cymba(Nicolet 1855)a | 1624 | EU432201 | sexual | clavate | strong |
| Scapheremaeus palustris(Sellnick 1924) | 1640 | EU433989 | sexual | clavate | strong | |
| Eremaeidae | Eueremaeus oblongus(Koch 1835)a | 1624 | EU432205 | sexual | clavate | strong |
| Eutegaeidae | Eutegaeus curviseta (Hammer 1966) | 1624 | EF081297 | sexual | non-clavate | strong |
| Liacaridae | Adoristes poppei (Oudemans 1906)a | 1624 | EU432202 | sexual | clavate | strong |
| Neoliodidae | Liodessp. (Heyden 1829) | 1625 | AF022035 | sexual | clavate | strong |
| Poroliodes farinosus(Koch 1839) | 1624 | EF203779 | sexual | clavate | strong | |
| Xenillidae | Xenillus discrepans(Grandjean 1936)a | 1624 | EU432203 | sexual | clavate | strong |
| Brachypylina (Poronota) | ||||||
| Achipteriidae | Achipteria coleoptrata (Linnaeus 1758) | 1624 | EF091418 | sexual | non-clavate | strong |
| Ceratozetidae | Oromurcia sudetica (Willmann 1939)a | 1625 | EU432194 | sexual | non-clavate | strong |
| Trichoribates trimaculatus(Koch 1835)a | 1625 | EU432195 | sexual | clavate | strong | |
| Chamobatidae | Chamobates pusillus (Berlese 1895)a | 1624 | EU432188 | sexual | non-clavate | strong |
| Chamobates subglobulus (Oudemans 1900)a | 1624 | EU432190 | sexual | non-clavate | strong | |
| Chamobates voigtsi (Oudemans 1902)a | 1624 | EU432189 | sexual | non-clavate | strong | |
| Eremaeozetidae | Eremaeozetessp. (Berlese 1913)a | 1639 | EU432187 | sexual | clavate | strong |
| Galumnidae | Galumna lanceata (Oudemans 1900)a | 1625 | EU432197 | sexual | non-clavate | strong |
| Humerobatidae | Humerobates rostrolamellatus(Grandjean 1936)a | 1624 | EU432196 | sexual | clavate | strong |
| Hydrozetidae | Hydrozetes lacustris (Michael 1882) | 1624 | EU433987 | thelytokous | non-clavate | intermediate |
| Oribatulidae | Phauloppia lucorum(Koch 1841)a | 1648 | EU432198 | sexual | clavate | strong |
| Oribatula tibialis (Nicolet 1855) | 1651 | EU433990 | sexual | non-clavate | strong | |
| Phenopelopsidae | Eupelops acromios(Hermann 1804)a | 1624 | EU432192 | sexual | clavate | strong |
| Eupelops plicatus (Koch 1835) | 1623 | EF091419 | sexual | non-clavate | strong | |
| Punctoribatidae | Mycobates parmeliae(Michael 1884)a | 1624 | EU432191 | sexual | clavate | strong |
| Symbioribatidae | Scheloribates ascendens | 1627 | EU432199 | sexual | clavate | strong |
| (Weigmann & Wunderle 1990)a | ||||||
| Tectocepheidae | Tectocepheus velatus (Michael 1880) | 1628 | EF093781 | thelytokous | clavate | intermediate |
| Tegoribatidae | Lepidozetes singularis(Berlese 1910)a | 1625 | EU432193 | sexual | clavate | strong |
aSpecies sequenced for this study.
(b). Sample preparation, PCR and sequencing
DNA was extracted from single individuals. Each mite was placed in an Eppendorf tube, frozen in liquid nitrogen and crushed with a plastic rod. Total DNA was extracted using Qiagen DNeasy Kit for animal tissues according to the manufacturer's protocol (elution was performed in 30 µl instead of 400 µl; Qiagen, Germany).
