Abstract
Gene duplication is the primary source of new genes, but the mechanisms underlying the functional divergence and retention of duplicate genes are not well understood. Because eukaryotic proteins are localized to subcellular structures and localization can be altered by a single amino acid replacement, it was recently proposed that protein subcellular relocalization (PSR) plays an important role in the functional divergence and retention of duplicate genes. Although numerous examples of distinct subcellular localizations of paralogous proteins have been reported, it is unknown whether PSR occurs more frequently after gene duplication than without duplication. By analyzing experimentally determined and computationally predicted genome-wide protein subcellular localization data of the budding yeast Saccharomyces cerevisiae and two other fungi (Schizosaccharomyces pombe and Kluyveromyces waltii), we show that even singleton genes have an appreciable rate of relocalization in evolution and that duplicate genes do not relocalize more frequently than singletons. These results suggest that subcellular relocalization is unlikely to have been a major mechanism for duplicate gene retention and functional divergence at the genomic scale.
Keywords: yeast, duplicate gene, singleton gene, subcellular localization, evolution
Protein Subcellular Relocalization as a Molecular Mechanism for Duplicate Gene Retention
Gene duplication is widely believed to be the primary source of new genes (Ohno 1970; Zhang 2003; Conant and Wolfe 2008). With a few exceptions, functional divergence is required for duplicate genes to be stably retained in a genome (Zhang 2003). Ohno (1970) proposed that functional divergence between duplicates mainly occurs by acquisition of new functions or neofunctionalization. However, because mutations creating new functions in a gene are presumably much rarer than mutations that destroy or inactivate the gene, a duplicate gene is unlikely to acquire new functions before becoming a pseudogene. For this reason, several authors independently proposed that subdivision of ancestral functions of the progenitor gene into daughter genes, or subfunctionalization, may be more important for the functional divergence and retention of duplicate genes (Hughes 1994; Force et al. 1999; Stoltzfus 1999). Analyzing genome-wide protein–protein interaction and gene expression data, He and Zhang (2005b) found evidence for rapid subfunctionalization accompanied by substantial and prolonged neofunctionalization in duplicate gene evolution, suggesting that the stable retention of duplicate genes is primarily owing to subfunctionalization, whereas new functions are gradually acquired in retained duplicates. There are potentially many molecular mechanisms for subfunctionalization and neofunctionalization, such as changes in the cis-regulatory motifs of a gene or in the binding sites for protein–protein interaction, but the relative importance of these and other molecular mechanisms remains elusive (Conant and Wolfe 2008).
Recently, protein subcellular relocalization (PSR) was proposed as an important molecular mechanism for the functional divergence and retention of duplicate genes (Byun-McKay and Geeta 2007). The proposal is based on the facts that 1) most eukaryotic proteins are localized to subcellular structures to perform their functions, 2) proteins may be directed to a different subcellular structure by a single amino acid change, and 3) proteins can have altered functions when relocalized due to altered microenvironments (Byun-McKay and Geeta 2007). Because of the relative ease of PSR, both subdivision of ancestral localizations and acquisition of new localizations may happen relatively quickly after gene duplication, and thus, PSR could play an important role in the functional divergence and retention of duplicate genes (Byun-McKay and Geeta 2007). Several paralogous proteins are known to differ in subcellular localization (Byun-McKay and Geeta 2007). More recently, Marques et al. (2008) conducted a genome-wide analysis in the budding yeast Saccharomyces cerevisiae and found that 24–37% of the duplicate gene pairs generated by the whole-genome duplication (WGD) ∼100 million years ago (Ma) now have distinct protein subcellular localizations. Although these observations are consistent with an appreciable PSR rate in duplicate gene evolution, it is unclear whether the PSR rate is higher in duplicate genes than in singletons, which would be expected if PSR is an important determinant of duplicate gene retention. Below, we estimate PSR rates in singleton and duplicate genes, using experimentally determined and computationally predicted genome-wide subcellular localization data from the budding yeast and two other fungi.
