Abstract
Eukaryotic genomes contain large amount of repetitive DNA, most of which is derived from transposable elements (TEs). Progress has been made to develop computational tools for ab initio identification of repeat families, but there is an urgent need to develop tools to automate the annotation of TEs in genome sequences. Here we introduce REPCLASS, a tool that automates the classification of TE sequences. Using control repeat libraries, we show that the program can classify accurately virtually any known TE types. Combining REPCLASS to ab initio repeat finding in the genomes of Caenorhabditis elegans and Drosophila melanogaster allowed us to recover the contrasting TE landscape characteristic of these species. Unexpectedly, REPCLASS also uncovered several novel TE families in both genomes, augmenting the TE repertoire of these model species. When applied to the genomes of distant Caenorhabditis and Drosophila species, the approach revealed a remarkable conservation of TE composition profile within each genus, despite substantial interspecific covariations in genome size and in the number of TEs and TE families. Lastly, we applied REPCLASS to analyze 10 fungal genomes from a wide taxonomic range, most of which have not been analyzed for TE content previously. The results showed that TE diversity varies widely across the fungi “kingdom” and appears to positively correlate with genome size, in particular for DNA transposons. Together, these data validate REPCLASS as a powerful tool to explore the repetitive DNA landscapes of eukaryotes and to shed light onto the evolutionary forces shaping TE diversity and genome architecture.
Keywords: transposable elements, transposons, repetitive elements, genome annotation, repeat classification
Introduction
The lower cost and increased pace of genome sequencing has created a need to develop new computational methods that will accelerate genome annotation and enhance biological discovery from raw sequence data. Many such tools have been developed to identify protein-coding exons ab initio and automate gene annotation (Jones 2006; Flicek 2007; Brent 2008; Ter-Hovhannisyan et al. 2008). However, protein-coding sequences represent only a small fraction of most eukaryotic genomes. Instead, the nuclear genome of most eukaryotes is replete with noncoding and repetitive DNA, a characteristic that has been appreciated for a long time (Britten and Davidson 1971) and reaffirmed by the analyses of draft genome sequences now available for a wide range of multicellular eukaryotes (e.g., Lander et al. 2001; Waterston et al. 2002; IRGSP 2005; Carlton et al. 2007; Clark et al. 2007; Mikkelsen et al. 2007; Nene et al. 2007). These studies have revealed that the bulk of repetitive DNA is composed of interspersed repeats that are derived predominantly from the past amplification of diverse forms of mobile or transposable elements (TEs). Hence, TEs and their remnants often represent a sizeable portion of eukaryotic genomes, for example, ∼22% in Drosophila melanogaster (Kapitonov and Jurka 2003), ∼35% in rice (IRGSP 2005), and nearly 50% in human (Lander et al. 2001).
Comparative and evolutionary genomic analyses have also revealed that TEs and other repetitive DNA account for the most rapidly evolving components of the genome, whereas (cellular) genes represent more conservative entities, with homologous and often orthologous genes being detectable across widely diverged species (e.g., Waterston et al. 2002). Thus, many of the protein-coding genes of an organism can be identified based on sequence similarity with genes annotated in other species. In contrast, such homology-based approaches can only capture a small fraction of TE content. Indeed, a relatively small amount of TEs are conserved among eukaryotic species, sometimes even at a close evolutionary distance, which makes TE identification and annotation a daunting task.
The dynamic turnover and complex evolutionary histories of TEs bestow these elements with an enormous potential as catalysts of lineage-specific genome evolution (Marino-Ramirez et al. 2005; Mikkelsen et al. 2007; Wang et al. 2007; Bourque et al. 2008; Feschotte 2008). Indeed, it is now well established that TEs are an important source of spontaneous mutations and evolutionary innovations and that they have been key players in the shaping of chromosomal architecture and gene regulation in eukaryotes (Kidwell and Lisch 2001; Eichler and Sankoff 2003; Kazazian 2004; Feschotte and Pritham 2007b; Belancio et al. 2008; Feschotte 2008). Therefore, knowing how many and what kind of TEs populate a genome is of fundamental interest to those studying genome structure and function, and TE annotation lays at the heart of many comparative and evolutionary genomic studies.
Eukaryotic TEs are divided into two classes according to their transposition intermediates (for review, Wicker et al. 2007). Class 1 elements, or retrotransposons, transpose via an RNA intermediate, whereas class 2 elements use a DNA intermediate. Each class is further divided into subclasses (or “orders” in Wicker et al. 2007) based on structural characteristics and mode of replication. Most class 1 elements fall into two subclasses, the long terminal repeat (LTR) retrotransposons, which are inserted by means of an element-encoded retroviral-like integrase, or the non-LTR retrotransposons, which include long and short interspersed elements (LINEs and SINEs) and use target-primed reverse transcription. A somewhat less common subclass of retrotransposons is represented by the DIRS-like elements, which use a tyrosine recombinase for integration. Class 2 elements can be divided into three major subclasses: the classic “cut-and-paste” DNA transposons, the rolling-circle Helitrons, and the self-replicating Maverick (or Polintons) elements (for review, Feschotte and Pritham 2007b). TE subclasses can be further split into superfamilies based on structural features and phylogenetic clustering. TE families are more difficult to delimit, but it is generally accepted that two different families occur when they are represented by consensus sequences that share no more than 80% nucleotide similarity. Thus, individual elements are generally grouped into the same family when they share more than 80% similarity to each other over at least 80% of their length and at least 80 bp of sequence (also known as the 80/80/80 rule in Wicker et al. 2007).
The process of TE annotation in a genome sequence can be broken down into three distinct steps: identification, classification, and masking (Feschotte and Pritham 2007a). Of these three steps, masking is currently the most straightforward as it consists of scanning the genome with sensitive algorithms for segments of the genome with significant similarity to one of several repeats precharacterized for the species and stored in a library of representative consensus sequences. The Repeatmasker software (http://www.repeatmasker.org/), which makes use of “manually–curated” reference libraries of consensus sequences (e.g., Jurka et al. 2005), has become the gold standard for masking. So far, the compilation of the reference libraries used for masking relies on the ability of a few experts to mine individual repeats, reconstruct consensus sequences and classify each TE family. Because of the explosion of sequence data and of the evolutionarily labile nature of TEs, ab initio approaches to repeat identification have become highly desirable to automate the construction of consensus TE library from complete or partial genome sequences. Ab initio repeat identification is theoretically challenging and computationally intensive, and software packages like RECON (Bao and Eddy 2002), RepeatScout (Price et al. 2005), Piler (Edgar and Myers 2005), and ReAS (Li et al. 2005) have been designed to automate this process. Individually, none of these programs is able to generate a comprehensive, “masking-ready” library of consensus repeats from an input genome sequence, but they produce a useful output representing the most abundant and homogeneous repeat families in a genome, especially when several programs are combined and integrated (Quesneville et al. 2005; Bergman and Quesneville 2007; Smith, Edgar, et al. 2007; Saha et al. 2008). The next step in TE annotation is to identify the diagnostic features of each consensus sequence, thereby inferring the biological classification of each repeat. Currently, there is no published application that can provide an automated biological classification of TEs at a relatively fine scale. Some repeat finding programs have implemented procedures to distinguish tandem from interspersed repeats (Edgar and Myers 2005) or class 1 versus class 2 TEs (i.e., retrotransposons vs. DNA transposons; (Andrieu et al. 2004). But until now, the classification of repeats into TE superfamilies and subclasses has been performed “manually,” one repeat family at a time, a painstaking task that requires an exquisite knowledge of the structure and characteristics of each type of TE. Even for TE experts, this undertaking can be tedious and extremely time-consuming because of the bewildering diversity of TEs (Wicker et al. 2007) and of the colossal output produced by ab initio repeat finding programs; typically thousands of individual consensus sequences for a medium-sized eukaryotic genome (Bao and Eddy 2002; Li et al. 2005; Price et al. 2005).
Here we introduce REPCLASS, a package that automates several steps in the annotation and classification of TEs. We show that REPCLASS can accurately diagnose all the major subclasses of TEs and accelerate TE annotation of eukaryotic genomes when combined to ab initio repeat finding programs. In addition, REPCLASS is able to identify a large number of previously undescribed TE families, even in the genomes of model organisms whose TE content has been extensively characterized. Finally, we exploit the ability of REPCLASS to produce a genome-wide profile of TE composition to gather new insights into the evolutionary dynamics of TE landscapes in nematode, fly, and fungi genomes.
Methods
Overview of REPCLASS Workflow
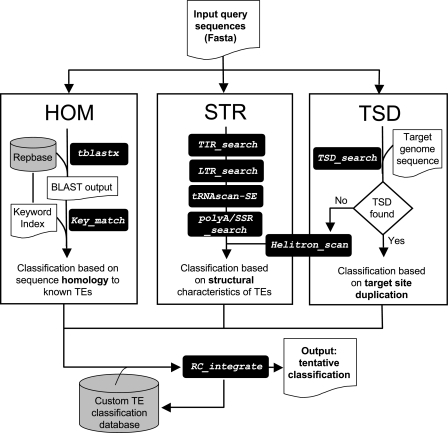
The workflow of REPCLASS is schematized in figure 1. The input file for the program is a single text file containing the DNA sequences to be classified in Fasta format. Each entry is then processed by the three classification modules: homology (HOM), structure (STR), and target site duplication (TSD). Each of the modules involves multiple steps and processes, which are described in detail below. The final step is an integration step that aims to compare, rank, and combine the results of the three modules providing a single tentative classification for each Fasta entry in the input file. The output of REPCLASS is a text file reporting the classification for each Fasta entry in the input file, if any classification is obtained. The classification terms are preceded by a letter code that indicates the modules that were used to produce the classification (H, S, or T). The classification is accompanied by a description of the structural features identified (e.g., length of TIRs, LTRs, and poly A terminus) and of the consensus length of the TSD, if any was identified. At the end of the output file, the total number of entries classified by REPCLASS, and the breakdown of this count by module or combination of modules, is given. Note that the user has also the option to run each of the modules of REPCLASS separately or in any pairwise combination (see user's guide and documentation).