Amplifications were performed using the primers 18Sforward (5′-TACCTGGTTGATCCTGCCAG-3′) and 18Sreverse (5′-TAATGATCCTTCCGCAGGTTCAC-3′) (modified after Turbeville et al. 1991) in 25 µl volumes containing 0.5–0.7 µl of each primer (100 pmol µl−1), 5–8 µl DNA and 12.5 µl HotStarTaq Mastermix (1.25 U HotStarTaq polymerase, 100 µM of each dNTP and 7.5 mM MgCl2 buffer solution; Qiagen). PCR conditions were as follows: initial activation at 95°C for 15 min, 34 amplification cycles (95°C for 45 s, 57°C for 1 min and 72°C for 1 min); final elongation at 72°C (10 min).
PCR products were visualized on 1 per cent agarose gels and purified using QIAquick PCR Purification Kit (Qiagen); PCR products were directly sequenced by Macrogen Inc. (Seoul, South Korea) using the additional primers 18S554f (5′-AAGTCTGGTGCCAGCAGCCGC-3′), 18S1282r (5′-TCACTCCACCAACTAAGAACGGC-3′), 18S1150f (5′-ATTGACGGAAGGGCACCACCAG-3′) and 18S614r (5′-TCCAACTACGAGCTTTTTAACC-3′) (modified after Turbeville et al. 1991). All sequences are available at GenBank (see table 1 for accession numbers).
(c). Alignment and phylogenetic analyses
DNA sequences of the ribosomal 18S region were aligned using the default settings in ClustalX (Thompson et al. 1994, 1997); the alignment was modified by eye since gaps occurred. The evolutionary model parameters were determined with Modeltest 3.7 (Posada & Crandall 1998) using a hierarchical likelihood ratio test. The model of evolution was GTR + I + G (Tamura & Nei 1993) with base frequencies A = 0.2567, C = 0.2246, G = 0.2611, gamma distribution shape parameter α = 0.5050 for four categories of among-site variation and fraction of invariant sites I = 0.4170. Substitution rates were estimated as A–C = 1.1382, A–T = 2.4404, C–G = 0.6364 and G–T = 1.0, A–G = 3.0285 and C–T = 4.8970. This model of evolution was used to construct the neighbour joining (NJ) and maximum-likelihood (ML) trees.
To test whether there is a phylogenetic signal in the dataset, we carried out the permutation tail probability (PTP) test (Faith & Cranston 1991) using PAUP* 4b10 (Swofford 1999) with 10 000 replicates. The use of the PTP test has been questioned (Peres-Neto & Marques 2000), but the test is still used in a number of recent studies (e.g. Simmons & Weller 2002).
Phylogenetic trees were constructed using NJ, maximum parsimony (MP) and ML as implemented in PAUP* 4b10. MP and ML trees were constructed with a heuristic search of 100 random additions, and the tree bisection–reconnection branch-swapping algorithm with the option to collapse zero branch length. Reliability of the branches was ascertained by bootstrap analyses for NJ (100 000 replicates), MP (1000 replicates) and ML (100 replicates) in PAUP* 4b10. Bayesian phylogenetic analysis was performed with MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001) using prior settings nst = 6 and rates=invgamma with four independent runs of 3 000 000 generations and five chains each; rate matrix, base frequencies and branch lengths were estimated and trees were sampled every 300 generations. A majority consensus tree was generated using a burn-in of 2000. Posterior probabilities were calculated based on the topology of the Bayesian tree.
History and the ancestral state of character evolution were reconstructed using parsimony algorithms of the StochChar package in Mesquite 2.5 (Maddison & Maddison 2008). A step matrix for each character was constructed under the following assumptions: the colonization of bark from soil-living oribatid mites is more likely than the reverse; the capitate sensillus is probably evolved from a non-capitate sensillus; sex is the ancestral mode of reproduction and was frequently lost, and the sclerotization of oribatid mites evolved from weaker to stronger sclerotized species.
We investigated whether tree living is correlated with the type of sensillus, the reproductive mode or the degree of sclerotization using Phylocom (Webb et al. 2008). Independent pairwise contrasts between tree living and the three traits, i.e. type of sensillus, mode of reproduction and degree of sclerotization, were calculated (Garland et al. 1999) with default values for number of randomizations (999 replicates).
3. Results
Phylogenetic analyses of the ribosomal 18S rDNA region were based on 1699 base pairs and 55 taxa in total. Of the 1699 positions, 1113 were conserved and 586 were variable with 379 positions being parsimony informative. Variable positions of the ingroup (four outgroup taxa excluded) were 474 with 278 parsimony informative positions. The average pairwise ML distance of the whole dataset averaged 7.8 per cent with a maximum value of 33 per cent (the model used to calculate the ML distance was the same as that used to construct the ML tree).