Duplicates Do Not Relocalize More Frequently Than Singletons: Analysis of Experimental Data
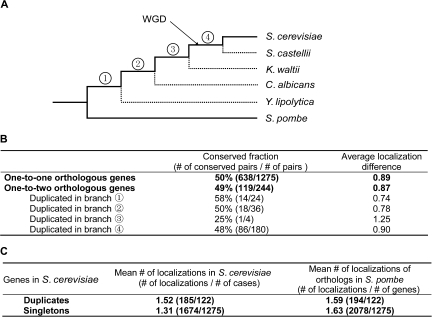
We compared protein subcellular localizations between S. cerevisiae and the fission yeast Schizosaccharomyces pombe (fig. 1A), which diverged from each other at least 300 Ma (Wapinski et al. 2007). Protein subcellular localization data generated from large-scale fluorescent imaging–based experiments are available for the two species, covering 75% and 90% of all genes in the two genomes, respectively (Huh et al. 2003; Matsuyama et al. 2006). We used the data from the large-scale experiments instead of those complied from individual small-scale experiments (e.g., the gene ontology [GO] database) because the large-scale experiments used the same criteria in identifying subcellular localizations for all genes. We retrieved fungal gene trees from an earlier study (Wapinski et al. 2007) to identify one-to-one orthologous gene pairs between S. pombe and S. cerevisiae. These genes have not duplicated on the lineages leading to the two species since their separation (fig. 1A). We similarly identified one-to-two orthologous gene trios between S. pombe and S. cerevisiae. These genes did not duplicate on the S. pombe lineage but duplicated once on the S. cerevisiae lineage since the species separation.
FIG. 1.—
Similar rates of subcellular relocalization between singletons and duplicates. (A) A phylogeny of fungi (Wapinski et al. 2007), where four interior branches and the WGD are marked. The branches connecting the two species under comparison are shown by solid lines, whereas other branches are shown by dotted lines. (B) PSRs in one-to-one and one-to-two orthologs between S. pombe and S. cerevisiae. (C) Subcellular localization numbers of duplicates and singletons. Duplicates refer to those with one copy in S. pombe but two copies in S. cerevisiae; the joint localization number is considered for the two copies in S. cerevisiae.
We found that among the 1,275 one-to-one orthologous pairs with protein subcellular localization information, 638 pairs (FC =50.0%) have exactly the same localizations between the two species (fig. 1B). There are 122 one-to-two orthologous gene trios with localization information. To make a fair comparison with one-to-one orthologs, we consider each trio as two orthologous pairs. That is, for a trio between S. pombe gene B and S. cerevisiae genes A1 and A2, we consider an orthologous pair between B and A1 and a second orthologous pair between B and A2 because the divergence time between B and A1 and that between B and A2 are both identical to the divergence time of one-to-one orthologs, which is the divergence time between S. pombe and S. cerevisiae. Of the 244 orthologous pairs of the 122 trios, 119 pairs (FC =48.8%) have exactly the same localizations between the two species (fig. 1B). Thus, there is no significant difference between singletons and duplicates in the percentage of genes with completely conserved protein subcellular localizations (P = 0.76, two-tailed chi-square test). Further, there is no significant difference between singletons (19%) and duplicates (21%) in the fraction of genes with completely different localizations in the two species (P = 0.52, two-tailed chi-square test). To examine whether the mean numbers of relocalization events are similar between singletons and duplicates, we calculated the number of subcellular localization differences (D) for each orthologous pair, which is the number of localizations found in S. pombe but not S. cerevisiae plus the number of localizations found in S. cerevisiae but not S. pombe. Again, we found that D is not significantly different between one-to-one (0.89) and one-to-two (0.87) orthologs (P = 0.92, two-tailed Mann–Whitney test; fig. 1B). Note that the mean number of localizations (N) in S. pombe is slightly higher for one-to-one (1.63) than one-to-two (1.59) orthologs, although the difference is not statistically significant (P = 0.74, two-tailed Mann–Whitney test; fig. 1C). We found no significant difference in f = D/N between one-to-one (0.52) and one-to-two (0.55) orthologs (P = 0.71, two-tailed Mann–Whitney test).