FIG. 1.—
Overview of the REPCLASS workflow. Subroutines are shown in italics in black boxes. Databases are shown in gray cylinders. Each input query sequence (typically a consensus) is analyzed by the three classification modules of REPCLASS. HOM: homology-based, searches similarity to known repeats deposited in Repbase using TBlastX and extract classification from keyword index file; STR: structure-based, several subroutines search for structural features characteristic of different group of TEs, such as terminal inverted repeats (TIR_search), LTRs (LTR_search), tRNA-like sequences (tRNAscan-SE), or polyA/SSRs (polyA/SSR_search); TSD: target site duplication, individual copies are extracted from the target genome sequence using BlastN and their flanking sequences are searched for TSD. If no TSD are found, the subroutine Helitron_scan is executed to look for structural features of Helitrons. The final step attempts to compare and integrate the results of the three modules, resulting in a tentative classification for each input sequence. For a complete description of the workflow and subroutines, see Results and Methods.
HOM Module
This module uses each entry sequence as a query in a TBlastX search (translated query against translated database) of all reference repeat libraries deposited at Repbase Update (Jurka et al. 2005) or any custom repeat library annotated and indexed as in Repbase. The latest version of Repbase Update used in this study was version 13.03, downloaded from http://www.girinst.org/. The TBlastX search is performed with default parameters using a local installation of WU-Blast version 2.0 (http://blast.wustl.edu/). We use TBlastX (rather than BlastN) as it provides increased sensitivity to detect conserved protein motifs, as well as short but significant matches in noncoding sequences. The user has the option to modify the source code to run any other applications of the WU-Blast suite.
The TBlastX output files are parsed, and the first x (default of 10) hits with an e value lesser than e−5 are chosen. The classification for these x TEs is retrieved from a keyword index file created for the Repbase database and analyzed using a subroutine called Key_match. This program extracts keywords and descriptions from Repbase Update in EMBL format for each of the hit (subject) TE sequences. The indexing tool searches for specific keywords such as subclass, superfamily, family, etc. The index consists of the Repbase-assigned ID for the TE, along with terms defining the classification: subclass (SC), superfamily (SF), family (FM), group (GP), subgroup (SG), and keywords (KW). For each keyword, two confidence scores, Pe and Pk, are calculated as follows. Pe is the weighted average of the e values for all the hits containing the keyword, after transforming each e value with the formula Pe = |ln (e value)|/100 and with e values < e−100 set to e−100. Pk is the weighted average of the occurrence of a particular keyword to the total number of hits, that is, Pk = keyword count/no. of hits. The program sorts the keywords by Pe and Pk scores and assigns a tentative classification based on the highest scoring keyword for both scores.
STR Module
This module consists of several subroutines designed to search for structural features characteristic of different subclass of elements. Four subroutines (described below) are executed independently, and REPCLASS reports the results for each subroutine along with descriptive statistics of the features found, if any. A fifth subroutine, Helitron_scan, is executed if no TSD have been identified through the TSD module (described below).
LTR_search
LTR_search scans for LTRs, using a sliding-window procedure, with an initial default window size of 10 bp, incremented by 1 bp upon match, and sliding in opposite direction from each terminus of the query sequence (+/−20 bp). A mismatch of 1 bp for every 10 bp is allowed. The user has the option to specify the initial window size. The program considers a region a putative LTR if the total length of the direct repeat is greater than 100 bp and starts/terminates within 20 bp of each termini of the query.
TIR_search
TIR_search uses a modified version of the einverted program, which is part of the EMBOSS 6.0 suite (Olson 2002), to identify the longest possible inverted repeats that occur within 30 bp of the termini of the query sequence. The parameters for einverted are gap = 12, threshold = 50, match = 3, mismatch = 4, and maxrepeat = 10,000. The program reports the size of the TIR, if any is identified and if it is >10 bp long.
tRNAscan-SE
The goal of this subroutine is to look for the presence of a tRNA-like secondary structure within the query sequence. Such structure is indicative of a SINE as most of them are derived from tRNA sequences. We use the program tRNAscan-SE version 1.23 (Lowe and Eddy 1997), whose UNIX source code is available at http://lowelab.ucsc.edu/tRNAscan-SE/. We apply tRNAscan-SE to each query sequence using the default parameters. The output of the program reports a number of statistics, including the number of tRNAs found and the number of tRNA pseudogenes. Our empirical testing suggested that tRNAscan-SE was able to recognize the tRNA-derived portion of many known SINEs, which were typically predicted as tRNA pseudogenes.
PolyA/SSR_search
This subroutine uses a simple sliding-window algorithm to detect the presence of simple sequence repeats (SSRs) with units ranging in size from 1 to 5 nt at or near the termini of the query sequence. The presence of these features at one (but not both) ends of the query is indicative of a potential non-LTR retrotransposon. For SSRs, we apply a variable threshold to retain only those with a minimum number of repeated units, depending on the length of the unit (at least 10 perfect units for mononucleotides [including polyA/T], 7 for dinucleotides, 5 for trinucleotides, 4 for tetranucleotides, and 3 for pentanucleotides). For each query (consensus) sequence, SSRs are searched for a sample of individual elements (1–10 depending on copy number) by extracting the first and last 50 nt matching the consensus plus 50 bp of flanking genomic sequences on each side, extracted from the target genome (see also TSD module, below). This is done because of inherent variation in the length of the SSR at each locus, which may prevent the inclusion of long SSRs in the consensus. The presence and average length of polyA/T tails is reported in the REPCLASS output file as it is strongly indicative of retroposed elements.
Helitron_scan
This program is designed to look for the terminal sequence features characteristic of Helitrons, which include conserved 5′-TC and CTRR-3′ (R = A or G) at their 5′ and 3′ termini, respectively, and a subterminal hairpin-like GC-rich motif (16- to 20-bp long with a 2- to 5-bp loop) located 10–12 nt from the CTRR-3′ terminus (Kapitonov and Jurka 2001). Helitrons do not create TSDs, but they insert preferentially between A and T nucleotides, resulting in an overall conserved terminal sequence arrangement (5′-A|TC…/x nt/…gcctgcggt/2–5 nt/accgcaggc…/2-8 nt/CTRR|T-3′).
Helitron_scan searches for terminal and hairpin motifs independently and synthesizes this information into a score indicative of the presence or absence of the structural hallmarks of Helitrons. Half of the score (H53) is based on the combined detection of the 5′ and 3′ terminal motifs within +/− 5 nt of the predicted boundaries of individual copies of the repeat. The detection of both motifs is designated as a hit. Individual copies are retrieved from a BlastN search of the target genome using the consensus repeat sequence as a query (for parsing strategy of the BlastN output below, see TSD module). This is done to search not only the termini of the consensus, which may not be perfectly defined, but also the flanking sequences immediately adjacent to individual copies. It also takes into account the structural heterogeneity among copies, a common phenomenon with Helitron families (Kapitonov and Jurka 2001; Brunner et al. 2005). The score for this part of the search is calculated based on the number of hits (∑i) to the total number of copies (Tc) examined using the formula: H53 = (∑i/Tc) × 0.5. The other half of the score is based on the detection of the subterminal hairpin motif. This step is accomplished by using the palindrome program of the EMBOSS 6.0 suite (Olson 2002) to find all possible hairpin-like motifs. The parameters for palindrome are minpallen = 5, maxpallen = 70, gaplimit = 70, nummismatches = 0, and “nooverlap.” The output of palindrome is parsed to retain only those motifs with no more than a 2- to 5-bp loop and located less than 5–12 nt from one of the termini of the repeat. A hairpin score (HP) is calculated based on the GC content (% GC) and length of the hairpin, as follows:
If several hairpin motifs are found in the same repeat, the highest scoring motif is retained. The final score HT for Helitron_scan is the sum of the H53 and HP scores. A HT score of 0.75 and above is taken as indicative of a Helitron.
TSD Module
This module is designed to identify potential TSDs created by insertion of individual TE sequences. With few exceptions (e.g., TA in Tc1/mariner elements), the sequence and/or length of the TSD are not conserved among individual elements (Wicker et al. 2007). Thus, the TSD is not generally included within the query (consensus) sequence but is found flanking each insertion. Therefore, the TSD_search subroutine first performs a BlastN search (via a local WU-Blast install) with each query against a nucleotide database of the target genome (as defined and uploaded by the user) in order to retrieve individual copies of the repeat. Next the BlastN output is parsed to retain only copies matching both ends of the query and extracting the first and last 10 bp of each element plus 50 bp of flanking genomic sequence on each side. A sliding-window algorithm is then used to scan 5′ and 3′ flanking sequences in opposite directions (starting with the end of the 5′ flank and the beginning of the 3′ flank) for sequence motifs of length >2 bp matching in direct orientation. We allow a mismatch of 1 bp/motif of 6–10 and 2 bp/motif of >10 bp. The inclusion of 10 bp of the element's terminal sequences allows the recovery of TSDs that are conserved in length and sequence and may have been included as part of the consensus. The first matching motif is interpreted as the potential TSD. If >50% of the elements examined have a potential TSD, the maximum number of elements having the same TSD length is retrieved and a consensus of those TSD sequences is generated. The sequence and length of the consensus are stored and reported in the REPCLASS output file. If TSDs are found in >50% of the copies examined, but no consensus TSD length can be reconstructed, the search reports “variable TSD length,” which is indicative of non-LTR elements. If TSDs are found in less than 50% of the copies examined, the element is considered to create no TSD. Because the lack of TSD is a characteristic of Helitrons, repeats with no TSD are then subject to an additional search for structural features of Helitrons (described above).