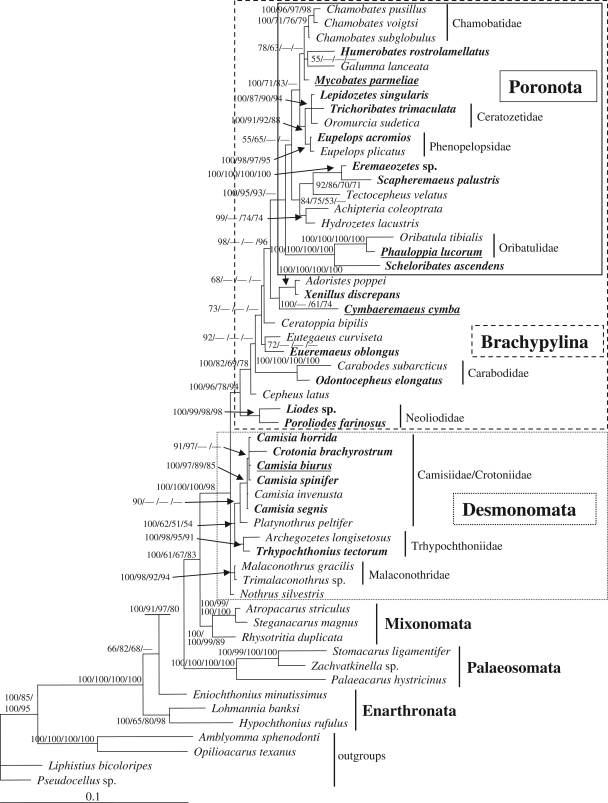
As each of the tree topologies of the phylogenetic algorithms, NJ, MP, ML and Bayesian methods, were almost identical, only the Bayesian tree is shown (figure 1). Bayesian inference has been shown to be most robust against model violations and recovers (known) correct trees in nearly all cases (Mar et al. 2005). The PTP test indicates that there is a strong phylogenetic signal in the dataset (p < 0.0001). Arboreal oribatid mite species were not randomly distributed in the phylogenetic tree but dominated among evolutionarily younger taxa, especially in the Poronota. Enarthronota (Eniochthonius minutissimus, Hypochthonius rufulus and Lohmannia banksi) were paraphyletic except in the ML analysis, where Hypochthonius and Lohmannia were sister taxa. Enarthronota were followed by Palaeosomata (Stomacarus ligamentifer, Palaeacarus hystricinus and Zachvatkinella sp.) and Mixonomata (Atropacarus striculus, Steganacarus magnus and Rhysotritia duplicata). The middle-derivative Desmonomata included 12 species, of which Trhypochthonius tectorum, Crotonia brachyrostrum and four species of the genus Camisia are arboreal. The Brachypylina (=Circumdehiscentiae) were always monophyletic with high statistical support. Basal in Brachypylina were the two arboreal species of Neoliodidae, Poroliodes farinosus and Liodes sp., followed by Cepheus latus and two Carabodidae, Carabodes subarcticus and Odontocepheus elongatus; most groups had high bootstrap and posterior probability support. The phylogenetic positions of the soil-living species Ceratoppia bipilis, Eutegaeus curviseta, Adoristes poppei and of the arboreal species Eueremaeus oblongus, Cymberemaeus cymba, Xenillus discrepans varied among different phylogenetic analyses, but were identical in the Bayesian and ML tree. Poronota s.l. (including Scapheremaeus palustris and Eremaeozetes sp.) were monophyletic with high bootstrap support and posterior probabilities and included the arboreal species Scheloribates ascendens, Phauloppia lucorum, Scapheremaeus palustris, Eremaeozetes sp., Eupelops acromios, Trichoribates trimaculata, Lepidozetes singularis, Mycobates parmeliae and Humerobates rostrolamellatus. Among arboreal oribatid mites, lichen feeding evolved at least four times, in the genus Camisia and in Cymberemaeus cymba, Phauloppia lucorum and Mycobates parmeliae (figure 1).