Because singletons and duplicates may have intrinsic differences in function (Marland et al. 2004; He and Zhang 2005a, 2006), we further examined whether there is significant difference in relocalization rates between singletons and duplicates of the same functional categories. We retrieved GO annotations of S. cerevisiae genes and examined the six GO categories for which the localization data exist for at least 30 one-to-one orthologs and 30 one-to-two orthologs. Again, no significant difference in D was found between one-to-one and one-to-two orthologs in any functional category (supplementary table S1, Supplementary Material online).
A previous study found that highly expressed duplicate genes have fewer between-paralog differences in localization than lowly expressed ones of the same ages, suggesting that protein expression levels impact the rate of relocalization in evolution (Marques et al. 2008). We found that duplicates generally have higher protein expressions than singletons (P = 0.02, two-tailed Mann–Whitney test). Nonetheless, even after the control of protein expression level, no significant difference in D was found between one-to-one and one-to-two orthologs (Table S2).
In our analysis, we have combined all one-to-two orthologs regardless of the time of gene duplication on the S. cerevisiae lineage. Although it is possible that an increase in relocalization occurs only in the early stage after gene duplication, it is also possible that it lasts for a long time. In the latter case, the signal of potentially accelerated relocalization would be easier to detect in genes that duplicated relatively earlier. Using fungal gene trees (Wapinski et al. 2007), we determined for each of the one-to-two trios the branch in the species tree on which the duplication occurred (fig. 1A). However, there is no significant difference in conservation fraction (FC) and localization difference (D) among groups of duplicate genes that duplicated at different times (P > 0.9, two-tailed chi-square test; fig. 1B). Using computer simulation, we found that an enhancement of relocalization in duplicates would have been detectable in our samples if duplication had led to relocalization events in each daughter gene equivalent to the quantity in 100 million years (My) of singleton gene evolution. These results suggest that, compared with singletons, relocalization in duplicates is either not increased or only increased for such a small extent that the signal is difficult to discern today.
Although on average a duplicate gene does not have more subcellular localizations than a singleton in S. cerevisiae (P = 0.18, two-tailed Mann–Whitney test), the joint number of subcellular localizations of a duplicate pair is significantly larger than a singleton in S. cerevisiae (P = 2.5 × 10−6, two-tailed Mann–Whitney test; fig. 1C), as was previously observed (Marques et al. 2008). This difference is not attributable to a potential difference in subcellular localization number between the progenitors of singletons and duplicates because there is no significant difference in localization number between the S. pombe genes of the one-to-one orthologous group and the one-to-two orthologous group (P = 0.74, two-tailed Mann–Whitney test; fig. 1C). Rather, it is caused by relocalization of duplicate genes that occurred after duplication. Nevertheless, as shown above, the rate of relocalization is not significantly higher in duplicates than in singletons. One then wonders why the joint number of subcellular localizations of a duplicate pair is significantly greater than the number of localizations of a singleton in S. cerevisiae. The answer is that loss of an ancestral subcellular localization in one member of a duplicate pair does not decrease the joint number of localizations for the pair, whereas the opposite is true for a singleton. Furthermore, even when the number of neolocalization is the same in a duplicate gene and a singleton gene, the number for a duplicate pair is twice that for a singleton. Thus, even though the relocalization rates are not different between singletons and duplicates, duplicates have a greater joint number of localizations than singletons.