Integration Step
The final step in the REPCLASS workflow is an integration process that interprets, compares, weights, and synthesizes the results of the three modules in the context of the current TE classification system to arrive at a tentative classification for each query sequence. To do this, we created a custom classification database that largely mirrors the “unified classification system for eukaryotic transposable elements” (Wicker et al. 2007). This relational database is used to integrate the different levels of classification and validate the results produced by the three upstream modules. For example, when two or three of the modules converge to the same subclass, this subclass is adopted as the final classification. If one of the modules produces a classification at the superfamily level, then this information is extracted and added to the subclass classification. The classification database is also used to augment or complete the information received from the modules. For example, the HOM module may report the superfamily but not the subclass or class. This is because the keyword index extracted from Repbase Update during the HOM search is not always complete or accurate, especially for older entries.
Another goal of the integration step is to resolve conflicting classifications that may be produced by the different modules. In this case, the integration program applies a hierarchical strategy based on a ranking of the three modules in decreasing level of confidence: HOM > STR > TSD (see also Results). The hierarchical rule is applied separately at each level of the classification. Our empirical testing showed that the ranking resolved most cases of conflicting classifications. The user may also find it useful to modify the ranking between modules or disable the integration step, which then allows the display of the classifications produced by each module, and let the user manually perform the integration of the results for each classified repeat.
Computing and Processing Time
Most of the results reported in this paper were obtained by running REPCLASS on the UT Arlington Distributed and Parallel Computing Cluster that consists of 81 dual processor 2.667 GHZ Xeon compute nodes with 2 GB memory each. The software was run on varying number of processors to measure computing performance in terms of scalability and load balancing (for more details, see Ranganathan et al. 2006). In brief, processing time was linearly correlated to the number of Fasta entries in the input file and to the number of processors used. For example, it took around 2 h with 2 processors or 40 min with 10 processors to run REPCLASS on the Caenorhabditis elegans Repbase Update library (116 entries) and 21 or 2 h using 2 and 10 processors, respectively, for the C. elegans RepeatScout unfiltered library (1,851 entries). Thus, for a relatively small genome with a filtered repeat library, REPCLASS can be executed on a standard desktop computer in just a few hours. For larger and repeat-rich genomes, turnaround time is significantly improved by using parallel cluster or Grid computing (Ranganathan et al. 2006).
Software Availability
REPCLASS 1.0 is available as a UNIX-based package downloadable at http://www3.uta.edu/faculty/cedric/repclass.htm, with complete documentation, including user's guide and instructions for installation, initial setup, and filtering. The package and source code are also available as open source software through http://sourceforge.net/projects/repclass/.
RepeatScout and Filtering
RepeatScout (RepeatScout; Price et al. 2005) version 1.0.5 was downloaded from http://bix.ucsd.edu/repeatscout/ and run with default parameters. The output of RepeatScout consists of a library of consensus sequences for each of the repeat families identified. Prior to running REPCLASS, three different filters are applied to the RepeatScout output. First, Tandem Repeats Finder version 4.0 (Benson 1999; http://tandem.bu.edu/trf/trf.html) and nseg (Wootton and Federhen 1996; ftp://ftp.ncbi.nih.gov/pub/seg/nseg) are used to remove consensus sequences predominantly or entirely composed of tandem repeats, SSR, and other low-complexity repeats. In this study, we discarded all sequences masked as SSR/low complexity for more than 70% of their length. Second, we filtered out repeat consensus sequences of length less than or equal to 100 bp because the size of known TEs generally exceeds 100 bp (see Results). We consider this cutoff to be the minimum threshold that should be applied to any genome, irrespective of genome size, and number of repeat consensus sequences. However, a higher threshold may be appropriate for genomes that are larger and contain a larger number of repeats. To facilitate the task of determining the most appropriate length threshold for the genomic landscape analyzed, REPCLASS generates a graph of repeat length distribution for the sequences compiled in the input query file. An example of the repeat length distribution for the RepeatScout library obtained for C. elegans is shown in supplementary figure 4 (Supplementary Material online). The third and last filter is based on copy number per repeat family. In principle, when RepeatScout is run with default parameters, repeats present in less than 10 copies are not reported. However, the repeat count determined by RepeatScout may include very small repeat fragments and may not accurately reflect the bona fide copy number of TE families. Hence, we apply a second filter based on a more stringent estimation of copy number based on a BlastN search of the target genome with each consensus repeat as a query using the WU-Blast package. We count all those hits as valid copies when they span at least half of the query sequence length with ≥80% nucleotide similarity. This cutoff is similar to the one used traditionally to define TE families (Feschotte and Pritham 2007a; Wicker et al. 2007). In order to assist the user in determining the copy number cutoff for this filtering step, REPCLASS generates a graph of the copy number distribution of the query sequences contained in the input library. An example of the graph obtained for the C. elegans RepeatScout library is shown in supplementary figure 5 (Supplementary Material online). In the present study, we only retained repeat families with copy number greater than 10. The cutoff value may vary depending on the genome size and overall repeat content of the genome analyzed.
Genome Sequence Data
Details on the genome sequences analyzed in this study are provided in supplementary table 1 (Supplementary Material online), including genome size, version of the assembly analyzed, whole genome shotgun (WGS) coverage, sequencing centers producing the sequence and assembly, and related references. All sequence assemblies were downloaded from the NCBI or the University of California–Santa Cruz (UCSC) Genome Browser or the Broad Institute.
Results
REPCLASS Design and Workflow
REPCLASS uses three different approaches to classify TEs that are implemented as independent modules (fig. 1). The first module (HOM for homology) attempts to detect sequence similarity with known, previously classified TEs. This homology-based approach works well when the elements contain coding sequences with conserved domains and motifs that can be used to classify elements at a relatively fine level (typically at the superfamily level). The second module (STR for structure) aims at recognizing the structural characteristics of some types of TEs, which are generally located at their termini. These features can be used to classify elements at the subclass level: non-LTR retrotransposons end in SSRs, LTR retrotransposons are characterized by LTRs, cut-and-paste DNA transposons have terminal inverted repeats, and rolling-circle transposons (Helitrons) have a short GC-rich palindromic stem loop structure near one end and a 5′-TC-3′ motif at the other end (for review, Wicker et al. 2007). The third module of REPCLASS is designed to determine the short duplication of host sequence induced upon chromosomal integration of individual elements. The length and sequence of the TSD reflect the mechanisms and properties of the enzymes catalyzing integration (Craig et al. 2002). Thus, TSD length is often diagnostic of specific subclasses or superfamilies. For example, non-LTR elements are flanked by TSD of variable length, LTR elements create 4–6 bp TSD, DNA transposons have TSDs that vary from 2 to 9 bp but are generally conserved in length for a given family and superfamily, and Helitrons create no TSD upon insertion but they insert between a 5′-A and a 3′-T (for review, Wicker et al. 2007). Hence, information on TSD can be useful to confirm or refine the classification based on other criteria. To execute this module, the user needs to upload a target genomic sequence where individual TE copies can be retrieved and examined for the presence/absence of TSD (see Methods).
The three modules of REPCLASS are run independently, and the output reports the results for each of the modules. The three modules are complementary, and, in principle, a sequence receiving the same classification by two or more modules should be more reliably classified. However, it is not expected that every TE family will return results for more than one module. For example, nonautonomous element families, which are common in many species, generally have no coding sequence and display little or no significant sequence similarity to other TEs (Feschotte et al. 2002; Wicker et al. 2007). For these families, the HOM module would return no results, and there is a chance that either the TSD or the STR modules would fail to return any informative results.
Because different modules might occasionally yield conflicting or uncertain classification, we implemented a final integration step that weights the results obtained by each module hierarchically based on empirical observations and other considerations. For example, the results returned by the HOM module, which typically yields highly confident classification, prevail over any conflicting results given by the other two modules, which are more sensitive to misclassification. In the absence of HOM classification, the results of the STR module prevail over the TSD module. We observed empirically that this simple hierarchy (HOM > STR > TSD) allowed the resolution of most cases of conflicting or ambiguous classification. For example, elements flanked by 5 bp TSD would be classified by the TSD module as LTR retrotransposons or DNA transposons. However, the latter are expected to be also classified by STR based on the presence of TIRs, whereas the former are expected to be classified either by HOM, if they are autonomous elements, or by TSD, if they are not. Other strategies implemented to facilitate the integration of the results of the three modules and enhance the interpretation of the REPCLASS output are described in Methods.