Figure 1.
Bayesian phylogeny of oribatid mites based on the ribosomal 18S gene using GTR + I + G as an evolutionary model. Numbers at nodes, respectively, represent Bayesian posterior probabilities and bootstrap support values for NJ, MP and ML. Arboreal oribatid mite species are in bold face and italics; lichen-feeding species on trees are additionally underlined.
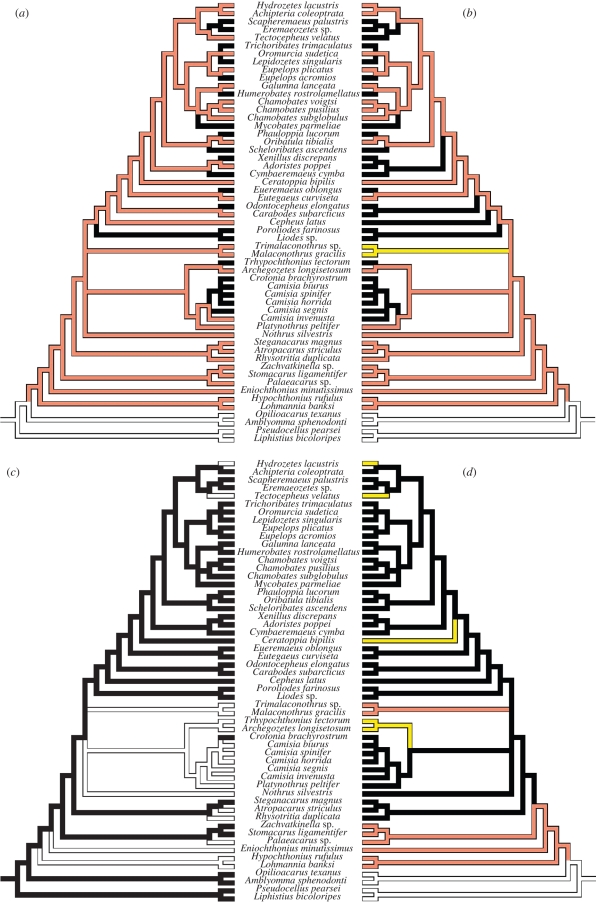
Ancestral character state reconstruction indicated that arboreal life evolved at least 15 times among the studied oribatid mites, (figure 2a). All studied arboreal (and very few soil living) oribatid mite species possess a clavate sensillus (figure 2b); the studied soil-living oribatid mites possess a non-clavate sensillus (e.g. pectinate, fusiform, setiform, bacilliform or ciliate) and two genera (Malaconothrus and Trimalaconothrus) have no sensillus at all (table 1). All studied arboreal oribatid mites, except the four species of Camisia, reproduce sexually (figure 2c; table 1). Furthermore, sclerotization is usually strong in arboreal and soil living species, except in phylogenetically older soil-living species, most of which are only weakly sclerotized (Enarthronota and Palaeosomata; figure 2d; table 1).
Figure 2.
Ancestral character state reconstruction of (a) living mode, (b) sensillus type, (c) reproductive mode and (d) degree of sclerotization as reconstructed with Mesquite 2.5 using parsimony algorithms. Bark living and a clavate sensillus are strongly correlated, whereas bark living is not strongly correlated with sexual reproduction and strong sclerotization. See text for details. (a) Black, bark; orange, soil; white, outgroups. (b) Black, clavate; orange, non-clavate; yellow, no sensillus; white, outgroups. (c) Black, sexual; white, thelytokous. (d) Black, strong; orange, weak; yellow, intermediate; white, outgroups.
Bark living was strongly correlated with a capitate sensillus (correlation coefficient R = 0.68) but only weakly correlated with a strong sclerotization (R = 0.21) and even less with sexual reproduction (R = 0.12), as indicated by the test for independent contrasts using Phylocom.
4. Discussion
The aim of this study was to investigate whether ecological factors, pre-adaptations or chance were responsible for the convergent evolution of arboreal life in oribatid mites. Phylogeny and model-based reconstruction of ancestral states indicated that arboreal life evolved at least 15 times in oribatid mites. As not all arboreal genera and species of oribatid mites were included, arboreal life certainly evolved more often. The arboreal oribatid mite taxa are not randomly distributed in the phylogenetic tree but cluster among the more derived Brachypylina, suggesting that higher oribatid mites may be pre-adapted to colonize trees.