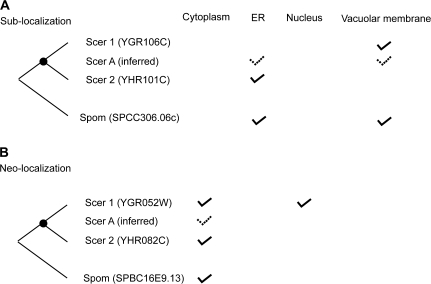
For each of the 122 one-to-two trios, it is possible to infer the localizations of the progenitor gene of the duplicate copies and differentiate between sublocalization and neolocalization after gene duplication (Marques et al. 2008), based on the parsimony principle. Here sublocalization is stringently defined by the observation that each daughter gene loses at least one ancestral localization that is kept in the other paralog, whereas neolocalization is defined by the acquisition of at least one new localization in one daughter gene. In 84 cases, neither copy relocalized after duplication. Of the remaining 38 cases, 11 cannot be classified as sublocalization or neolocalization, 1 had sublocalization, 26 had neolocalization, and 0 had both (fig. 2). When the 122 cases are separated into two groups based on whether the duplication occurred in branch #4 (the younger group) or earlier branches (the older group; fig. 1A), the younger group has a higher fraction (74.4%) than the older group (53.1%) of duplicates where neither copy relocalized (P = 0.029, two-tailed Fisher's exact test), as expected. Further, the fraction of trios with neolocalization is greater in the older group (36.4%) than in the younger group (15.2%, P = 0.014, two-tailed Fisher's exact test), suggesting that neolocalization is a slow process, similar to the acquisition of new protein interactions and new expression sites (He and Zhang 2005b). However, contrary to the patterns of protein interaction and expression site changes (He and Zhang 2005b), sublocalization after gene duplication appears rare.
FIG. 2.—
Examples of (A) sublocalization and (B) neolocalization after gene duplication. The solid check marks indicate experimental data, whereas the dashed check marks indicate parsimony-based ancestral state inferences.
Duplicates Do Not Relocalize More Frequently Than Singletons: Analysis of Predicted Data
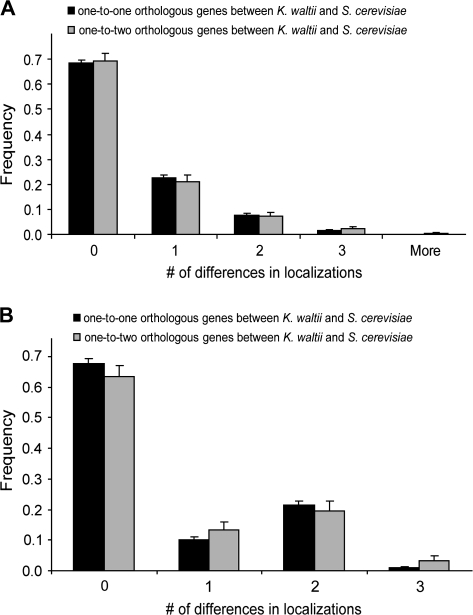
Because the majority (180/244 = 74%) of the genes in the one-to-two group analyzed above duplicated after the separation of S. cerevisiae and Kluyveromyces waltii (fig. 1), the power of detecting potential acceleration in relocalization after gene duplication is expected to be greater in an S. cerevisiae−K. waltii comparison than in an S. cerevisiae−S. pombe comparison. Furthermore, when comparing the more closely related S. cerevisiae and K. waltii, we could use additional genes that were either lost or duplicated in the S. pombe lineage and unsuitable for the earlier analysis. In fact, the majority of one-to-two orthologous genes between K. waltii and S. cerevisiae are those retained from the WGD that occurred on the S. cerevisiae lineage shortly after its separation from the K. waltii lineage (Wolfe and Shields 1997; Kellis et al. 2004). We decided to focus on this group of genes because they were all duplicated at the same time and because they spent most of their time in the S. cerevisiae lineage as duplicates. However, there are no experimentally determined subcellular localization data from K. waltii, and we thus use computer programs to predict the localizations in K. waltii.
