Abstract
Pathogen genes involved in interactions with their plant hosts are expected to evolve under positive Darwinian selection or balancing selection. In this study a single copy avirulence gene, AvrP4, in the plant pathogen Melampsora lini, was used to investigate the evolution of such a gene across species. Partial translation elongation factor 1-alpha sequences were obtained to establish phylogenetic relationships among the Melampsora species. We amplified AvrP4 homologues from species pathogenic on hosts from different plant families and orders, across the inferred phylogeny. Translations of the AvrP4 sequences revealed a predicted signal peptide and towards the C-terminus of the protein, six identically spaced cysteines were identified in all sequences. Maximum likelihood analysis of synonymous versus non-synonymous substitution rates indicated that positive selection played a role in the evolution of the gene during the diversification of the genus. Fourteen codons under significant positive selection reside in the C-terminal 28 amino acid region, suggesting that this region interacts with host molecules in most sequenced accessions. Selection pressures on the gene may be either due to the pathogenicity or avirulence function of the gene or both.
Keywords: Melampsora, rust fungi, pathogen, avirulence, effector, selection
1. Introduction
The gene-for-gene model of plant–pathogen interactions was proposed by Flor (1942, 1955) over 60 years ago, and has shaped our current thinking about host–pathogen coevolution in both animal and plant systems. This paradigm underlies much of the theoretical literature on disease evolution (e.g. Frank 1993; Damgaard 1999; Thrall & Burdon 1999, 2002; Sasaki 2000; Agrawal & Lively 2002). Under this model, host resistance (R) genes control recognition of pathogens carrying specific avirulence (Avr) genes leading to disease resistance. Many plant R genes have been cloned and most encode proteins of a conserved class that together represent the pathogen recognition components of plant innate immunity (Dangl & Jones 2001). Pathogen Avr proteins on the other hand are highly diverse, and are generally thought to act as pathogenicity effectors that promote infection in the absence of recognition by corresponding R proteins. Many such Avr/effectors appear specific to particular pathogen classes, and recent genome sequencing of two Phytophthora species has shown very little overlap between their predicted effector protein repertoires (Tyler et al. 2006). Studies of Avr gene evolution have focused on genetic variation within single host species, with an emphasis on the presence or absence and characteristics of gene homologues among races and pathotypes, e.g. Pseudomonas syringae Avr genes (Stevens et al. 1998; Arnold et al. 2001), weighing up the role of Darwinian selection versus balancing selection in introducing and maintaining polymorphisms (Holub 2001; Tellier & Brown 2007). Loss of gene function through deletions, gene disruption or point mutations has been shown to play a role in overcoming host resistance (Farman et al. 2002; Tosa et al. 2005), and in some cases Avr gene diversification correlates with the ability to overcome host resistance (Allen et al. 2004; Dodds et al. 2006).
The interaction between flax rust (Melampsora lini) and cultivated flax (Linum usitatissimum) has been a useful model system for understanding the molecular basis of the gene-for-gene interaction (Ellis et al. 2007; Lawrence et al. 2007). To date, 19 flax rust resistance genes have been isolated in flax and four Avr genes have been isolated from the rust. All four Avr genes, AvrL567, AvrM, AvrP4 and AvrP123 encode small secreted proteins that are expressed in haustoria, and are thought to be transported into the host cytoplasm (Dodds et al. 2004; Catanzariti et al. 2006). Dodds et al. (2006) showed that for the AvrL567 locus in M. lini, and the L5, L6 and L7 resistance specificities in L. usitatissimum, direct protein interactions underlie gene-for-gene specificity. Differences in recognition specificity between corresponding R and Avr protein variants, and evidence for diversifying selection acting on these genes suggest a sustained gene-specific coevolutionary interaction between L. usitatissimum and M. lini. Considerable sequence variation and evidence for diversifying selection also occurs at the AvrM, AvrP4 and AvrP123 loci (Catanzariti et al. 2006; P. N. Dodds 2007, unpublished data).
Much of the theoretical and empirical work on gene-for-gene interactions has focused on within-species gene evolution and diversity. Sackton et al. (2007) investigated the evolution of innate immune-related genes across species of Drosophila, however in plants little work has been done to characterize variation in pathogen avirulence genes across species boundaries, despite the importance of such a perspective for understanding the evolutionary history of these genes both within and among host–pathogen interactions. For example, determination of whether homologues are present in sister taxa or within a genus or family can answer questions about sequence variation and selective constraints in gene sequences, and whether specific areas of the genes are prone to selection or relaxed selective constraint. The potential for conservation in function of host resistance and pathogen avirulence genes among species remains unclear and unexplored. In contrast to systems where deletion of a locus renders the pathogen virulent, all characterized isolates of M. lini from both cultivated flax and native Linum marginale populations contain intact copies of each of the Avr genes (Catanzariti et al. 2006; Dodds et al. 2006; P. N. Dodds & L. G. Barrett 2006, unpublished data). Avr gene sequence diversification, rather than major gene disruptions such as deletions, suggests an important functional role. It is possible that these genes remain conserved at a higher phylogenetic level, raising the question of whether genes related to M. lini Avr genes are present in the genomes of related rust species.
In this study, we examine the evolution of the flax rust AvrP4 gene within the rust genus Melampsora. Melampsora includes species with heteroecious (alternating between two unrelated host plants) and autoecious (completing the entire life cycle on one host plant) life cycles. All heteroecious Melampsora taxa have their dikaryotic stage on a Populus or Salix (Salicaceae) host. The haplont hosts are found among diverse plant families both in the gymnosperms (Pinaceae) and angiosperms (Araceae, Alliaceae, Orchidaceae, Papaveraceae, Grossulariaceae, Saxifragaceae, Celastraceae, Clusiaceae, Euphorbiaceae, Linaceae and Violaceae). The autoecious taxa are mainly found on hosts in the families Clusiaceae, Euphorbiaceae, Linaceae (all Malpighiales) and Pinaceae. Little information is available on specific relationships among the 80 plus species (Hawksworth et al. 1995) of Melampsora, but molecular phylogenies of the Uredinales (rust fungi) support Melampsora as a monophyletic genus (Maier et al. 2003; Wingfield et al. 2004).