High correlation of bark living and a capitate sensillus indicates that the sensillus co-evolved with the arboreal lifestyle of oribatid mites. Most arboreal oribatid mite species possess a capitate sensillus that is most probably an adaptation for arboreal life (Aoki 1973; Alberti et al. 1994). Presumably, this typical sensillus is an air-current receptor (anemoreceptor) that has this typical shape to avoid overstimulation (Norton & Palacios-Vargas 1982). The compact shape could limit the sensitivity of the receptor under the higher air flow of exposed situations when compared with soil. Sensilli of soil species are usually thinner and longer; they often have cilia or other ornamentations that increase sensitivity to air currents. Alternatively, the large distal ball and thin stalk of capitate sensilli could serve as a gravity receptor in arboreal species (Alberti et al. 1994). This idea is supported by the fact that capitate sensilli in some arboreal species (Crotoniidae and Camisia abdosensilla; Olszanowski et al. 2002) are entirely protected from air currents by being almost completely enclosed in a covered bothridium.
A capitate sensillus is not characteristic of all derived oribatid mites, indicating that it evolved several times in taxa that permanently colonized trees. In contrast to sexual reproduction and strong sclerotization (which were pre-adaptations of oribatid mites before they colonized the trees; see below), the capitate sensillus evolved convergently after the trees were colonized and is therefore a true adaptation to arboreal life.
The low correlation of bark living with sexual reproduction as well as strong sclerotization indicates that these traits already existed before the trees were colonized. The most important pre-adaptation for arboreal life in Brachypylina probably was the sexual mode of reproduction. In contrast to basal oribatid mite lineages, Brachypylina are predominantly sexual. The importance of the reproductive mode for arboreal oribatid mites can be inferred from the arboreal genus Crotonia that re-evolved sexual reproduction from a previously soil-living and parthenogenetic taxon, the Camisiidae/Crotoniidae (Domes et al. 2007b). It is not known why sexual reproduction is advantageous for arboreal species but it is probably related to food resources. While soil-living taxa predominantly feed on little defended food substrates, such as dead organic material, arboreal species predominantly feed on algae and lichens that at least in part are heavily defended (Seyd & Seaward 1984; Erdmann et al. 2007). Sexual reproduction therefore may be necessary for the co-evolutionary arms race between predators and prey (Red Queen hypothesis; Hamilton 1980).
The second important pre-adaptation of tree living oribatid mites probably was the strongly sclerotized body of the adults. Most adult oribatid mites are sclerotized, but the sclerotization of arboreal taxa is often even stronger. The strong sclerotization of bark-living oribatid mite species probably functions as predator defence. This also applies to oribatid mites in soil and litter (Sanders & Norton 2004), but this feature is probably less important in soil than on the bark of trees owing to the opaqueness of the soil habitat, which renders prey location more difficult. This hypothesis is supported by the stronger sclerotization of juvenile oribatid mite species living on the bark of trees when compared with juveniles of soil-living species. While many oribatid mites are sclerotized, Brachypylina are unique among them in possessing an extensive tracheal system, which may be evolutionarily linked to the difficulty of respiring through a sclerotized cuticle (Norton & Alberti 1997). The combination of a hard cuticle, a water-resistant epicuticle and an internalized respiratory surface could have been an effective pre-adaptation of Brachypylina to life in desiccating environments such as tree bark.
Oribatid mite species have a number of morphological characters that can be used to test whether arboreal species are really adapted to arboreal life or just colonized the trees permanently without evolving specific adaptations. The bark of trees is a permanent habitat for a large number of (mainly sexual) oribatid mite species (Proctor et al. 2002; Erdmann et al. 2006; Lindo & Stevenson 2007). Only a few ubiquitous parthenogenetic species such as Tectocepheus velatus and Oppiella nova live on the bark of trees and also in soil. This indicates a clear niche differentiation between soil and arboreal oribatid mite species.