We identified in K. waltii and S. cerevisiae one-to-one orthologous genes and one-to-two orthologous genes generated by WGD. To predict subcellular localization, we excluded programs that use homology information because such predictions will not reveal relocalizations in evolution. We tested five commonly used programs, WoLF PSORT, Cello, MultiLoc, Proteome Analyst, and pTarget, by comparing the predicted subcellular localizations of S. cerevisiae proteins with the experimentally determined localizations. WoLF PSORT performed the best, with an accuracy of 45%. That is, 45% of proteins have their predicted localizations match exactly the experimentally determined localizations, when subcellular structures that are considered by the computer program are concerned. We thus used WoLF PSORT in subsequent analysis. For each one-to-one or one-to-two orthologous pair, we predicted subcellular localizations in K. waltii using a threshold above which the program is known to make correct predictions for the S. cerevisiae ortholog. We found no significant difference in subcellular relocalization rate between one-to-one and one-to-two orthologous genes (fig. 3A). To confirm the above results, we also used the second best prediction program, MultiLoc, which has an accuracy of 36% in predicting subcellular localizations of S. cerevisiae proteins. Virtually identical results were obtained (fig. 3B). We further confirmed that the same conclusion can be drawn by using predicted localizations of S. pombe proteins (supplementary fig. S1, Supplementary Material online). Thus, experimentally determined and computationally predicted localization data both show similar rates of relocalization in singleton and duplicate genes.
FIG. 3.—
Duplicate genes generated from WGD did not relocalize more frequently than singletons. (A) Comparison based on WoLF PSORT-predicted localizations in K. waltii. (B) Comparison based on MultiLoc-predicted localizations in K. waltii. Error bars show one standard error. No significant difference in relocationization rate between one-to-one and one-to-two orthologs is found (P = 0.94 for panel [A] and 0.31 for panel [B], two-tailed Mann–Whitney test).
Caveats and Implications
Our analysis has several potential caveats. First, the difficulty of computational prediction of subcellular localization is well recognized (Lei and Dai 2005; Chou and Shen 2007). Although we set stringent criteria in applying the computational methods, the stringency potentially lowered the power of detecting small differences in relocalization rates between singletons and duplicates. Second, false-positive and false-negative errors may also exist in the experimentally determined subcellular localization data. In both budding yeast and fission yeast, the florescent protein tags were inserted into the C-terminus of the protein and may affect the subcellular localization if the localization signal is at the C-terminus (Huh et al. 2003). However, this systematic bias should not affect the comparison between singletons and duplicates of the two species. Third, an important difference between the experiments in the two yeasts is that the endogenous promoter was used in expressing each florescent protein–tagged gene at the original chromosomal location in budding yeast, whereas florescent protein–tagged genes are expressed from a very strong common promoter on plasmids in fission yeast (Matsuyama et al. 2006). This difference could result in 1) protein mistargeting in S. pombe due to an increase in protein amount and 2) different sensitivities in identifying subcellular localizations of orthologous proteins in the two yeasts, which could generate artificial differences between the localizations of orthologs. Consistent with the above prediction, we found one-to-one orthologous proteins to have more localizations (1.63) in S. pombe than in S. cerevisiae (1.31; P < 10−28, two-tailed Mann–Whitney test; fig. 1C). However, this problem is expected to affect singletons and duplicates equally and should not bias our results, although it may reduce the power in detecting the difference between singletons and duplicates if only a small difference exists.