The AvrP4 gene encodes a protein of 95-amino acids including a 28-amino acid cleavable secretion signal peptide and six cysteine (cys) residues, spaced according to the consensus of a cystine-knot, found in the C-terminal 28 amino acids of the protein (Catanzariti et al. 2006). While the function of the protein is unknown, a cys knot is commonly found in toxins and in inhibitors of receptors and proteases (Pallaghy et al. 1994). Here we assess the extent of sequence maintenance of the AvrP4 locus across a range of Melampsora species, and evaluate selection pressures acting on the AvrP4 gene by investigating the non-synonymous versus synonymous rate ratio using maximum-likelihood methods. This is compared to a phylogeny obtained using the translation–elongation factor 1-alpha ‘house-keeping’ gene sequence.
2. Material and methods
(a). Collection of rust accessions
Infected host material was obtained for 17 species (26 specimens) of Melampsora, including the aggregate species M. lini and Melampsora epitea (which includes M. aff. capraearum, Melampsora repentis and Melampsora reticulatae). The position of the Caeoma sp. is unclear, although the habitat indicates that it alternates with Salix and is a member of the M. epitea aggregate. The material represents autoecious and heteroecious life cycles, and pathogenicity on a range of diverse plant families. For all species, the dikaryont host is a member of the order Malpighiales among the eurosids I, while the haplont hosts include members of Asparagales (monocots) and Saxifragales (core eudicots) (APG II 2003). Hosts and life cycle information is given in figure 1 and table S1 in the electronic supplementary material. Additional information on each isolate is available in table S1 in the electronic supplementary material. Two out-group taxa, Chrysomyxa woronini (Coleosporiaceae) and Pucciniastrum epilobii (Pucciniastraceae) were included. Owing to quarantine regulations most DNA extractions were conducted on spore material stored and shipped in ethanol (see table S1 in the electronic supplementary material for type of material used), using a modified CTAB method described by Van der Merwe et al. (2007). Melampsora lini isolates from three Hesperolinon species were collected, but it is not clear whether host specific isolates have evolved (Springer 2007; Y. P. Springer 2006, personal communication).
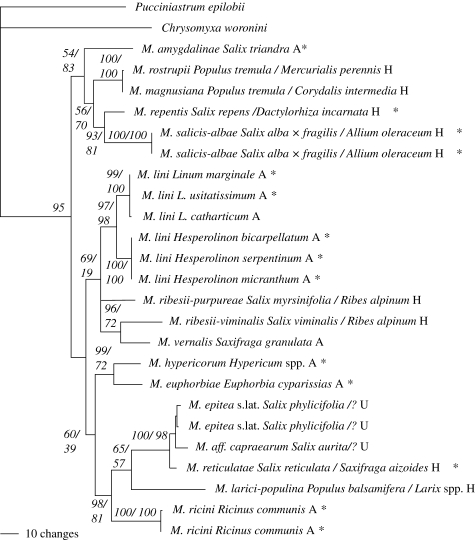
Figure 1.
A partial translation elongation factor 1-alpha gene sequence Bayesian phylogeny with posterior probabilities and ML bootstrap support values indicated at the nodes (in this order). Chrysomyxa and Pucciniastrum were used as out-groups. The telial and aecial host names are given after the rust species name in this order followed by the type life cycle (A=autoecious and H=heteroecious and U=unknown). The AvrP4 locus was amplified from those accessions indicated with an asterisk.
(b). PCR amplification
Partial sequences of the translation elongation factor 1-alpha (TEF) gene were obtained for the accessions following the methods of Van der Merwe et al. (2007).
We designed primers to amplify 180 bp of 5′ flanking sequence, 285 bp of open reading frame (ORF), and 103 bp of 3′ flanking sequence for the AvrP4 gene (F: CATCAAAATCTAACCCGTAC and R: GTAGCATTGAGATCCATGG). Polymerase chain reactions (PCRs) were carried out in 50 μl reactions with 10 pmol of each primer, 1 unit of Immolase polymerase (a high-fidelity polymerase from Bioline), 1.2 μl Betaine (Sigma), 5 μl PCR buffer (supplied with the polymerase), 2.5 mM dNTPs and 2 mM MgCl2. Best amplification was found when using a touch-down PCR with the following conditions: 7 min at 97°C followed by 10 cycles of 30 s at 94°C; 40 s at 55°C (with a temperature decrease of 1°C with every cycle) and 1 min 30 s at 70°C; followed by 28 cycles of 30 s at 94°C; 30 s at 54°C and 1 min at 70°C; and a final extension of 8 min at 72°C.
PCR fragments were purified using the Qiagen PCR clean-up system and sequenced using the PCR primers. Standard sequencing reactions using BigDye v. 3.1 were performed and run on an ABI3730 at ACRF Biomolecular Resource Facility (ANU). The pGEM-T easy vector system (Promega) was used for cloning where initial sequencing indicated the presence of multiple variant copies. Ten colonies were sequenced from each cloned PCR product and colony PCRs were carried out using M13 primers. To test for the level of PCR and sequencing error, we amplified and cloned the AvrP4 gene from the same isolates used by Catanzariti et al. (2006). No sequence variation was detected between our sequences and those obtained in their study.
Amplification attempts were made on all accessions in figure 1. Accessions from eight Melampsora species and out-group taxa failed to amplify, leaving AvrP4 sequence data from nine species of Melampsora available for analysis. Unsuccessful amplification may have resulted from poor sample quality, gene deletions or mutations within the primer sites.