Overall, our data indicate that the frequent convergent evolution of arboreal life in oribatid mites was driven in part by chance, as the arboreal species cluster randomly in higher taxa. However, the major driving force for the colonization of trees by oribatid mites was the ecological factor supporting the adaptionist view of evolution (Johannesson 2003; Morris 2003, 2006). Pre-adaptations such as sexual reproduction and strong sclerotization presumably facilitated the arboreal life of oribatid mites, and characters such as the clavate sensillus evolved later during tree colonization. We conclude that ecological forces swamp chance events such as drift and historical contingencies during evolution, supporting the ‘adaptionist programme’.
Acknowledgements
We thank Ina Schaefer for help using Mesquite. Thanks to Heinrich Schatz for the collection of several oribatid mite species, Martin Rosenberger for assistance in the molecular work and Barbara Fischer for helpful information. We also thank the German Research Foundation (DFG) for financial support. Finally, we thank two anonymous referees and Zoë Lindo for useful comments and suggestions on the manuscript.
References
- Alberti G., Moreno A. I., Kratzmann M.1994The fine structure of trichobothria in moss mites with special emphasis on Acarogalumna longipluma (Berlese, 1904) (Oribatida, Acari, Arachnida). Acta Zool. 75, 57–74 [Google Scholar]
- Aoki J. I.1973Soil mites (oribatids) climbing trees. Proc. 3rd Int. Congr. Acarol. Academia, Prague, Czechoslovakia, pp. 59–65 [Google Scholar]
- Behan-Pelletier V. M., Winchester N. N.1998Arboreal oribatid mite diversity: colonizing the canopy. Appl. Soil Ecol. 9, 45–51 (doi:10.1016/S0929-1393(98)00052-3) [Google Scholar]
- Behan-Pelletier V. M., St. John M. G., Winchester N.2007Canopy oribatida: tree specific or microhabitat specific? Eur. J. Soil Biol. 44, 220–224 (doi:10.1016/j.ejsobi.2007.06.002) [Google Scholar]
- Cianciolo J., Norton R. A.2006The ecological distribution of reproductive mode in oribatid mites, as related to biological complexity. Exp. Appl. Acarol. 40, 1–25 (doi:10.1007/s10493-006-9016-3) [DOI] [PubMed] [Google Scholar]
- Domes K., Scheu S., Maraun M.2007aResources and sex: soil re-colonization by sexual and parthenogenetic oribatid mites. Pedobiologia 51, 1–11 (doi:10.1016/j.pedobi.2006.11.001) [Google Scholar]
- Domes K., Norton R. A., Maraun M., Scheu S.2007bRe-evolution of sex in oribatid mites breaks Dollo's law. Proc. Natl Acad. Sci. USA 104, 7139–7144 (doi:10.1073/pnas.0700034104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F.1981Similar amino-acid-sequences: chance or common ancestry? Science 214, 149–159 (doi:10.1126/science.7280687) [DOI] [PubMed] [Google Scholar]
- Erdmann G., Floren A., Linsenmair K. E., Scheu S., Maraun M.2006Effect of forest age on oribatid mites from the bark of trees. Pedobiologia 50, 433–441 (doi:10.1016/j.pedobi.2006.08.004) [Google Scholar]
- Erdmann G., Otte V., Langel R., Scheu S., Maraun M.2007The trophic structure of bark-living oribatid mite communities analysed with stable isotopes (15N; 13C) indicates strong niche differentiation. Exp. Appl. Acarol. 41, 1–10 (doi:10.1007/s10493-007-9060-7) [DOI] [PubMed] [Google Scholar]
- Faith D. P., Cranston P. S.1991Could a cladogram this short have arisen by chance alone? On permutation tests for cladistic structure. Cladistics 7, 1–28 (doi:10.1111/j.1096-0031.1991.tb00020.x) [Google Scholar]
- Garland T., Midford P. E., Ives A. R.1999An introduction to phylogenetically based statistical methods, with a new method for confidence intervals on ancestral values. Am. Zool. 39, 374–388 (doi:10.1093/icb/39.2.374) [Google Scholar]
- Gould S. J.1989Wonderful life. The Burgess Shale and the nature of history New York, NY: W.W. Norton and Company [Google Scholar]