We found that only 50% one-to-one orthologs between S. cerevisiae and S. pombe have exactly the same subcellular localizations (or 63% if the localizations in S. pombe are predicted by WoLF PSORT). The corresponding number is 68% between S. cerevisiae and K. waltii. Although the low conservation may in part arise from the differential experimental sensitivities mentioned above and/or errors in computational prediction of subcellular localization, it is also possible that even one-to-one orthologous proteins change their subcellular localization with an appreciable rate in evolution. For example, the protein composition of the mitochondrion varies among species (Heazlewood et al. 2003; Meisinger et al. 2008); <80% of human mitochondrial proteins with yeast homologs are localized to the mitochondrion in yeast (Prokisch et al. 2006). This observation is unlikely to have arisen from under-detection of yeast mitochondrial proteins because comprehensive identifications of mitochondrial proteins have been conducted in yeast using different methods (Kumar et al. 2002; Steinmetz et al. 2002; Huh et al. 2003). Rather, it is consistent with the fact that the same subcellular compartment may have different functions in different species. For example, the mitochondrion is more important for apoptosis in human than in budding yeast (Heazlewood et al. 2003). Our results are also broadly consistent with the recent observation that a considerable fraction of one-to-one orthologous genes differ in function or functional importance between closely related species such as human and mouse (Liao and Zhang 2008). Together, these findings are consistent with the contention that duplicates do not necessarily diverge faster than orthologs in function (Studer and Robinson-Rechavi 2009).
If PSR contributes significantly to the retention of duplicate genes at the genomic scale, duplicate genes should show more relocalizations than singletons for the same amount of evolutionary time considered. Hence, our finding of a lack of significant difference in relocalization rates between singletons and duplicates in yeast evolution suggests that subcellular relocalization is not an important determinant of duplicate gene retention at the genomic scale. There are multiple aspects of gene function (gene expression, protein localization, molecular function, physiological function, etc.) that can be altered after duplication. The first of these functional changes brought about by mutation is likely the one that makes the greatest contribution to duplicate gene retention. Although PSR requires as few as one amino acid replacement, the replacement required is usually specific in type and position. Furthermore, signals for protein targeting to dual destinations are often nonmodular (Karniely and Pines 2005), which may explain the scarcity of sublocalization after gene duplication. Together, it is quite possible that mutations causing relocalization are not as common as other types of mutations, such as those altering gene expression level or pattern. Consequently, relocalization plays a less important role than other functional changes in duplicate gene retention. Indeed, there is circumstantial evidence for gene expression divergence being a major contributor to duplicate gene retention (Force et al. 1999; Gu et al. 2002, 2004; Huminiecki and Wolfe 2004; He and Zhang 2005b). That said, our results do not exclude subcellular relocalization as an important factor in the retention of some duplicate genes, as has been demonstrated recently (Byun-McKay and Geeta 2007; Rosso, Marques, Reichert, and Kaessmann 2008; Rosso, Marques, Weier, et al. 2008).
Materials and Methods
Fungal one-to-one and one-to-two orthologs were identified from published fungal gene trees (Wapinski et al. 2007). One-to-one orthologs are those genes with neither duplication nor gene loss on the S. cerevisiae and S. pombe lineages since their separation. One-to-two orthologs have neither duplication nor gene loss on the S. pombe lineage but only one duplication on the S. cerevisiae lineage. Saccharomyces cerevisiae duplicates generated from the WGD were previously published (Kellis et al. 2004). Protein sequences of S. cerevisiae were downloaded from the Saccharomyces genome database (SGD; http://www.yeastgenome.org/) and those of K. waltii were obtained from its published genome sequence (Kellis et al. 2004). GO functional category annotations for yeast genes were downloaded from SGD. Saccharomyces cerevisiae protein expression levels (molecules per cell) were previously determined (Ghaemmaghami et al. 2003). Protein subcellular localization data of S. cerevisiae (Huh et al. 2003) and S. pombe (Matsuyama et al. 2006) were previously published. Subcellular compartments in a data set that do not have counterparts in the other data set were not considered. Eighteen and 14 subcellular compartments were considered in S. cerevisiae and S. pombe, respectively, and their homologous relationships are depicted in supplementary figure S2 (Supplementary Material online) based on a previous definition (Matsuyama et al. 2006). For example, the “bud neck” in S. cerevisiae and the “septum” in S. pombe are regarded as homologous. In S. pombe, protein subcellular localization data included the relative fluorescence intensities among the subcellular locations indicated by “=,” “>,” and “≫” signs. We disregarded the locations after the “≫” sign. For all analyses, only orthologous locations were considered. When comparing a pair of orthologs, we considered them to have the same subcellular localizations only when their localizations match each other completely. For those pairs with different localizations, we calculated the minimal number of evolutionary changes (i.e., gains or losses of localizations) required to explain their distinct localizations.