(c). Sequence analysis and data exploration
(i) Translation elongation factor 1
Sequences were aligned using MEGA v. 3.1. Phylogenetic trees inferred using Bayesian methods were constructed using MrBayes (Huelsenbeck & Ronquist 2001; Ronquist & Huelsenbeck 2003). Data was partitioned into the intronic and exonic regions with the latter further partitioned according to the first, second and third codon positions. A general time reversible (GTR) model was implemented as determined by ModelTest v. 3.7 (Posada & Crandall 1998). Two million Markov chain Monte Carlo generations were run and trees were sampled every 100th generation. The first 20 per cent of sampled trees were removed and a 50 per cent majority rule consensus tree was constructed. Maximum likelihood (ML) analysis was conducted at the Cipres Portal v. 1.13 (available at http://www.phylo.org/) using the RAxML algorithm for finding the best known likelihood tree from the data and from 1000 non-parametric bootstrap datasets (Stamatakis et al. 2008). The data were partitioned into first, second and third codon positions and non-coding sequence with a distinct GTR+Γ substitution model for each partition.
(ii) AvrP4
Sequences were manually aligned in MEGA v. 3.1 (Kumar et al. 2004) by using conserved amino acid motifs as reference points. Bayesian and ML methods similar to the TEF data above were implemented. ML analysis were conducted in PAUP (Swofford 2001) and RAxML with the AvrP4 data partitioned according to the three codon positions. Substitution rates for each codon position were estimated from the data and included in the GTR model with invariable sites and a gamma distribution (GTR +I+G) using a heuristic search method. The resultant ML tree was used in the CODEML analysis. Bootstrap support values were obtained by analysing 100 replicates without codon partitioning.
(iii) Recombination
Four tests available in RDP3 (Martin et al. 2005) were implemented to detect possible recombination blocks with the following settings: RDP (Martin & Rybicki 2000)—six nucleotide windows with no reference sequences used; GENECONV (Padidam et al. 1999)—sequence triplets with a scale of 1; BOOTSCAN (Maynard Smith 1992)—100 nucleotide windows with a 20 nucleotide step distance scanned, using a 70 per cent bootstrap cut-off and 100 bootstrap replicates; and CHIMAERA (Maynard Smith 1992) set to 30 variable sites per window, 10 permutations with a maximum permutation p-value of 0.01.
(d). Detection of positive selection
The non-synonymous/synonymous rate ratio parameter ω was estimated using the program CODEML (Yang 1997) in phylogenetic analysis by ML v. 3.15 (Yang 1997). To investigate variation in selection among branches we applied a model that estimated ω for each branch in the phylogeny (ML tree obtained with PAUP). Tests for positive selection were performed using the site class models implemented in CODEML that estimate ω for amino acid sites. Neutral sites have ω=1, while those under purifying selection have ω<1 and those under positive selection, ω>1 (Neilsen & Yang 1998; Yang et al. 2000). Likelihood ratio tests were performed using the likelihood values λ 0 and λ 1 from the nested models M7 and M8 by comparing the test statistic 2Δλ=2(λ 1−λ 0), with the Χ 2 distribution (d.f.=2). The null model M7 allowed for 10 site classes that provide a discrete approximation of the beta distribution of site-wise ω-values between 0 and 1. Model M8 added an 11th class with ω>1 and estimates the proportion of sites within this class and the value of ω for that class. Simulations have suggested that the beta distribution null model provides greater power to detect positive selection than simpler models implemented in CODEML (Anisimova et al. 2001). For model M8, the Bayes empirical Bayes (Yang et al. 2005) procedure estimated the mean ω-value for each codon site, and the posterior probability that the site is under positive selection.
3. Results
(a). Translation elongation factor 1-alpha
Total length of the sequence alignments for the TEF data was 696 bp, including 150 codons and 246 bp of intronic sequence (GenBank accessions: EF487218 to EF487243). The average pairwise nucleotide divergence was 8 per cent, ranging between 0.2 and 13 per cent. Since no evidence for recombination was detected, the sequences were used for phylogenetic data analysis (figure 1). Topologies obtained from the best ML and Bayesian trees were identical. Despite poor support for relationships along the backbone, the following relationships received significant support from both the ML and Bayesian analysis: (i) all M. lini isolates, (ii) M. repentis and Melampsora salicis-albae, (iii) Melampsora hypericorum and Melampsora euphorbiae, (iv) M. reticulatae and the M. epitea complex in a clade with Melampsora larici-populina and Melampsora ricini, (v) Melampsora rostrupii and Melampsora magnusiana, and (vi) Melampsora ribesii-viminalis and Melampsora vernalis.
(b). Identification of AvrP4 homologues in Melampsora species
Primers flanking the AvrP4 gene of M. lini were used to amplify homologous sequences (GenBank accessions EF639296 to EF 639314) from 9 of the 17 species of Melampsora (including the Caeoma sp.) occurring across the TEF phylogeny. The sequence length varied between 509 and 526 bp. Average pairwise nucleotide divergence was 20 per cent, ranging between 2 and 32 per cent. No significant evidence for recombination was detected among the sequences. Each sequence contained an ORF of between 91 and 98 codons that corresponded to the predicted coding region of the M. lini AvrP4 gene (figure 2). The first methionine codon conserved in all sequences corresponded to amino acid five of the predicted M. lini sequence. Beginning at the conserved methionine, the 24 amino acids corresponding to the predicted signal peptide for the M. lini isolates (Catanzariti et al. 2006) were highly conserved. Following the predicted signal peptide was a variable region of nine amino acids, followed by a conserved region with the sequence I(Q/R)GFS (figure 2). This motif was followed by a variable region of eight amino acids, which was followed by another highly conserved region of five amino acids with a consensus sequence L(E/Q)E(D/E)S. The last 49 amino acids in the alignment were highly variable apart from six cys residues present in all sequences.