- Gould S. J., Lewontin R. C.1979The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptionist program. Proc. R. Soc. Lond. B 205, 581–598 (doi:10.1098/rspb.1979.0086) [DOI] [PubMed] [Google Scholar]
- Grandjean F.1969Considérations sur le classement des oribates: leur division en 6 groupes majeurs. Acarologia 11, 127–153 [Google Scholar]
- Hamilton W. D.1980Sex versus non-sex versus parasite. Oikos 35, 282–290 (doi:10.2307/3544435) [Google Scholar]
- Hardison R. C.1996A brief history of hemoglobins: plant, animal, protist, and bacteria. Proc. Natl Acad. Sci. USA 93, 5675–5679 (doi:10.1073/pnas.93.12.5675) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F.2001MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- Johannesson K.2003Evolution in Littorina: ecology matters. J. Sea Res. 49, 107–117 (doi:10.1016/S1385-1101(02)00218-6) [Google Scholar]
- Jones G., Teeling E. C.2006The evolution of echolocation in bats. Trends Ecol. Evol. 21, 149–156 (doi:10.1016/j.tree.2006.01.001) [DOI] [PubMed] [Google Scholar]
- Karasawa S., Hijii N.2004Morphological modifications among oribatid mites (Acari: Oribatida) in relation to habitat differentiation in mangrove forests. Pedobiologia 48, 383–394 (doi:10.1016/j.pedobi.2004.05.003) [Google Scholar]
- Langerhans R. B., DeWitt T. J.2004Shared and unique features of evolutionary diversification. Am. Nat. 164, 335–349 (doi:10.1086/422857) [DOI] [PubMed] [Google Scholar]
- Lenski R. E.2003The eyes have it. Nature 425, 767–768 (doi:10.1038/425767a) [Google Scholar]
- Lindo Z., Stevenson S. K.2007Diversity and distribution of oribatid mites (Acari: Oribatida) associated with arboreal and terrestrial habitats in interior Cedar Hemlock forests, British Columbia, Canada. Northwest Sci. 81, 305–315 (doi:10.3955/0029-344X-81.4.305) [Google Scholar]
- Lindo Z., Winchester N. N.2006A comparison of microarthropod assemblages with emphasis on oribatid mites in canopy suspended soils and forest floors associated with ancient western red cedar trees. Pedobiologia 50, 31–41 (doi:10.1016/j.pedobi.2005.09.002) [Google Scholar]
- Maddison W. P., Maddison D. R.2008Mesquite: a modular system for evolutionary analysis, version 2.5. http://mesquiteproject.org
- Mar J. C., Harlow T. J., Ragan M. A.2005Bayesian and maximum likelihood phylogenetic analyses of protein sequence data under relative branch-length differences and model violation. BMC Evol. Biol. 5, 8 (doi:10.1186/1471-2148-5-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraun M., Salamon J. A., Schneider K., Schaefer M., Scheu S.2003Oribatid mite and collembolan diversity, density and community structure in a moder beech forest (Fagus sylvatica): effects of mechanical disturbances. Soil Biol. Biochem. 35, 1387–1394 (doi:10.1016/S0038-0717(03)00218-9) [Google Scholar]
- Marks C. O.2007The causes of variation in tree seedling traits: the roles of environmental selection versus chance. Evolution 61, 455–469 (doi:10.1111/j.1742-4658.2007.00021.x) [DOI] [PubMed] [Google Scholar]
- Morris S. C.2003Life's solution. Inevitable humans in a lonely universe Cambridge, UK: Cambridge University Press [Google Scholar]
- Morris S. C.2006Evolutionary convergence. Curr. Biol. 16, 826–827 (doi:10.1016/j.cub.2006.08.077) [DOI] [PubMed] [Google Scholar]
- Norton R. A., Alberti G.1997Porose integumental organs of oribatid mites (Acari, Oribatida). 3. Evolutionary and ecological aspects. Zoologica 146, 115–143 [Google Scholar]
- Norton R. A., Palacios-Vargas J. G.1982Nueva Belba (Oribatei: Damaeidae) de musgos epifitos de Mexico. Folia Entomol. Mex. 52, 61–73 [Google Scholar]
- Norton R. A., Bonamo P. M., Grierson J. D., Shear W. A.1988Oribatid mite fossils from a terrestrial Devonian deposit near Gilboa, New York. J. Paleontol. 62, 259–269 [Google Scholar]