Validation with Reference Repeat Libraries
To assess the performance of REPCLASS, we first examined the ability of the program to classify a variety of previously characterized TEs. To do this, we used the reference Repbase repeat libraries for C. elegans and D. melanogaster as input (Jurka et al. 2005) together with the latest genome sequence assemblies available for these species (listed in supplementary table 1, Supplementary Material online). These manually curated libraries are the result of more than a decade of TE mining, and they have been used for genome annotation in conjunction with Repeatmasker. The rationale for selecting the repeat libraries of C. elegans and D. melanogaster for these control experiments was 3-fold. First, together these two libraries provide a wide assortment of TEs largely representative of the diversity of TEs in eukaryotes (Wicker et al. 2007). Second, the two species offer complementary, but very contrasting, TE landscapes both in terms of TE types and structure. The C. elegans genome hosts a rich and diverse population of DNA transposons that are represented primarily by short (<500 bp) nonautonomous elements (e.g., Surzycki and Belknap 2000). This is reflected by consensus sequences that lack coding capacity but bear the structural hallmarks (TIRs and TSD) of their respective superfamilies. In contrast, D. melanogaster TE content is dominated by retrotransposons (both LTR and non-LTR), which are represented by consensus sequences of large (>3 kb) elements with coding capacity (Kaminker et al. 2002). Thus, these two divergent libraries allow us to assess the efficiency of the different classification modules implemented in REPCLASS and the ability of the program to accurately classify a variety of TEs.
Prior to running REPCLASS, we removed from the two control libraries all unclassified repeats and non-TE repeats (simple repeats, tandem repeats, and satellites). In Repbase, the LTRs and internal coding sequences of LTR retrotransposons are listed as separate entries to facilitate masking. Although we expected the internal regions to be classified by the HOM module of REPCLASS, we suspected that isolated LTR sequences could not be easily classified. Therefore, when both LTRs and internal regions of the same family were listed as separate entries in Repbase, for the sake of simplicity, we only retained the internal region in the library (note however that this procedure would prevent detection of the LTRs by the STR module). Lastly, in order to avoid systematic classification by self-homology during these control experiments, the two libraries analyzed were removed from the collection of Repbase libraries queried by the HOM module of REPCLASS (see fig. 1).
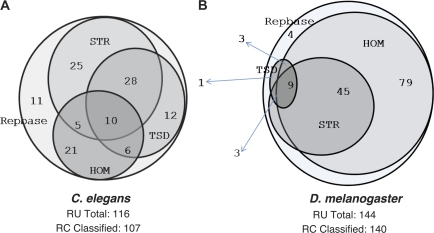
The results of the REPCLASS analysis on each of the two control libraries (fig. 2) showed that the program was able to classify 107 of 116 (92%) consensus sequences in the C. elegans library and 140 of 144 (96%) consensus sequences in the D. melanogaster library. In both experiments, we evaluated the accuracy of classification to 96%, as judged by the matching of Repbase and REPCLASS classification at least at the subclass level. Thus, out of 260 different TE families cataloged in the two reference libraries, only 13 were not classified by REPCLASS. These unclassified TEs did not belong to any particular subclass (2 non-LTR, 3 LTR, 6 DNA, and 2 Helitrons), but we noted that some of them lack the sequence or structural hallmarks of their subclass, which might explain in part the inability of REPCLASS to classify them unambiguously.
FIG. 2.—
Validation of REPCLASS with Repbase libraries. Venn diagrams showing the number of consensus sequences in the Repbase Update (RU) library of (A) C. elegans (n = 116) and (B) D. melanogaster (n = 144) classified by the different modules of REPCLASS.
As expected based on the contrasting TE landscape of the two species, we observed that the STR module was most efficient at classifying the TEs of C. elegans (fig. 2A), whereas the HOM module was by far the best at classifying TEs in D. melanogaster (fig. 2B). The TSD module was more useful in C. elegans than in D. melanogaster, in particular to assign DNA transposons to specific superfamilies. The abundance of non-LTR elements in D. melanogaster (for which TSDs are sometimes not created or difficult to detect automatically) might explain in part the relatively meager output produced by the TSD module in this genome (fig. 2B). Another explanation lays in our artificial removal of LTR and retention of only internal sequences for some of the LTR elements in the library, as explained above. This procedure not only prevented detection of the LTRs by the STR module but also of the TSD normally flanking the LTRs. Finally, it is important to note that for both species only a small fraction of TEs (7.3%) was classified by all three modules but 42% were classified by at least two modules (fig. 2). These data emphasize the need to combine all 3 modules to effectively classify TEs, a critical asset of REPCLASS. These results also demonstrate that the program is able to recognize virtually all known types of TEs provided that their consensus sequences have been precisely defined.
TE Annotation by Combining REPCLASS with Ab Initio Repeat Finding
The primary motivation for developing REPCLASS is the need to automate the classification of TEs in repeat libraries generated ab initio from raw sequence data. Several software packages have been developed to create such repeat libraries. Although it is clear that no single algorithm can generate a consensus repeat library comparable to reference libraries curated manually, RepeatScout (RepeatScout; Price et al. 2005) is emerging as one of the most reliable and computationally economical tools currently available (Saha et al. 2008). Thus, we explored how REPCLASS performed on libraries assembled by RepeatScout from raw sequence data. As a preliminary experiment, we ran RepeatScout with default parameters on the genome assemblies of C. elegans and D. melanogaster (see supplementary table 1, Supplementary Material online). The number of repeat consensus sequences compiled by RepeatScout for each species (1,851 and 1,844, respectively) far exceeded the number of repeat families cataloged in Repbase for the same species (144 for D. melanogaster and 116 for C. elegans). This was not unanticipated because the output produced by RepeatScout contained not just TEs but all kinds of repeats, including tandem and low-complexity repeats, gene families, and segmental duplications.
To decrease the complexity of the RepeatScout output, limit false positives, and minimize computing time, we devised several filtering steps to apply to RepeatScout output prior to running REPCLASS. First, tandem and low-complexity repeats were filtered out using Tandem Repeat Finder (Benson 1999) and nseg (Wootton and Federhen 1996), respectively (see Methods). Second, all consensus sequences of less than 100 nt were discarded because known TEs are typically longer than this threshold. For example, the smallest C. elegans and D. melanogaster TE cataloged in Repbase are 150 and 175 bp long, respectively, and only 12 of 260 consensus sequences in the two species are less than 200 bp long. Lastly, we removed all repeat families with copy number less or equal to 10 copies, reasoning that this threshold should allow us to retain most TE families but filter out low-copy number gene families and segmental duplications, which might yield false positives due to the potential inclusion of TE copies embedded within them. Applying these filtering steps considerably reduced the complexity of the RepeatScout output, leaving a total of 445 consensus sequences for C. elegans and 810 for D. melanogaster.
We next ran REPCLASS on each of these filtered RepeatScout libraries and compared the output with repeats cataloged in the cognate Repbase libraries. For C. elegans, REPCLASS classified 146 TE families out of 445 repeats identified and 57 of those matched one of the 116 TE consensus deposited in the C. elegans reference library (>85% identity over >50% of consensus length). Fifty of these 57 TEs (87.7%) were classified accurately by the program, whereas seven were misclassified or classified ambiguously. Out of the 59 TEs cataloged in Repbase but not classified by REPCLASS, we found that 39 had no close match in the filtered RepeatScout output. These may be low-copy number families that had been removed during our filtering step or they may have escaped RepeatScout identification in the first place. The remaining 20 families did have a close match in the filtered RepeatScout library but had not been classified by REPCLASS. Apparently, this was caused by an inaccurate or incomplete definition of consensus sequences by RepeatScout. Inspection of these 20 RepeatScout consensus sequences showed that they were noncoding and/or were severely truncated at one or both ends compared with their matching Repbase consensus (data not shown), precluding classification by either of the three modules implemented in REPCLASS. Interestingly, the program was able to classify an additional 89 repeats that were identified by RepeatScout in the C. elegans genome but have no close match in the C. elegans Repbase library, potentially representing novel TE families (see below).
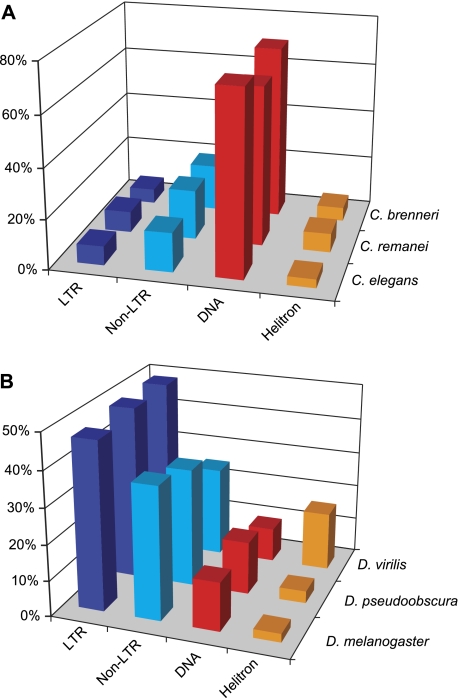
A breakdown of all classified TEs by subclass (fig. 3A) produces a composition profile predominated by DNA transposons, as noticed previously for this species (Surzycki and Belknap 2000; Stein et al. 2003; Feschotte and Pritham 2007b). The remaining 289 repeats identified by RepeatScout, which have no match in Repbase and are not classifiable by REPCLASS, deserve closer inspection as they might comprise some novel types of TEs with unconventional features.
FIG. 3.—
TE composition profiles generated by REPCLASS for (A) three Caenorhabditis species and (B) three Drosophila species. The profile depicts the percentage of families falling within one of the four TE subclasses (LTR retrotransposons, non-LTR retrotransposons, cut-and-paste DNA transposons, and Helitrons).