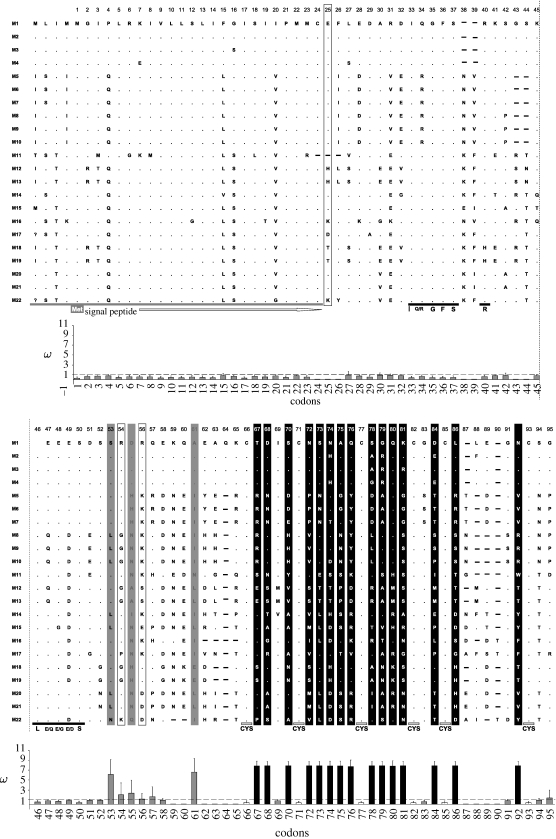
Figure 2.
An amino acid sequence alignment of the AvrP4 homologues identified in this study with a bar diagram indicating the value of ω calculated with CODEML for each codon underneath. The putative signal peptide and conserved motifs I(Q/R)GFS, L(E/Q)E(E/D)S and cys residues are indicated. The dashed line across the bar diagram indicates the ω-value of 1. Amino acids and ω-values in black indicate the codons that were found to be under significant positive selection; those in grey indicate codons found to be under selection but not with significant ω-values. The boxed amino acids indicate the codons found to be under selection when M. amygdalinae were removed from the data analysis. Dots represent similar amino acids while the dashes indicate a gap. M1–M22 correspond to the labels in figure 3.
The spacing of the six cys residues in the predicted amino acid alignment (Cx4Cx5Cx4Cx2Cx4-7C; figure 2) was in accordance with the consensus sequence of an inhibitor cys-knot structure (Cx3-7Cx4-6Cx0-5Cx1-4Cx4-10C). All sequences in the alignment contained a termination codon at an equivalent position with the exception of Melampsora amygdalinae, which contained an additional four codons (PIVQ). Within the 123 bp 5′ non-coding region, 45 variable nucleotide sites and one indel were present. There were 35 variable nucleotide sites in the 97 bp non-coding region at the 3′ end. The overall mean (Nei's) genetic distance between sequences was 0.231 (std 0.018).
(c). Occurrence of AvrP4 variants within species
Two AvrP4 variants were amplified from each of three accessions of M. lini pathogenic on Hesperolinon and the one accession of M. reticulatae (A and B in figure 3). The AvrP4 A variant sequences from accessions pathogenic on Hesperolinon bicarpellatum and Hesperolinon serpentinum were identical, as were the B variant sequences (figure 3), but differed from the A and B variant sequences obtained from Hesperolinon micranthum (three non-synonymous changes among the A copies and one non-synonymous and one synonymous change among the B copies). Sequence variation between the A and B variants for these rusts involved 45 substitutions in the coding region (20 amino acids). The two M. reticulatae variants differed from each other by 28 amino acid substitutions and two indels (four codons), but unlike the Hesperolinon rusts, these two sequences were not sister to each other on the AvrP4 phylogeny (figure 3).
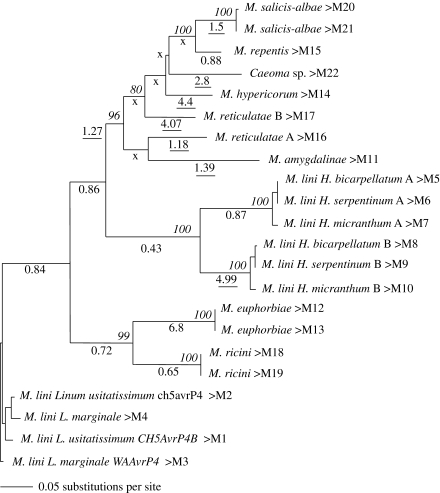
Figure 3.
An unrooted maximum-likelihood tree constructed from 22 AvrP4 sequences from 18 isolates. Bootstrap support values higher than 70% are given in italics adjacent to the nodes on top of the branches. Values of ω are given underneath each branch with those greater than 1 underlined and those branches for which ω could not be calculated are indicated with X. The codes (M1–M22) after the names correspond to the amino acid sequences in figure 2.
(d). Evidence for positive selection driving AvrP4 evolution within the genus Melampsora
We investigated positive selection in the AvrP4 phylogeny by examining the rates of synonymous and non-synonymous nucleotide substitutions using ML methods. The results from pairwise comparisons are given in table S2 in the electronic supplementary material. The sequence of M. amygdalinae contained unique deletions, and as CODEML requires the removal of codons corresponding to gaps in the alignment, we conducted the analyses with and without the M. amygdalinae data. Including M amygdalinae allowed 78 codons to be used, and the subsequent analysis indicated 17 codons to be under positive selection (14 highly significant (p>0.99), one significant (p>0.95) and two not significant). All occurred after the L(E/Q)E(E/D)S motif (figure 2) and all with significant posterior probabilities were in the 3′ region encoding the six conserved cys residues. In a likelihood ratio test (see §2), model M8 provided a significantly better fit to the data (p<0.001) than model M7 (table 1). Parameter estimates from the two models are given in table 1. For model M8, the Bayes empirical Bayes (Yang et al. 2005) procedure estimated ω for each codon site (figure 2). When M. amygdalinae was excluded from the analysis, the dataset contained 81 codons of which 21 were found to be under positive selection (figure 2).
Table 1.