- Olszanowski Z., Clayton M. R., Humble L. M.2002New species of the genus Camisia (Acari: Oribatida): an arboreal mite with enclosed sensilli. Can. Entomol. 134, 707–721 [Google Scholar]
- O'Riain M. J., Jarvis J. U. M., Alexander R., Buffenstein R., Peeters C.2000Morphological castes in a vertebrate. Proc. Natl Acad. Sci. USA 97, 13 194–13 197 (doi:10.1073/pnas.97.24.13194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres-Neto P. R., Marques F.2000When are random data not random, or is the PTP test useful? Cladistics 16, 420–424 (doi:10.1006/clad.2000.0140) [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K. A.1998Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818 (doi:10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- Proctor H. C., Montgomery K. M., Rosen K. E., Kitching R. L.2002Are tree trunks habitats or highways? A comparison of oribatid mite assemblages from hoop-pine bark and litter. Aust. J. Entomol. 41, 294–299 (doi:10.1046/j.1440-6055.2002.00309.x) [Google Scholar]
- Sanders F. H., Norton R. A.2004Anatomy and function of the ptychoid defensive mechanism in the mite Euphthiradarus cooki (Acari: Oribatida). J. Morphol. 259, 119–154 (doi:10.1002/jmor.10183) [DOI] [PubMed] [Google Scholar]
- Schatz H.2002Die Oribatidenliteratur und die beschriebenen Oribatidenarten (1758–2001)—Eine Analyse. Abh. Ber. Natkdmus. Görlitz 74, 37–45 [Google Scholar]
- Seyd E. L., Seaward M. R. D.1984The association of oribatid mites with lichens. Zool. J. Linn. Soc. 80, 369–420 (doi:10.1111/j.1096-3642.1984.tb02552.x) [Google Scholar]
- Simmons R. B., Weller S. J.2002What kind of signal do mimetic tiger moths send? A phylogenetic test of wasp mimicry systems (Lepidoptera: Arctiidae: Euchromiini). Proc. R. Soc. Lond. B 269, 983–990 (doi:10.1098/rspb.2002.1970) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair B. J., Vernon P., Klok C. J., Chown S. L.2003Insects at low temperatures: an ecological perspective. Trends Ecol. Evol. 18, 257–262 (doi:10.1016/S0169-5347(03)00014-4) [Google Scholar]
- Swofford D.1999PAUP*: phylogenetic analysis using parsimony (and other methods), version 4.0. Sunderland, MA: Sinauer Associates [Google Scholar]
- Tamura K., Nei M.1993Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzee. Mol. Biol. Evol. 10, 512–526 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J.1994Clustal W: improving the sensitivity of progressive multiple alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucl. Acids Res. 22, 4673–4680 (doi:10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G.1997The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 24, 4876–4882 (doi:10.1093/nar/25.24.4876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbeville J. M., Pfeifer D. M., Field K. G., Raff R. A.1991The phylogenetic status of arthropods, as inferred from 18S ribosomal RNA sequences. Mol. Phylogenet. Evol. 8, 669–686 [DOI] [PubMed] [Google Scholar]
- Walter D. E., Behan-Pelletier V.1999Mites in forest canopies: filling the size distribution shortfall? Annu. Rev. Entomol. 44, 1–19 (doi:10.1146/annurev.ento.44.1.1) [DOI] [PubMed] [Google Scholar]
- Walter D. E., Proctor H. C. Mites: ecology, evolution and behaviour. Wallingford, UK: CABI Publishing; 1999. [Google Scholar]
- Webb C. O., Ackerly D. D., Kembel S. W.2008Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100 (doi:10.1093/bioinformatics/btn358) [DOI] [PubMed] [Google Scholar]
- Weigmann G.2006Hornmilben (Oribatida) Keltern, Germany: Goecke and Evers [Google Scholar]
- Zhang J. Z.2006Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat. Genet. 38, 819–823 (doi:10.1038/ng1812) [DOI] [PubMed] [Google Scholar]
- Zhang J. Z., Kumar S.1997Detection of convergent and parallel evolution at the amino acid sequence level. Mol. Biol. Evol. 14, 527–536 [DOI] [PubMed] [Google Scholar]