In D. melanogaster, the filtered RepeatScout library contained 810 consensus sequences and 464 (57%) were classified by REPCLASS (table 1B). Out of the 464, 361 were matching 92 unique TE consensus sequences in the D. melanogaster Repbase library, and 86 of these (93%) were classified correctly by the program. Thus, 103 consensus sequences were classified as TEs by REPCLASS but had no close match in the D. melanogaster Repbase library, potentially representing novel TE families (see below). The breakdown of all classified TEs by subclass recapitulated the TE profile typical of D. melanogaster, with a predominance of LTR and non-LTR retrotransposons (fig. 3B).
Table 1.
Genome Statistics and Annotation of TEs in Caenorhadbitis and Drosophila species
|
Caenorhadbitis elegans |
Caenorhadbitis remanei |
Cenorhabditis brenneri |
||||
| DNA analyzed (Mb) | 100.3 | 138.4 | 170.4 | |||
| WGS coverage | n/a | 9.2X | 9.5X | |||
| Number of contigs | Chromosomes | 12,680 | 13,589 | |||
| Average contigs length (bp) | n/a | 10,915 | 12,545 | |||
| Number of families identified by RepeatScout | 445 | 1,368 | 1,477 | |||
| Number of families classified by REPCLASS | 146 | 331 | 362 | |||
| Number of families | Average consensus length (bp) | Number of families | Average consensus length (bp) | Number of families | Average consensus length (bp) | |
| DNA | 107 | 649 | 212 | 654 | 254 | 558 |
| Helitron | 5 | 891 | 25 | 674 | 20 | 1,038 |
| LTR | 11 | 1,403 | 28 | 1,070 | 21 | 1,073 |
| Non-LTR | 23 | 707 | 66 | 788 | 67 | 478 |
| Drosophila melanogaster | Drosophila pseudoobscura | Drosophila virilis | ||||
| DNA analyzed (Mb) | 137.7 | 146 | 189.2 | |||
| WGS coverage | n/a | 9.1X | 8.0X | |||
| Number of contigs | Chromosomes | 4,896 | 13,530 | |||
| Average contigs length (bp) | n/a | 29,832 | 13,984 | |||
| Number of families identified by RepeatScout | 810 | 1,673 | 1,743 | |||
| Number of families classified by REPCLASS | 464 | 855 | 868 | |||
| Number of families | Average consensus length (bp) | Number of families | Average consensus length (bp) | Number of families | Average consensus length (bp) | |
| DNA | 63 | 508 | 127 | 330 | 83 | 519 |
| Helitron | 11 | 433 | 29 | 444 | 142 | 619 |
| LTR | 218 | 1,411 | 415 | 766 | 424 | 1,222 |
| Non-LTR | 172 | 906 | 284 | 519 | 219 | 1,077 |
REPCLASS-Assisted Discovery of New TE Families in C. elegans and D. melanogaster
The application of REPCLASS to repeat libraries generated ab initio by RepeatScout yielded a surprisingly large number of apparently new TE families in C. elegans (89 families) and D. melanogaster (103 families; Fasta sequences available in supplementary files 1 and 2, Supplementary Material online). This result was unforeseen because the genomes of these two species have been subject to intensive TE mining for over a decade and they rank among the best-annotated eukaryotic genomes.
To corroborate these findings, we selected randomly (i.e., following the order provided in the RepeatScout output) 50 of the potentially novel families in each of the two species for further inspection. For each family, we used the consensus sequence constructed by RepeatScout as a query in a BLAT search of the corresponding genome to retrieve 5–10 copies with at least 20 bp of flanking sequences, built a multiple alignment, refine the consensus sequences when necessary, and manually examined sequence features (coding and noncoding) and TSD diagnostics for TE classification. In addition, we checked whether the chromosomal positions of the individual copies overlap with those of known TEs annotated by Repeatmasker in the latest annotation of the corresponding genome assembly available at the UCSC Genome Browser (see supplementary table 1, Supplementary Material online). We observed four categories: 1) no overlap with any annotated TE; 2) partial overlap over a short region of the consensus (much less than 80% of length), with generally weak similarity (<80%) to the Repbase consensus; 3) complete or nearly complete overlap but weak similarity (<80%) with the Repbase consensus; and 4) complete or almost complete overlap and high similarity (>80%) with the Repbase consensus (see supplementary tables 2 and 3, Supplementary Material online). We consider cases (1) to (3) as indicative of newly discovered TE families because they fulfill the 80/80/80 rule proposed by Wicker et al. 2007 (for a definition of TE family, see Introduction), with case (1) representing the strongest argument for validation as a novel family. In principle, case (4) should not occur because these families should have been eliminated during our initial BlastN search for matches to Repbase. Nevertheless, we did retrieve a few instances that had passed through our parsing strategy, apparently because of an inaccurate or incomplete definition of the consensus produced by RepeatScout. Anyhow, we did not consider these as new families.
Out of the 50 repeat families inspected in C. elegans, we were able to validate 43 (86%) as novel TE families. All 43 families had been classified correctly by REPCLASS, including 42 DNA transposon families (all with TIRs) and one family of CR1-like LINE (supplementary table 2, Supplementary Material online). Most of the new DNA transposon families displayed structural features of known superfamilies (e.g., Tc1/mariner, hAT, MuDR), but several appeared to represent novel eukaryotic superfamilies as judged by the length of their TSD (2, 4, 5, or 6 bp; see supplementary table 2, Supplementary Material online) and the lack of sequence similarity between their TIRs and those of known autonomous DNA transposons (data not shown). Based on the copy numbers of these families, we estimate that these novel TEs cover about 1.15% of the genome. The seven other families that we could not validate as novel TE families were two satellite repeats, one F-box gene family, and four close variants of known C. elegans TEs (described above as case [4]). Based on the false discovery rate of new TE families (∼14% in this example), we can predict the discovery of ∼75 families previously not reported in Repbase, which would increase the number of TE families known in C. elegans by ∼66%.
In D. melanogaster, 32 of 50 families inspected manually were confirmed as new TE families (supplementary table 3, Supplementary Material online). Out of the 18 families not validated, 15 had been classified correctly by REPCLASS but had extensive sequence similarity (>80%) over most of their length to TE sequences in the D. melanogaster Repbase library and thus fell within case (4) described above. These families are not false positives sensu stricto as they should have been included in the set of repeats matching known TEs, but they do not represent new families. Among the 32 confirmed new families, 26 were LTR retrotransposons, 3 were non-LTR, 2 were DNA transposons, and 1 was a new Helitron family (supplementary table 3, Supplementary Material online). We noticed that several of the RepeatScout consensus sequences identified as LTR retrotransposons represented nonoverlapping fragments of the same TE family rather than distinct families as they were found to colocalize in the genome (data not shown) and they were most similar (in their coding regions) to the same known LTR retrotransposon family (see supplementary table 3, Supplementary Material online). Such fragmentation is likely to artificially inflate the number of newly discovered LTR retrotransposon families in D. melanogaster. Thus, we considered those repeats that colocalize in the genome and had homology with the same retrotransposon as a single family. This reduced the number of new LTR retrotransposon families from 26 to 15. This fragmentation issue did not appear to affect the counts for the other types of TEs. Together, we can therefore estimate that 21 of the 50 repeat families examined represent newly identified TE families (listed in supplementary table 3, Supplementary Material online). Extrapolating this ratio to the entire data set suggests that the application of the RepeatScout/REPCLASS suite to the D. melanogaster genome yielded a crop of ∼40 new TE families from all 4 major subclasses of TEs. This increases by ∼30% the number of TE families recognized in D. melanogaster.
Comparative TE Profiling of Caenorhabditis and Drosophila Genomes
Having demonstrated the accuracy of REPCLASS and the utility of the program in combination with ab initio repeat mining, we next applied the RepeatScout/REPCLASS suite to explore the TE landscape of species that have not yet been subject to systematic TE annotation. First, we focused on Caenorhabditis brenneri and Caenorhabditis remanei, two nematode species distantly related to C. elegans (Cutter 2008), and then on Drosophila pseudoobscura and Drosophila virilis, two fly species that diverged from each other and from D. melanogaster ∼55 Ma (Tamura et al. 2004). The choice of these species was motivated by several considerations. First, all these genomes are of relatively small size, which facilitates computational processing and subsequent data analysis. Second, because TEs and other forms of repetitive DNA represent the major obstacle for genome assembly (e.g., most contigs will terminate in variably truncated repeats), we were curious to see how RepeatScout and REPCLASS performed on nonmodel species with lower quality assemblies (see supplementary table 1, Supplementary Material online). Third, we were interested to see if the diametrically opposed TE composition of C. elegans and D. melanogaster would be conserved in their distant relatives. We analyzed all species by applying the same parameters and filters as described above.