Log-likelihoods, (lnL) estimates of the model parameters, and codon sites identified as under positive selection obtained from the site class models M7 and M8 in CODEML when all sequences were included in the analysis. (The estimated model parameters are the transition/transversion rate ratio parameter κ, and the parameters of the beta distribution p and q, describing the distribution of ω-values among codon sites. Also listed are p1, the fraction of sites estimated to fall within the class of sites with ω>1 and the mean ω-value for that class. Sites identified as under positive selection by CODEML are numbered as in figure 2, those with a Bayesian posterior probability greater than 0.99 of being under positive selection are in italics.)
| model M7 | model M8 | ||||
|---|---|---|---|---|---|
| parameters | lnL | parameters | lnL | LRT | codon sites with ω>1 |
| κ=1.17263, | −1795.494 | κ=1.67594, | −1722.054 | p<0.001 | 53, 55, 61, 67, 68, 70, 72, 73, 74, 75, 76, 78, 79, 80, 81, 84, 86, 92 |
| p=0.30466, | p=0.37015, | ||||
| q=0.2934 | q=0.22905, | ||||
| p1=0.21379, | |||||
| ω=7.90969 | |||||
The AvrP4 gene tree (figure 3) is divided into four clades: (i) the M. lini clade that includes the rusts on flax and other Linum species, (ii) a clade with M. ricini and M. euphorbiae, (iii) a clade with the six sequences amplified from the M. lini accessions derived from Hesperolinon, and (iv) a clade consisting of sequences from rusts that are mostly pathogenic on various Salix species. High values of ω were estimated in several Melampsora species lineages (nine branches with ω>1), although for several internal branches, the presence of only non-synonymous changes and no synonymous changes precluded the estimation of ω (indicated by X in figure 3). The remaining branches for which ω could be estimated indicated conservation of the protein sequence, with ω estimated to be between 0.4 and 1.
4. Discussion
We have demonstrated that highly variable AvrP4 homologues are present in a range of species in the rust genus Melampsora across a phylogenetic tree constructed from the TEF gene. The species are pathogenic on a variety of hosts from distantly related families. Five characteristics of the alignment (present in all sequences), support the new sequences as homologues of the M. lini AvrP4 locus: (i) the highly conserved flanking regions, (ii) the conserved signal peptide sequence, (iii) two highly conserved amino acid motifs, I(Q/R)GFS and L(E/Q)E(E/D)S, (iv) the six cys residues towards the C-terminus of the predicted protein (similar to a cys-knot structure), and (v) the termination codons, adjacent to the six cys codons. AvrP4 is one of a class of secreted proteins expressed in haustoria of M. lini and translocated into host cells during infection (Catanzariti et al. 2006), where they are presumed to enhance pathogenicity. The presence of the AvrP4 locus across the TEF phylogenetic spectrum indicates that this gene arose prior to the diversification of the genus Melampsora, and is not specific to the system in which it was first identified. This contrasts with the host specific appearance and disappearance of an Avr gene in rice specific Magnaporthe strains (Tosa et al. 2005). Although gene deletions may have occurred in the evolution of the genus, reflected in the non-amplification of certain accessions, it is possible that divergence in the primer target sites precluded amplification of AvrP4 homologues from these species. Conservation of AvrP4 homologues across the Melampsora genus suggests that the AvrP4 gene product is important for the biology of these rust species.
Although the focus of this study was not phylogenetic relationships, it is important to note that the gene trees are neither fully resolved nor are they congruent and lack support for the deeper nodes. This may be due to poor species sampling or rapid speciation during the early diversification of the genus. Good support was found for a sister relationship between the AvrP4 genes of the two autoecious species on Euphorbiaceae (M. ricini and M. euphorbiae) reflecting host relationships. TEF, however, placed M. euphorbiae closer to M. hypericorum (host family Clusiaceae) (figures 1 and 3; and S2 in the electronic supplementary material). The close relationship between M. lini from Hesperolinon and those from Linum inferred from the TEF gene, beta-tubulin 1 sequence data (M. M. Van der Merwe 2006, unpublished data) and morphology (Y. P. Springer 2005, personal communication) were not reflected in the AvrP4 phylogeny. The relationship between M. repentis and M. salicis-albae, with monocot haplont hosts (Orchidaceae and Alliaceae) were supported by both genes. Although, there seems to be a tendency for the heteroecious species to form a supported clade on the AvrP4 phylogeny, it would be premature to comment on the evolutionary significance of hosts/life cycles and the AvrP4 gene.
(a). Selection on the AvrP4 gene
The high ω-values estimated for 15 codons indicate that the AvrP4 gene is under strong positive selection within Melampsora. The ω estimated along the branches of the gene tree imply that strong positive selection has occurred throughout the evolutionary history of the gene. Diversifying selection has previously been observed among allelic variants of AvrP4 in M. lini, associated with differences in recognition by the corresponding host R genes, and suggesting that R gene-imposed selection has driven the accumulation of sequence diversity in this species (Catanzariti et al. 2006; L. G. Barrett 2006, unpublished data). Such R gene-imposed selection is a likely explanation for the rapid divergence of AvrP4 homologues among Melampsora species. However, we do not know whether the AvrP4 proteins from other Melampsora species function as avirulence factors that are recognized by R genes in their host plants. The role of AvrP4, similar to that of bacterial Avr proteins (Tsiamis et al. 2000; Alfano & Collmer 2004; Janjusevic et al. 2006) may be as an effector of pathogen virulence, promoting pathogen growth or suppressing host defence pathways. Thus, AvrP4 may be under selection to maintain a host specific pathogenicity function across different host species, and evade specific host resistance mechanisms. This group of Melampsora species infects diverse host plants, so it is possible that variation in the host target proteins may have driven AvrP4 divergence. Between species variation indicative of co-phylogenetic patterns between hosts and pathogens may reflect co-speciation, involving selection on the effector function of a gene, whereas strong selection acting on Avr genes within host specific lineages is a strong indication that recognition by R genes drives selection. Positive selection detected within the AvrP4 locus across species may have been driven by the effector function of the locus, or the interaction between R and Avr genes or a combination of both.