We used computer simulation to evaluate the power of our analysis in detecting a relocalization rate difference between one-to-one and one-to-two orthologs of S. pombe and S. cerevisiae. We first estimated the mean number (D) of subcellular localization difference of one-to-one orthologs. Let the divergence time between S. pombe and S. cerevisiae be 2T years. Let the extra relocalization per daughter gene that is associated with duplication be equivalent to t years of relocalization without duplication and let f = t/(2T). For each one-to-two ortholog, we added to its number of localization difference a Poisson random number with the mean of Df. We then compared the localization differences of the 1,275 one-to-one orthologs with the augmented localization differences of the 244 one-to-two orthologs, using a two-tailed Mann–Whitney test. We found that when f = 1/6 (i.e., t = 100 My if T = 300 My), the test is significant at P = 0.05 in 60.6% of the 1,000 simulation replications and significant at P = 0.10 in 90.1% of the simulation replications. This result suggests that our test could detect a significant difference in relocalization rate between singletons and duplicates if f is not smaller than 1/6.
Five computer programs were used to predict the subcellular localizations of K. waltii proteins: WoLF PSORT (Horton et al. 2007), Cello (Yu et al. 2006), MultiLoc (Hoglund et al. 2006), Proteome Analyst (Szafron et al. 2004), and pTarget (Guda 2006). These programs were chosen because they do not use homology information in prediction and because they consider at least seven subcellular locations. The procedure was as follows. First, each of the five programs was tested for prediction accuracy in S. cerevisiae. If more than one subcellular localization was predicted by a program with different confidences, we ranked the localizations from high to low levels of confidence and examined whether there was a confidence level above which all the subcellular localizations matched experimental data. If this level existed, we considered the prediction to be correct above a threshold. The threshold was calculated as the mean of 1) the lowest confidence level above which all predictions were correct and 2) the highest confidence level for which the prediction was wrong (0 if all predictions were correct). For a given gene, if the prediction with the highest confidence was incorrect, the prediction for the gene was considered completely incorrect. WoLF PSORT and MultiLoc were found to have higher prediction accuracies than other programs and were used in subsequent analysis. For an S. cerevisiae gene whose subcellular localizations were correctly predicted (at least partially), we used the aforementioned confidence threshold to predict localizations for its K. waltii ortholog. That is, only predictions above the threshold were considered. We did not predict protein subcellular localizations for K. waltii when the localization predictions of its S. cerevisiae ortholog were completely incorrect. Subcellular localizations of 1,154 one-to-one orthologs and 224 one-to-two orthologs were predicted for K. waltii by WoLF PSORT and, 912 and 178, respectively, by MultiLoc. Subcellular localizations of 659 one-to-one orthologs and 137 one-to-two orthologs were predicted for S. pombe by WoLF PSORT.
Funding
US National Institutes of Health (R01GM067030) to J.Z.
Supplementary Material
Supplementary figures S1 and S2 and table S1 and S2 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Supplementary Material
Acknowledgments
We thank Meg Bakewell, Ashley Byun-McKay, Henrik Kaessmann, and Zhi Wang for valuable comments.