For C. remanei and C. brenneri, the filtered RepeatScout output contained 1,368 and 1,477 consensus sequences, respectively (table 1A). These counts were ∼3-fold higher than in C. elegans (n = 445), even though we applied the same filtration parameters for all three species. The number of families classified by REPCLASS was elevated proportionally; 331 in C. remanei and 362 in C. brenneri versus 146 in C. elegans. These results raised the question of whether these differences were an artifact of increased fragmentation of consensus sequences by RepeatScout in the two genome assemblies of lesser quality (C. remanei and C. brenneri) or whether they reflected true biological differences in the amount and diversity of TEs among the three nematodes. To address this question, we compared the mean lengths of the consensus sequences for each of the major TE subclasses in each of the species (table 1A), reasoning that increased fragmentation would result in shorter consensus sequences. We found no consistent shortening of consensus lengths in C. remanei and C. brenneri compared with C. elegans, except for LTR retrotransposons, which were slightly (about 1.4 times) shorter in both C. remanei and C. brenneri. Typically, LTR elements are much longer than elements from the other subclasses and therefore are more likely to be artificially fragmented by RepeatScout. Because this subclass accounts for only a small fraction of TEs in all three nematodes, these data indicate that the issue of fragmentation alone is unlikely to explain the overall increase in the number of TE families retrieved for C. remanei and C. brenneri. Furthermore, we observed that the increase in TE families in these two species was accompanied by a roughly proportional increase in the overall copy number of non-LTR, DNA, and Helitron elements (supplementary fig. 1, Supplementary Material online). Thus, the larger number of TE families observed in C. remanei and C. brenneri does not appear to be an artifact of consensus fragmentation but rather reflects an increased diversity of non-LTR, DNA, and Helitron elements in these two species. The data also imply that a larger fraction of the C. remanei and C. brenneri genomes is occupied by TEs, which is consistent with the larger genome size of these two species compared with C. elegans (1.4- and 1.7-fold larger, respectively, see table 1A). Thus, the difference in genome size among these species can be largely accounted for by variation in the amount of TEs and other repetitive DNAs, as noticed previously for C. briggsae (Stein et al. 2003). Furthermore, the REPCLASS analysis suggests that the increased amount of repetitive DNA in C. remanei and C. brenneri does not merely result from elevated copy number in one or a few TE families but rather from a wholesale expansion in the number of DNA, Helitron, and non-LTR families. This phenomenon, however, does not fully explain the larger genome size of C. brenneri (1.2-fold that of C. remanei) because the amount of repeats identified by RepeatScout and the number of TE families classified by REPCLASS in this species are only slightly higher than in C. remanei (table 1A). It appears that DNA transposons have reached significantly higher copy number in C. brenneri than in the other two nematode species (supplementary fig. 1, Supplementary Material online), which may explain its larger genome size.
Despite the variation in the number of TE families across the three nematodes, the relative representation of the four TE subclasses was strongly conserved, with an overwhelming predominance of DNA transposons in all three species (see table 1A; fig. 3A). As in C. elegans, the DNA transposons of C. remanei and C. brenneri were mostly represented by an abundance of small nonautonomous element families affiliated with diverse superfamilies (data not shown). However, we found that there was very little, if any, sequence similarity between the consensus sequences retrieved in the three species, which stems from the lack of coding sequences in most of the TEs and the rapid turnover of repeats in these genomes. Indeed, as in C. elegans, most of the TE families identified in C. remanei and C. brenneri were classified through the STR and TSD modules but not by homology (data not shown). This observation emphasizes the necessity of ab initio repeat identification and the utility of REPCLASS to capture the TE content of these organisms.
The results for D. pseudoobscura and D. virilis revealed a significant increase in the total number of families identified by RepeatScout in both species, as well as those classified by REPCLASS, when compared with D. melanogaster (table 1B). This was unexpected at least for D. pseudoobscura because the genome size of this species is comparable to D. melanogaster and the total amount of DNA analyzed was indeed similar (table 1B). To test for the effect of consensus fragmentation, we examined the length of the consensus sequence reconstructed by RepeatScout and classified by REPCLASS for each TE subclasses (table 1B). The mean length of the consensus was significantly shorter (1.5- to 2-fold) in D. pseudoobscura for all TE subclasses except Helitron, compared with D. melanogaster and D. virilis. Assuming that TEs from the same subclass have comparable size in all Drosophila species, these data suggest that the rate of consensus fragmentation is about twice as high in D. pseudoobscura as in the other two genomes. This difference can largely account for the apparent increase in the number of TE families in this species. In contrast, the mean consensus lengths in D. melanogaster and D. virilis were similar for all four TE subclasses (table 1B), which suggests that the increase in the number of TE families identified in D. virilis (about twice as many families detected by RepeatScout and classified by REPCLASS) reflect a bona fide expansion of TE diversity in this species. These data may explain the enlarged genome size of D. virilis (about 1.3-fold) compared with the other two Drosophila species. The number of families in D. virilis is larger in all four TE subclasses compared with D. melanogaster, but the most dramatic expansion (over 10-fold), both in number of families (table 1B) and in total copy numbers (supplementary fig. 2, Supplementary Material online), involves Helitrons. This is consistent with the recent report of lineage-specific amplification of DINE-1, a nonautonomous family of Helitrons, across 12 Drosophila genomes, including a nearly 10-fold expansion in D. virilis compared with D. melanogaster (Yang and Barbash 2008).
Regardless of the absolute number of TE families, we observe a striking conservation of TE composition in the three Drosophila species examined, with both LTR and non-LTR retrotransposons prevailing over DNA transposons in terms of number of TE families (table 1B; fig. 3B). These data indicate that the increased fragmentation of repeats in the RepeatScout output did not affect the ability of REPCLASS to recapitulate the TE profile characteristic of Drosophila (Clark et al. 2007).
Evolution of TE Landscape in 10 Model Fungal Genomes
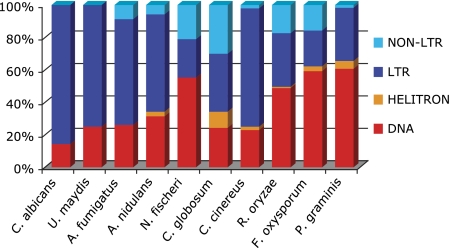
To further demonstrate the utility of REPCLASS, we performed a comparative analysis of TE content in several fungal genomes for which draft genome assemblies are available. We selected 10 different species representing a broad range of genome size and covering wide taxonomic diversity, including six ascomycetes, three basidiomycetes, and one zygomycete (table 2, supplementary table 1 [Supplementary Material online], and for a phylogeny, Fitzpatrick et al. 2006). Ascomycetes were represented by one saccharomycetale (Candida albicans), three closely related Eurotiomycetes (Aspergillus fumigatus, Aspergillus nidulans, and Neosartorya fischeri), and two Sordariomycetes (Chaetomium globosum and Fusarium oxysporum). Basidiomycetes were represented by Ustilago maydis, Coprinus cinereus, and Puccinia graminis. Finally, Rhizopus oryzae was the only Zygomycete available with a draft WGS assembly. Among these fungi, genome size varies from 14.3 Mb in C. albicans to 81.5 Mb in P. graminis (table 2). Because the WGS sequencing coverage is comparable for these genome projects, the broad variation in genome size among the species implies substantial variation in the quality of the WGS assembly. This is reflected by the total number of contigs and average contig size, which tend to be positively and inversely correlated to genome size, respectively, with the exception of A. fumigatus, which has a relatively better WGS assembly (table 2). TE content has been investigated previously in four of these species (Goodwin and Poulter 2000; Jones et al. 2004; Galagan et al. 2005; Kamper et al. 2006), although not always comprehensively in terms of TE diversity. For the other six species, little or nothing is known on their genome-wide TE content (Daboussi and Capy 2003).
Table 2.
Genome Statistics and Annotation of TEs of 10 Fungal Species
| Species | Phylum | Genome Size (Mb) | WGS Coverage | Number of Contigs | Average Contig Length (kb) | Number of Repeat Families Identified | Number of TE Families Classified |
| Candida albicans | Ascomycete | 14.3 | 10.9X | 8 | 1,787.2 | 37 | 7 |
| Ustilago maydis | Basidiomycete | 19.7 | 10X | 274 | 71.8 | 25 | 4 |
| Aspergillus fumigatus | Ascomycete | 29.4 | 10.5X | 8 | 3,673.1 | 31 | 23 |
| Aspergillus nidulans | Ascomycete | 30 | 13X | 248 | 121.2 | 49 | 35 |
| Neosartorya fischeri | Ascomycete | 32.5 | 11X | 976 | 33.3 | 124 | 38 |
| Chaetomium globosum | Ascomycete | 34.3 | 7X | 1,245 | 27.6 | 176 | 70 |
| Coprinus cinereus | Basidiomycete | 36.2 | 10X | 431 | 84.1 | 178 | 48 |
| Rhizopus oryzae | Zygomycete | 45.3 | 12X | 389 | 116.3 | 496 | 127 |
| Fusarium oxysporum | Ascomycete | 59.9 | 6.8X | 1,362 | 44.0 | 516 | 204 |
| Puccinia graminis | Basidiomycete | 81.5 | 7.8X | 4,557 | 17.9 | 2,085 | 430 |
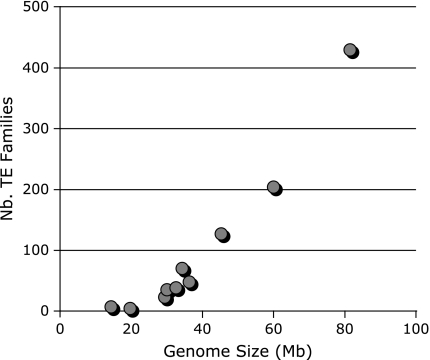
After running RepeatScout and filtering out repeat families of less than 100 bp and 5 copies/genome, the numbers of repeat families identified by RepeatScout in the 10 fungi species differ by up to two orders of magnitude, ranging from 25 in U. maydis to 2,085 in P. graminis. Overall, there is a positive linear relationship (R2 = 0.81) between genome size and the number of repeat families identified by RepeatScout (supplementary fig. 3, Supplementary Material online). This correlation becomes even stronger (R2 = 0.93) when the number of TE families classified by REPCLASS is plotted against genome size (fig. 4). It is well established that the total amount of repetitive DNA is, in general, positively correlated with genome size in eukaryotes (Gregory 2005). Our data are consistent with this trend and, furthermore, reveal that in fungi the increase in genome size is accompanied by an increase in TE diversity (as defined by the number of TE families per genome). The percentage of repeat families classified by REPCLASS out of the filtered RepeatScout output varied greatly, ranging from 16% in U. maydis to 74% in A. fumigatus, but the average (36.4%) was intermediate between that for Caenorhabditis (25.5%) and Drosophila (51.2%).