If the AvrP4 gene does function as an avirulence elicitor across these pathogen species, it raises an important question regarding host defence systems. Do these hosts have a single conserved homologous defence system, or do the hosts each have their own independently evolved mechanisms for recognizing the AvrP4 gene products? Kruijt et al. (2005) have suggested that the interaction between Cladosporium fulvum and host species in the genus Lycopersicon pre-dates speciation in Lycopersicon, since several functional homologues of R genes (Cf-4 and Cf-9) from multiple species in Lycopersicon interact with Avr4 and Avr9 elicitors from C. fulvum. However, the AvrB protein from P. syringae pv. glycinea can act as an avirulence factor in both Arabidopsis thaliana and Glycine max (phylogenetically diverse plants), interacting with the unrelated R genes Rpm1 and Rpg1b, respectively (Ong & Innes 2006).
Strong diversifying selection has been found in several fungal and oomycete Avr genes within a single pathogen species such as ATR1 and ATR13 of Hyaloperonospora parasitica (Allen et al. 2004; Rehmany et al. 2005), scr74 in Phytophthora infestans (Liu et al. 2005), the flax rust AvrL567, AvrM, AvrP123 and AvrP4 genes (Catanzariti et al. 2006; Dodds et al. 2006), and Avr4 in C. fulvum (Stergiopoulos et al. 2007), consistent with R-Avr gene coevolution. Other Avr loci are characterized by presence and absence polymorphisms within species (see DeWit & Stergiopoulos in press). Few studies have observed variation in Avr gene homologues across species. Avr3 of Fusarium oxysporum f.sp. Lycopersicon is restricted to tomato infecting isolates of this species and does not occur in other forma speciales that infect different hosts (Rep et al. 2004). Comparisons of the complete genomes of three oomycetes Phytophthora ramorum, Phytophthora sojae and H. parasitica infecting oak, soya bean and Arabidopsis, respectively, have been more informative (Win et al. 2007; Jiang et al. 2008). Oomycetes encode a class of host-translocated effectors that are defined by the presence of an N-terminal RxLR uptake motif that occurs shortly after the secretion signal peptide. Approximately 400 such genes were predicted in the two Phytophthora genomes and approximately 100–200 in H. parasitica. Only approximately 10 per cent of the H. parasitica effectors have homologues in the Phytophthora species, while about a third of the Phytophthora effectors are common to both species. This class of effector genes seems to have been subject to a birth and death evolutionary process, with the rapid emergence of new effectors and loss of older ones since speciation. Thus gene conservation, such as AvrP4 across Melampsora species, is relatively uncommon in the limited datasets currently available and suggests that these genes perform important functions in rust infection.
(b). The cys knot is structurally conserved yet under strong positive selection
The conserved cys-knot structure (six cys residues and their spacing) indicates that these residues may be important for gene function across these accessions. Because sequence differences in the C-terminal cys-rich domain of the M. lini isolates resulted in non-recognition by the host, Catanzariti et al. (2006) suggested that this region is important for R gene recognition. As codons with highly significant ω-values were concentrated in this region (figure 2, black bars), we speculate that the corresponding region interacts with the host defence system, and this interaction drives the diversification of the gene. The rapid accumulation of non-synonymous changes detected in the cys knot compared with the rate of synonymous changes indicates that fixation of new mutations played a more important role in the accumulation of sequence variation than stochastic processes (Tellier & Brown 2007). This suggests that Darwinian selection outweighed balancing selection in the gene-for-gene interaction in the AvrP4 locus. It is possible that the N-terminus of AvrP4 is subject to proteolytic processing, as occurs in the C. fulvum cys-knot protein Avr9 (Vervoort et al. 1997), which would explain the relaxed selection in this region. Likewise, diversifying selection is confined to the C-terminal regions of ATR1 and ATR13 (Allen et al. 2004; Rehmany et al. 2005). Win et al. (2007) found a similar uneven pattern of diversification in many effector gene families in Phytophthora. In these cases, it is the N-terminal RxLR motif that is protected from diversification, presumably due to its conserved function in effector transport. It is possible that the N-terminal I(Q/R)GFS and L(E/Q)E(E/D)S motifs in AvrP4 have a conserved role such as in protein processing or transport into host cells.
(c). Within accession variation of the AvrP4 locus
Contrasting patterns of selection were observed for the accessions where two variant sequences were amplified. In the M. lini accessions from the Hesperolinon hosts (M5–M10; figures 2 and 3), the higher ω-values along the B copy branch contrasted markedly with those along the A copy branch (figure 3), indicating that positive selection may only be acting on the one copy (B). While multiple copies may act as reservoirs of mutational variation, selection may not act on all copies. Values of ω close to 1 may also indicate that the gene or a specific copy is non-functional. However, maintenance of conserved areas most probably indicate that the copy has been functional in the recent past.
In contrast to the Hesperolinon rusts, for the two sequences from M. reticulatae, values of ω>1 for both branches leading to the terminal nodes indicate positive selection driving amino acid substitutions (M16 and M17 figures 2 and 3) in both variants. The origin of divergent A and B variants may not be the same for the two species. Processes such as incomplete lineage sorting, hybridization, allelic diversity and gene-duplication could all result in divergent copies of a gene from one species (Maddison 1997).
5. Conclusions
Investigation of genetic variation in a known avirulence gene has revealed a locus that has been under strong positive selection in a number of rusts occurring on highly diverse hosts. The high values of ω across the gene phylogeny supports the idea that the AvrP4 gene is an avirulence elicitor across the genus Melampsora and that it has an important function that has been maintained through structurally conserved regions within the gene. Selection may have acted on these sequences because of the effector function of the locus or in order to escape recognition by host resistance factors or both. These findings are important for understanding the evolution of genes involved in the interaction between pathogens and their hosts and highlight that such genes could have originated and diversified long before a specific host–pathogen interaction.
Acknowledgements
The authors acknowledge the financial support of the Grains Research Development Corporation of Australia, Uri Springer for supplying material of the Hesperolinon rusts, John Walker for rust accessions and Celeste Linde (Australian National University, School of Botany and Zoology) for computer and office facilities, and the valuable comments of anonymous reviewers.