References
- Byun-McKay SA, Geeta R. Protein subcellular relocalization: a new perspective on the origin of novel genes. Trends Ecol Evol. 2007;22:338–344. doi: 10.1016/j.tree.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. Large-scale plant protein subcellular location prediction. J Cell Biochem. 2007;100:665–678. doi: 10.1002/jcb.21096. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gu Z, Nicolae D, Lu HH, Li WH. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 2002;18:609–613. doi: 10.1016/s0168-9525(02)02837-8. [DOI] [PubMed] [Google Scholar]
- Gu Z, Rifkin SA, White KP, Li WH. Duplicate genes increase gene expression diversity within and between species. Nat Genet. 2004;36:577–579. doi: 10.1038/ng1355. [DOI] [PubMed] [Google Scholar]
- Guda C. pTARGET: a web server for predicting protein subcellular localization. Nucleic Acids Res. 2006;34:W210–W213. doi: 10.1093/nar/gkl093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Gene complexity and gene duplicability. Curr Biol. 2005a;15:1016–1021. doi: 10.1016/j.cub.2005.04.035. [DOI] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005b;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Higher duplicability of less important genes in yeast genomes. Mol Biol Evol. 2006;23:144–151. doi: 10.1093/molbev/msj015. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Millar AH, Day DA, Whelan J. What makes a mitochondrion? Genome Biol. 2003;4:218. doi: 10.1186/gb-2003-4-6-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund A, Donnes P, Blum T, Adolph HW, Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics. 2006;22:1158–1165. doi: 10.1093/bioinformatics/btl002. [DOI] [PubMed] [Google Scholar]
- Horton P, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Huminiecki L, Wolfe KH. Divergence of spatial gene expression profiles following species-specific gene duplications in human and mouse. Genome Res. 2004;14:1870–1879. doi: 10.1101/gr.2705204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniely S, Pines O. Single translation–dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 2005;6:420–425. doi: 10.1038/sj.embor.7400394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kumar A, et al. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Dai Y. An SVM-based system for predicting protein subnuclear localizations. BMC Bioinformatics. 2005;6:291. doi: 10.1186/1471-2105-6-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci USA. 2008;105:6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marland E, Prachumwat A, Maltsev N, Gu Z, Li WH. Higher gene duplicabilities for metabolic proteins than for nonmetabolic proteins in yeast and E. coli. J Mol Evol. 2004;59:806–814. doi: 10.1007/s00239-004-0068-x. [DOI] [PubMed] [Google Scholar]
- Marques AC, Vinckenbosch N, Brawand D, Kaessmann H. Functional diversification of duplicate genes through subcellular adaptation of encoded proteins. Genome Biol. 2008;9:R54. doi: 10.1186/gb-2008-9-3-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Sickmann A, Pfanner N. The mitochondrial proteome: from inventory to function. Cell. 2008;134:22–24. doi: 10.1016/j.cell.2008.06.043. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin (Germany): Springer-Verlag; 1970. [Google Scholar]
- Prokisch H, et al. MitoP2: the mitochondrial proteome database—now including mouse data. Nucleic Acids Res. 2006;34:D705–D711. doi: 10.1093/nar/gkj127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso L, Marques AC, Reichert AS, Kaessmann H. Mitochondrial targeting adaptation of the hominoid-specific glutamate dehydrogenase driven by positive Darwinian selection. PLoS Genet. 2008;4:e1000150. doi: 10.1371/journal.pgen.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso L, et al. Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Biol. 2008;6:e140. doi: 10.1371/journal.pbio.0060140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, et al. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- Stoltzfus A. On the possibility of constructive neutral evolution. J Mol Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- Studer RA, Robinson-Rechavi M. How confident can we be that orthologs are similar, but paralogs differ? Trends Genet. 2009;25:210–216. doi: 10.1016/j.tig.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Szafron D, et al. Proteome Analyst: custom predictions with explanations in a web-based tool for high-throughput proteome annotations. Nucleic Acids Res. 2004;32:W365–W371. doi: 10.1093/nar/gkh485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.