FIG. 4.—
Relationship between genome size and the number of TE families classified by REPCLASS in 10 fungal genomes.
For these genomes, we were able to recover all subclasses of TEs currently recognizable by the modules of REPCLASS. After manually inspecting a random sample of consensus sequences classified by the program, we found the rate of false positives to be extremely low, except for a small set of repeats dubiously classified as non-LTR elements by the STR module due to the presence of an SSR at one of their termini (data not shown). Although this feature is, indeed, a structural characteristic of non-LTR elements, we reasoned that it might not be sufficient for reliable classification of non-LTR elements as it may fortuitously occur at the termini of other repeats. Thus, we dismissed repeats classified as non-LTR elements, unless they were classified through the HOM and/or the TSD modules, which are based on more reliable characters. The resulting TE composition profiles (fig. 5) for the 10 fungal genomes reveal several interesting trends. First, the two smallest genomes, C. albicans and U. maydis, display the least diverse assortment of TEs, containing only six and three LTR families, respectively, and a single DNA transposon family (fig. 5). These TE profiles closely resemble those reported for the similarly compact genomes of Saccharomyces cerevisiae (Kim et al. 1998) and Schizosaccharomyces pombe (Bowen et al. 2003). In these two yeasts, only a handful of LTR retrotransposon families have persisted, probably by virtue of their ability to target heterochromatin and other chromosomal “safe heavens,” which mitigates their disruptive effects (Bushman 2003). The detection of a DNA transposon family in C. albicans is interesting as it indicates that DNA elements must have been recently active in this yeast species, in contrast to S. cerevisiae and S. pombe. The fact that U. maydis, a basidiomycete species, has a similar profile suggests the possibility of convergent reduction of TE diversity in extremely compact fungi genomes, as observed in widely diverged yeasts. It would be interesting to see if the elements subsisting in U. maydis have also adopted targeting strategies.
FIG. 5.—
TE composition profiles generated by REPCLASS for 10 fungal genomes. The species are ranked by increasing genome size from left to right. For taxonomic information, see table 2 and supplementary table 1 (Supplementary Material online), and for a phylogenetic relationships, see Fitzpatrick et al. (2006).
As genome size increases, both the number and diversity of TE families classified by REPCLASS increase (fig. 5). The proportion of DNA transposons tends to grow as genome size increases and accounts for half or more of all TE families in the three species (representing three phyla) with genomes over 100 Mb. This trend suggests another type of convergence in the fungal TE landscape, whereby larger genomes harbor a greater diversity of DNA transposons. Together, these data evoke a relatively simple pattern of TE evolution in fungi, where genome contraction is associated with the elimination of DNA transposons, whereas genome expansion is associated with increasing amount and diversity of DNA transposons.
Discussion
Here we have introduced REPCLASS, a tool that automates the classification of TE sequences and allows for a rapid evaluation of TE content in diverse eukaryotic species. In principle, the program can be used to annotate any DNA sequences, whether it is a collection of consensus sequences generated through ab initio repeat discovery or genomic sequence. Although REPCLASS requires a target genomic sequence as input to execute the TSD module, the HOM and STR modules do not. If no target genomic sequence is provided, the program will still run but the TSD module will return no results and the classification will only rely on HOM and STR modules. The user has also the option to run each module as a standalone application (see Methods). Thus, REPCLASS makes the complex task of classifying TEs manageable for the non-TE expert. When used in conjunction with ab initio repeat finding tools, REPCLASS can assist in large-scale genome annotation. Although the current version of REPCLASS is geared toward the classification of eukaryotic TEs, the design of the program should be readily able to classify prokaryotic mobile elements, and in particular, Insertion Sequences as these share most characteristics of eukaryotic DNA transposons, including transposase, TIRs, and TSD. The availability of several excellent reference databases for prokaryotic TEs (Leplae et al. 2004; Siguier et al. 2006) should allow the future development of a version of REPCLASS that can efficiently handle prokaryotic genomes.
We have demonstrated that REPCLASS can accurately classify most of the known types of TEs when run on manually curated libraries, such as the Repbase reference libraries. When combined with RepeatScout, an ab initio repeat finding program, to search the C. elegans and D. melanogaster genomes, REPCLASS was able to correctly classify 64.5% and 93% of the repeats identified by RepeatScout that are annotated in Repbase. The difference in the efficiency of REPCLASS between the two genomes was largely attributable to the fact that the consensus sequences produced by RepeatScout for C. elegans were incomplete and therefore lacked the defining structural characteristics of the corresponding TEs. This fragmentation issue affected the consensus sequences generated by RepeatScout for D. melanogaster but did not hinder classification by REPCLASS because most fragmented consensus sequences still retained coding regions with homology to other TE proteins. Regardless, these results suggest that REPCLASS is sensitive to the quality of the consensus library and it reemphasizes the necessity to combine (Quesneville et al. 2005; Smith, Edgar, et al. 2007) as well as improve (Saha et al. 2008) ab initio repeat finding methods if one wants to fully automate the annotation of TEs in genome sequences. The false-positive rate of REPCLASS, which is defined as the proportion of repeats that are correctly identified as TEs but incorrectly classified by REPCLASS, was estimated in three independent approaches (table 3) as ranging from 2% to 9% and averaging 5% for a total of nearly 500 TEs examined.
Table 3.
Data Summary on REPCLASS False-Positive Rate
| Data Seta | Number of False Positivesb | Number of TE Examined | False-Positive Rate |
| Control librariesc | 10 | 247 | 0.04 |
| Ab initio—“known families”d | 14 | 150 | 0.09 |
| Ab initio—“new families”e | 2 | 100 | 0.02 |
| Total | 26 | 497 | 0.05 |
For each data set, the results obtained for C. elegans and D. melanogaster were combined.
Number of TE families misclassified by REPCLASS, based on comparison of the classification given by REPCLASS to the one provided by Repbase (b and c) or by “manual” inspection (d).
See section “Validation with reference repeat libraries.”
See section “TE annotation by combining REPCLASS with ab initio repeat finding.”
See section “REPCLASS-assisted discovery of new TE families in C. elegans and D. melanogaster.”
We were surprised to discover a substantial number of new TE families when applying RepeatScout and REPCLASS to the genomes of D. melanogaster and C. elegans, two species that have been the subject of intense TE mining over the past two decades. Some of these families, in particular in D. melanogaster (see supplementary table 3, Supplementary Material online), are clearly related to known families because individual copies in the genome show positional overlap with segments masked as TE by Repeatmasker in the most recent genome assembly available at the UCSC Genome Browser. Still these copies have weak similarity (<80%) to the Repbase consensus but high similarity (>90%) with our RepeatScout consensus. Hence, these families may not be considered entirely novel but distant relatives of known families. Another subset, most commonly encountered in C. elegans (see supplementary table 3, Supplementary Material online), represents TE families that are unrelated to previously described families (except sometimes for short conserved motifs in their TIRs or coding sequences). Indeed, individual copies from these families do not overlap significantly with segments masked as TE by Repeatmasker (for some examples, see supplementary tables 2 and 3, Supplementary Material online). These findings substantially augment the TE repertoire of these model species and highlight the power of REPCLASS for TE discovery.
Our analysis of the lesser quality genome assemblies of other Caenorhabditis and Drosophila species revealed that the RepeatScout output suffers more consensus fragmentation but mostly when the repeats are relatively long (e.g., LTR retrotransposons). In genomes that are dominated by relatively short TEs, like those of Caenorhabditis, the quality of the assembly had little impact on TE profiling by REPCLASS. In Drosophila genomes, which are populated by relatively long retrotransposons, the issue of fragmentation did not strongly affect the accuracy of classification but often artificially inflated the numbers of TE families because the same family may be classified multiple times. This issue was probably exacerbated by the peculiar genomic compartmentalization of TEs in Drosophila, which are largely concentrated in heterochromatic areas where TEs pile up densely and form complex nested arrangements (Pimpinelli et al. 1995; Bergman et al. 2006; Hoskins et al. 2007). These regions are likely to be misassembled, disrupted by gaps, and ultimately confined to short contigs, if not completely discarded from draft WGS assemblies. Although progress has been made in assembling heterochromatic regions in D. melanogaster (Hoskins et al. 2007; Smith, Shu, et al. 2007), these areas are likely to be poorly resolved in the D. pseudoobscura and D. virilis draft assemblies, which further increases the likelihood of consensus fragmentation by RepeatScout. Thus, although REPCLASS is efficient at capturing the overall TE composition profile characteristic of a species even in low-coverage WGS sequences, one should be cautious at interpreting interspecific variations in TE content when the input genome sequences are of variable quality.
We have shown that combining RepeatScout with REPCLASS effectively recapitulates the contrasting TE profiles of Caenorhabditis and Drosophila. Our analysis of fungal genomes yielded TE composition profiles consistent with those previously observed, indicating that the RepeatScout/REPCLASS suite should work well to characterize the TE content of a broad range of eukaryotic species. As another example, we recently applied the RepeatScout/REPCLASS suite to the draft genome sequence of the crustacean Daphnia pulex, a species where only a single TE family has been described previously (Penton et al. 2002). It took REPCLASS less than a day to screen 10,597 consensus sequences compiled by RepeatScout and classify 1,668 of them as TEs, including 1,198 with high level of confidence (i.e., by at least two modules or by HOM; Pritham E, Keswani U, Feschotte C, unpublished data). It would take weeks or months for any qualified individual to sift through such a colossal output manually. REPCLASS provides a much-needed addition to the genomicist's toolbox that will significantly accelerate TE discovery and genome annotation.