References
- Agrawal A., Lively C.M.2002Infection genetics: gene-for-gene versus matching allele models, and all points in between. Evol. Ecol. Res 4, 112 [Google Scholar]
- Alfano J.R., Collmer A.2004Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol 42, 385–414doi:10.1146/annurev.phyto.42.040103.110731 [DOI] [PubMed] [Google Scholar]
- Allen R.L., Bittner-Eddy P.D., Grenville-Briggs L.J., Meitz J.C., Rehmany A.P., Rose L.E., Beynon J.L.2004Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306, 1957–1960doi:10.1126/science.1104022 [DOI] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group II (APG II) 2003An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc 141, 399–436doi:10.1046/j.1095-8339.2003.t01-1-00158.x [Google Scholar]
- Anisimova M., Bielawski J.P., Yang Z.2001Accuracy and power of likelihood ratio tests in detecting adaptive molecular evolution. Mol. Biol. Evol 18, 1585–1592 [DOI] [PubMed] [Google Scholar]
- Arnold D.L., Jackson R.W., Fillingham A.J., Goss S.C., Taylor J.D., Mansfield J.W., Vivian A.2001Highly conserved sequences flank avirulence genes: isolation of novel avirulence genes from Pseudomonas syringae pv. pisi. Microbiol. SGM 147, 1171–1182 [DOI] [PubMed] [Google Scholar]
- Catanzariti A., Dodds P.N., Lawrence G.J., Ayliffe M.A., Ellis J.G.2006Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. The Plant Cell 1, 243–256doi:10.1105/tpc.105.035980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard C.1999Coevolution of a plant host-pathogen gene-for-gene system in a metapopulation model without cost of resistance or cost of virulence. J. Theor. Biol 201, 1–12doi:10.1006/jtbi.1999.1007 [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D.2001Plant pathogens and integrated defence responses to infection. Nature 411, 826–833doi:10.1038/35081161 [DOI] [PubMed] [Google Scholar]
- DeWit P. J. G. M., Stergiopoulus I. Fungal effector proteins. Annu. Rev. Phytopath. In press. [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.M., Ayliffe M.A., Ellis J.G.2004The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16, 755–768doi:10.1105/tpc.020040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.M., Teh T., Wang C.I.A., Ayliffe M.A., Kobe B., Ellis J.G.2006Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl Acad. Sci. USA 103, 8888–8893doi:10.1073/pnas.0602577103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J., Dodds P., Lawrence G.2007Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu. Rev. Phytopathol 45, 289–306doi:10.1146/annurev.phyto.45.062806.094331 [DOI] [PubMed] [Google Scholar]
- Farman M.L., Eto Y., Tosa Y., Nakayashiki H., Mayama S., Leong S.A.2002Analysis of the structure of the AVR1-CO39 avirulence rice-infecting isolates of Magnaporthe grisea. Mol. Plant Microbe Interact 15, 6–16doi:10.1094/MPMI.2002.15.1.6 [DOI] [PubMed] [Google Scholar]
- Flor H.H.1942Inheritance of pathogenicity in Melampsora lini. Phytopathology 32, 653–669 [Google Scholar]
- Flor H.H.1955Host-parasite interaction in flax rust: its genetics and other implications. Phytopathology 45, 680–685 [Google Scholar]
- Frank S.A.1993Coevolutionary genetics of plants and pathogens. Evol. Ecol 7, 45–75doi:10.1007/BF01237734 [Google Scholar]
- Hawksworth D.L., Kirk P.M., Sutton B.C., Pegler D.N.Dictionary of the fungi. 8th edn.1995Wallingford, UK:CAB International [Google Scholar]
- Holub E.B.2001The arms race is ancient history in Arabidposis, the wildflower. Nat. Rev. Genet 2, 516–527doi:10.1038/35080508 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F.2001MrBayes: Bayesian inference of phylogeny. Bioinformatics 17, 754–755doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Janjusevic R., Abramovitch R.B., Martin G.B., Stebbins C.E.2006A bacterial inhibitor of host defenses is an E3 ubiquitin ligase. Science 311, 212–225doi:10.1126/science.1120131 [DOI] [PubMed] [Google Scholar]
- Jiang R.H.Y., Tripathy S., Govers F., Tyler B.M.2008RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl Acad. Sci. USA 105, 4874–4879doi:10.1073/pnas.0709303105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijt M., Kip D.J., Joosten M., Brandwagt B.F., deWit P.2005The Cf-4 and Cf-9 resistance genes against Cladosporium fulvum are conserved in wild tomato species. Mol. Plant Microbe Interact 18, 1011–1021doi:10.1094/MPMI-18-1011 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M.2004MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinformatics 5, 150–163doi:10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Lawrence G.J., Dodds P.N., Ellis J.G.2007Rust of flax and linseed caused by Melampsora lini. Mol. Plant Path 8, 349–364doi:10.1111/j.1364-3703.2007.00405.x [DOI] [PubMed] [Google Scholar]
- Liu Z., et al. 2005Patterns of diversifying selection in the phytotoxin-like scr74 gene family of Phytophthora infestans. Mol. Biol. Evol 22, 659–672doi:10.1093/molbev/msi049 [DOI] [PubMed] [Google Scholar]
- Maddison W.P.1997Gene trees in species trees. Syst. Biol 46, 523–536doi:10.2307/2413694 [Google Scholar]
- Maier W., Begerow D., Weiß M., Oberwinkler F.2003Phylogeny of the rust fungi: an approach using nuclear large subunit ribosomal DNA sequences. Can. J. Bot 81, 12–23doi:10.1139/b02-113 [Google Scholar]
- Martin D., Rybicki E.2000RDP: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563doi:10.1093/bioinformatics/16.6.562 [DOI] [PubMed] [Google Scholar]
- Martin D.P., Williamson C., Posada D.2005RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21, 260–262doi:10.1093/bioinformatics/bth490 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J.1992Analyzing the mosaic structure of genes. J. Mol. Evol 34, 126–129 [DOI] [PubMed] [Google Scholar]
- Neilsen R., Yang Z.1998Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148, 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong L.E., Innes R.W.2006AvrB mutants lose both virulence and avirulence activities on soybean and Arabidopsis. Mol. Microbiol 60, 951–962doi:10.1111/j.1365-2958.2006.05162.x [DOI] [PubMed] [Google Scholar]
- Padidam M., Sawyer S., Fauquet C.M.1999Possible emergence of new Gemini-viruses by frequent recombination. Virology 265, 218–225doi:10.1006/viro.1999.0056 [DOI] [PubMed] [Google Scholar]
- Pallaghy P.K., Nielsen K.J., Craik D.J., Norton R.S.1994A common structural motif incorporating a cystine knot and a triple-stranded beta-sheet in toxic and inhibitory polypeptides. Protein Sci 3, 1833–1839doi:10.1002/pro.5560031022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A.1998ModelTest: testing the model of DNA substitution. Bioinformatics 14, 817–818doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rehmany A.P., et al. 2005Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. The Plant Cell 17, 1839–1850doi:10.1105/tpc.105.031807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M., Van der Does H.C., Meijer M., Van Wijk R., Houterman P.M., Dekker H.L., De Koster C.G., Cornelissen B.J.C.2004A small cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3 mediated resistance in tomato. Mol. Micobiol 53, 1373–1383doi:10.1111/j.1365-2958.2004.04177.x [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P.2003MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sackton T.B., Lazzaro B.P., Schlenke T.A., Evans J.D., Hultmark D., Clark A.G.2007Dynamic evolution of the innate immune system in Drosophila. Nat. Genet 39, 1461–1468doi:10.1038/ng.2007.60 [DOI] [PubMed] [Google Scholar]
- Sasaki A.2000Host-parasite coevolution in multi-locus gene-for-gene system. Proc. R. Soc. Lond. B 267, 2183–2188doi:10.1098/rspb.2000.1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer Y.P.2007Clinal resistance structure and pathogen local adaptation in a serpentine flax–flax rust interaction. Evolution 61, 1812–1822doi:10.1111/j.1558-5646.2007.00156.x [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J.2008A rapid bootstrap algorithm for the RAxML Web-Servers. Syst. Biol 75, 758–771doi:10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I., De Kock M.J.D., Lindhout P., DeWit P.G.J.M.2007Allelic variation in the effector genes of the tomato pathogen Cladosporium fulvum reveals different modes of adaptive evolution. Mol. Plant Microbe Interact 20, 1271–1283doi:10.1094/MPMI-20-10-1271 [DOI] [PubMed] [Google Scholar]
- Stevens C., Bennett M.A., Athanassopoulos E., Tsiamis G., Taylor J.D., Mansfield J.W.1998Sequence variations in alleles of the avirulence gene avrPphE.R2 from Pseudomonas syringae pv. phaseolicola lead to loss of recognition of the AvrPphE protein within bean cells and a gain in cultivar-specific virulence. Mol. Microbiol 29, 165–177doi:10.1046/j.1365-2958.1998.00918.x [DOI] [PubMed] [Google Scholar]
- Swofford D.L.PAUP*, phylogenetic analysis using parsimony (*and other methods), v. 4. 2001Sunderland, MA:Sinauer Associates [Google Scholar]
- Tellier A., Brown J.K.M.2007Stability of genetic polymorphism in host-parasite interactions. Proc. R. Soc. B 274, 809–817doi:10.1098/rspb.2006.0281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall P.H., Burdon J.J.1999The spatial scale of pathogen dispersal: consequences for disease dynamics and persistence. Evol. Ecol. Res 1, 681–701 [Google Scholar]
- Thrall P.H., Burdon J.J.2002Evolution of gene-for-gene systems in metapopulations: the effect of spatial scale of host and pathogen dispersal. Plant Pathol 51, 169–184doi:10.1046/j.1365-3059.2002.00683.x [Google Scholar]
- Tosa Y., Osue J., Eto Y., Oh H., Nakayashiki H., Mayama S., Leong S.A.2005Evolution of an avirulence gene, AVR1-CO39, concomitant with the evolution and differentiation of Magnaporthe oryzae. Mol. Plant Microbe Interact 18, 1148–1160doi:10.1094/MPMI-18-1148 [DOI] [PubMed] [Google Scholar]
- Tsiamis G., et al. 2000Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J 19, 3204–3214doi:10.1093/emboj/19.13.3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler B.M., et al. 2006Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266doi:10.1126/science.1128796 [DOI] [PubMed] [Google Scholar]
- Van der Merwe M., Ericson L., Walker J., Thrall P.H., Burdon J.J.2007Evolutionary relationships among species of Puccinia and Uromyces (Pucciniaceae, Uredinales) inferred from partial protein coding gene phylogenies. Mycol. Res 111, 163–175doi:10.1016/j.mycres.2006.09.015 [DOI] [PubMed] [Google Scholar]
- Vervoort J., Van den Hooven H.W., Berg A., Vossen P., Vogelsang R., Joosten M.H.A.J., De Wit P.J.G.M.1997The race-specific elicitor AVR9 of the tomato pathogen Cladosporium fulvum: a cystine knot protein. Sequence-specific 1H NMR assignments, secondary structure and global fold of the protein. FEBS Lett 404, 153–158doi:10.1016/S0014-5793(97)00117-8 [DOI] [PubMed] [Google Scholar]
- Win J., Morgan W., Bos J., Krasileva K.V., Cano L.M., Chaparro-Garcia A., Ammar R., Staskawicz B.J., Kamoun S.2007Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. The Plant Cell 19, 2349–2369doi:10.1105/tpc.107.051037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield B.D., Ericson L., Szaro T., Burdon J.J.2004Phylogenetic patterns in the Uredinales. Aust. Plant Path 33, 327–335doi:10.1071/AP04020 [Google Scholar]
- Yang Z.1997PAML: a computer package for phylogenetic analysis by mazimum likelihood. Comp. Appl. Biosci 13, 555–556 [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., Goldman N., Pedersen A.M.K.2000Codon-substitution models for heterogenous selection pressure at amino acid sites. Genetics 155, 431–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wong W.S.W., Nielsen R.2005Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol 22, 1107–1118doi:10.1093/molbev/msi097 [DOI] [PubMed] [Google Scholar]