To further illustrate the utility of the program, we applied the RepeatScout/REPCLASS suite to explore the repetitive DNA landscape of several genomes whose TE contents had not been thoroughly investigated previously. Here we highlight several interesting biological findings that have emerged from these genomic explorations. First, we found that distantly related species of Caenorhabditis and Drosophila display highly conserved TE composition profiles within each genus. Drosophila genomes are dominated by LTR and non-LTR retrotransposons (fig. 3B), although we note a significant increase in the number of Helitrons in D. virilis (supplementary fig. 2, Supplementary Material online), as noticed previously for one Helitron family (Yang and Barbash 2008). These results are in agreement with an initial analysis of TEs in 12 Drosophila genomes (Clark et al. 2007), which suggested a broad conservation of TE diversity, despite rapid turnover of TE sequences and significant variations in the total amount of TEs across the Drosophila phylogeny. The TE content of C. remanei and C. brenneri has not been examined previously, and only a brief analysis of repeat content has been published for C. briggsae (Stein et al. 2003). Our study indicates that the genomes of C. remanei and C. brenneri, like those of C. elegans (Surzycki and Belknap 2000) and C. briggsae (Stein et al. 2003), are dominated by a diverse assortment of DNA transposons (fig. 3B; supplementary fig. 1, Supplementary Material online). Together, these data indicate that the contrasting TE compositions of Caenorhabditis and Drosophila are not the result of stochastic variations caught by random snapshots in time but rather have been shaped by selective forces over evolutionary time. Interestingly, those forces appear to have acted in opposite directions in the Drosophila and Caenorhabditis genera as the former is dominated by long retrotransposons, whereas the latter contains mainly small DNA transposons. These divergent patterns cannot be simply explained by constraints associated with different genome size as nematodes and fruit flies both fall within the small end of the spectrum of invertebrate genome sizes (Gregory 2005). Furthermore, TE composition remains conserved among Caenorhabditis and among Drosophila species even though the total amount of repetitive DNA appears to vary within each genus as a function of genome size (Clark et al. 2007; Stein et al. 2003; and this study, see table 1). Our analysis also shows that the larger amounts of repetitive DNA in C. brenneri and C. remanei compared with C. elegans and in D. virilis compared with D. melanogaster do not result simply from elevated copy number in one or a few TE families but rather from an increase in the number of TE families in almost all subclasses (table 1; supplementary figs. 1 and 2, Supplementary Material online). The result is an overall conservation of TE composition despite significant variations in the sheer number of TEs and TE families among these species. Taken together, these results suggest that TE composition in nematodes and flies is constrained by selective forces, either adaptive or nonadaptive, possibly reflecting divergent life-history traits (Lynch 2007).
Our REPCLASS analysis of fungi genomes offers a comparison of TE composition at a much broader evolutionary scale (>500 million years of evolution) than our exploration of nematodes and flies. We observed that TE composition varies widely across the fungi “kingdom” and appears to strongly correlate with genome size (figs. 4 and 5). Smaller genomes tend to have low TE content and reduced TE diversity, with a predominance of LTR retrotransposons. As genome size increases, so does the number of TE families and a more diverse assortment of TE types becomes apparent, with a notable enrichment in DNA transposons. These findings again point to the existence of powerful, but as yet mysterious, forces underlying TE composition patterns. The ever-increasing pace of genome sequencing and the development of REPCLASS will make it possible to rapidly characterize TE landscapes in a large and diverse sample of eukaryotic species, an opportunity that should yield insights onto the evolutionary and ecological principles influencing TE composition and ultimately genome architecture.
Funding
National Institutes of Health (R01GM77582 to C.F.); National Science Foundation (EIA-0216500).
Supplementary Material
Supplementary figures 1–5, tables 1–3, and files 1 and 2 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Supplementary Material
Acknowledgments
We thank Assiatu Barrie, Clément Gilbert, Ellen Pritham, and Sarah Schaack for useful comments during the preparation of the manuscript. We are grateful to Patrick McGuigan of the UT Arlington Distributed and Parallel Computing Cluster for assistance with computing resources. We acknowledge the genome sequencing centers listed in supplementary table 1 (Supplementary Material online) for providing access to sequence data and especially John Spieth and The Genome Center at Washington University School of Medicine in St Louis for permission to use the C. brenneri and C. remanei assemblies prior to publication and James Galagan and the Fungal Genome Initiative at the Broad Institute.
References
- Andrieu O, Fiston AS, Anxolabehere D, Quesneville H. Detection of transposable elements by their compositional bias. BMC Bioinform. 2004;5:94. doi: 10.1186/1471-2105-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Eddy SR. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H. Discovering and detecting transposable elements in genome sequences. Brief Bioinform. 2007;8:382–392. doi: 10.1093/bib/bbm048. [DOI] [PubMed] [Google Scholar]
- Bergman CM, Quesneville H, Anxolabehere D, Ashburner M. Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol. 2006;7:R112. doi: 10.1186/gb-2006-7-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL. Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 2003;13:1984–1997. doi: 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent MR. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat Rev Genet. 2008;9:62–73. doi: 10.1038/nrg2220. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Brunner S, Pea G, Rafalski A. Origins, genetic organization and transcription of a family of non-autonomous helitron elements in maize. Plant J. 2005;43:799–810. doi: 10.1111/j.1365-313X.2005.02497.x. [DOI] [PubMed] [Google Scholar]
- Bushman FD. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–138. doi: 10.1016/s0092-8674(03)00760-8. [DOI] [PubMed] [Google Scholar]
- Carlton JM, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Craig NL, Craigie R, Gellert M, Lambowitz A. Mobile DNA II. Washington (DC): American Society for Microbiology Press; 2002. [Google Scholar]
- Cutter AD. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol. 2008;25:778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- Daboussi MJ, Capy P. Transposable elements in filamentous fungi. Annu Rev Microbiol. 2003;57:275–299. doi: 10.1146/annurev.micro.57.030502.091029. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Myers EW. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21(Suppl 1):i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. Computational analysis and paleogenomics of interspersed repeats in eukaryotes. In: Stojanovic N, editor. Computational genomics: current methods. Hethersett (Norwich): Horizon Scientific Press; 2007a. pp. 31–53. [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007b;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Zhang X, Wessler S. Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington (DC): American Society for Microbiology Press; 2002. pp. 1147–1158. [Google Scholar]
- Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P. Gene prediction: compare and CONTRAST. Genome Biol. 2007;8:233. doi: 10.1186/gb-2007-8-12-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. Multiple LTR-retrotransposon families in the asexual yeast Candida albicans. Genome Res. 2000;10:174–191. doi: 10.1101/gr.10.2.174. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Synergy between sequence and size in large-scale genomics. Nat Rev Genet. 2005;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRGSP. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Jones SJ. Prediction of genomic functional elements. Annu Rev Genomics Hum Genet. 2006;7:315–338. doi: 10.1146/annurev.genom.7.080505.115745. [DOI] [PubMed] [Google Scholar]
- Jones T, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kaminker JS, et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3:RESEARCH0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamper J, et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci USA. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci USA. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evol Int J Org Evol. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Leplae R, Hebrant A, Wodak SJ, Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res. 2004;32:D45–D49. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. ReAS: recovery of ancestral sequences for transposable elements from the unassembled reads of a whole genome shotgun. PLoS Comput Biol. 2005;1:e43. doi: 10.1371/journal.pcbi.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sunderland (MA): Sinauer; 2007. [Google Scholar]
- Marino-Ramirez L, Lewis KC, Landsman D, Jordan IK. Transposable elements donate lineage-specific regulatory sequences to host genomes. Cytogenet Genome Res. 2005;110:333–341. doi: 10.1159/000084965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson SA. EMBOSS opens up sequence analysis. European Molecular Biology Open Software Suite. Brief Bioinform. 2002;3:87–91. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- Penton EH, Sullender BW, Crease TJ. Pokey, a new DNA transposon in Daphnia (cladocera: crustacea) J Mol Evol. 2002;55:664–673. doi: 10.1007/s00239-002-2362-9. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S, et al. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci USA. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Quesneville H, et al. Combined evidence annotation of transposable elements in genome sequences. PLoS Comput Biol. 2005;1:166–175. doi: 10.1371/journal.pcbi.0010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan N, Feschotte C, Levine D. Cluster- and grid-based classification of transposable elements in eukaryotic genomes. Proc Sixth IEEE Int Symp Cluster Comput Grid. 2006;2:45–52. [Google Scholar]
- Saha S, Bridges S, Magbanua ZV, Peterson DG. Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 2008;36:2284–2294. doi: 10.1093/nar/gkn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, et al. Improved repeat identification and masking in Dipterans. Gene. 2007;389:1–9. doi: 10.1016/j.gene.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Shu S, Mungall CJ, Karpen GH. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316:1586–1591. doi: 10.1126/science.1139815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surzycki SA, Belknap WR. Repetitive-DNA elements are similarly distributed on Caenorhabditis elegans autosomes. Proc Natl Acad Sci USA. 2000;97:245–249. doi: 10.1073/pnas.97.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- Yang HP, Barbash DA. Abundant and species-specific DINE-1 transposable elements in 12 Drosophila genomes. Genome Biol. 2008;9:R39. doi: 10.1186/gb-2008-9-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.