Abstract
Prochlorococcus is a genus of marine cyanobacteria characterized by small cell and genome size, an evolutionary trend toward low GC content, the possession of chlorophyll b, and the absence of phycobilisomes. Whereas many shared derived characters define Prochlorococcus as a clade, many genome-based analyses recover them as paraphyletic, with some low-light adapted Prochlorococcus spp. grouping with marine Synechococcus. Here, we use 18 Prochlorococcus and marine Synechococcus genomes to analyze gene flow within and between these taxa. We introduce embedded quartet scatter plots as a tool to screen for genes whose phylogeny agrees or conflicts with the plurality phylogenetic signal, with accepted taxonomy and naming, with GC content, and with the ecological adaptation to high and low light intensities. We find that most gene families support high-light adapted Prochlorococcus spp. as a monophyletic clade and low-light adapted Prochlorococcus sp. as a paraphyletic group. But we also detect 16 gene families that were transferred between high-light adapted and low-light adapted Prochlorococcus sp. and 495 gene families, including 19 ribosomal proteins, that do not cluster designated Prochlorococcus and Synechococcus strains in the expected manner. To explain the observed data, we propose that frequent gene transfer between marine Synechococcus spp. and low-light adapted Prochlorococcus spp. has created a “highway of gene sharing” (Beiko RG, Harlow TJ, Ragan MA. 2005. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci USA. 102:14332–14337) that tends to erode genus boundaries without erasing the Prochlorococcus-specific ecological adaptations.
Keywords: marine cyanobacteria, horizontal gene transfer, introgression, quartet decomposition, supertree, genome evolution
Introduction
Discovered only 20 years ago (Chisholm et al. 1988), members of genus Prochlorococcus are now known to be some of the most abundant organisms on Earth, playing a vital role in global carbon cycle. Their closest relatives, members of marine Synechococcus clade A (Waterbury et al. 1979) (hereafter referred as marine Synechococcus), are also very abundant, with different but overlapping geographic and depth distribution (Zwirglmaier et al. 2008). Despite a close relationship originally indicated by comparative analyses of 16S ribosomal RNA (rRNA) genes, Prochlorococcus is distinguished from the Synechococcus among other things by a unique set of photosynthetic pigments, different light-harvesting apparatus, tiny size, and better ability to grow in oligotrophic waters (Partensky et al. 1999). From 16S rRNA, rpoC1 gene, and 16S–23S rRNA internal transcribed spacer (ITS) region analyses, Prochlorococcus appears as a sister clade to marine Synechococcus (Palenik and Haselkorn 1992; Urbach et al. 1992; Rocap et al. 2002). The great observed diversity within Prochlorococcus spp. was hypothesized to comprise multiple ecotypes (i.e., groups adapted to different environmental conditions, based on their physiology), two most distinguishable divisions being low-light adapted and high-light adapted ecotypes (Moore and Chisholm 1999), with further division into more refined subgroups (Ahlgren et al. 2006). This division is fuzzy: although there are correlations of certain environmental parameters (such as nutrient availability, temperature, light) with ecotypes, Coleman and Chisholm (2007) remark that “recognition of clades and clusters, and their interpretation in light of ecological factors, depends on the scale of observation.” Whether or not there exists a one-to-one mapping between Prochlorococcus and Synechococcus niches and their genomic content remains largely unresolved.
The availability of sequenced genomes from multiple isolates of both marine Synechococcus and Prochlorococcus provided more insights into the evolution of these organisms. It was noticed that Prochlorococcus spp. tend to have much smaller genomes with lower GC content (cf., table 1). Although these properties often characterize genomes under reduced selection, the ratio of rates of synonymous to nonsynonymous substitutions is higher in the lower-GC Prochlorococcus spp. than in the marine Synechococcus (Hu and Blanchard 2009). As a consequence of GC composition, protein-coding genes in lower GC genomes have skewed codon usage (Dufresne et al. 2005). Despite having >96% 16S rRNA identity, a remarkable genome divergence was observed between (and within) Prochlorococcus and marine Synechococcus as measured by average nucleotide identity (ANI) and average amino acid identity (AAI) (cf., supplementary tables 1 and 2, Supplementary Material online; for ANI analyses within marine Synechococcus, see also Dufresne et al. 2008). A notable exception is the single hyperconserved protein described by us (Zhaxybayeva et al. 2007). Genome dot plots revealed multiple rearrangements, especially among marine Synechococcus spp. (Dufresne et al. 2008), and presence of numerous genomic islands (Coleman et al. 2006; Dufresne et al. 2008). The rearrangements, flanked by transfer RNAs (tRNAs) or genomic islands, as well as the islands themselves, suggested the importance of horizontal (or lateral) gene transfer (HGT) and recombination in shaping diversity of these genomes (e.g., Rocap et al. 2003; Coleman et al. 2006).
Table 1.
Summary of Metadata for 19 Marine Cyanobacteria Used in This Study
| Genome | Isolation Location, Depth (m)a | Number of ORFs | Cladeb | GC Content, % |
| Prochlorococcus marinus strain MIT 9312 | Gulf Stream, 135 | 1,809 | HL, II | 31.2 |
| Prochlorococcus marinus strain MIT 9313 | Gulf Stream, 135 | 2,265 | LL, IV | 50.7 |
| Prochlorococcus marinus strain MIT 9303 | Sargasso Sea, 100 | 2,997 | LL, IV | 50.0 |
| Prochlorococcus marinus strain MIT 9515 | Equatorial Pacific, 15 | 1,906 | HL, I | 30.8 |
| Prochlorococcus marinus strain AS9601 | Arabian Sea, 50 | 1,921 | HL, II | 31.3 |
| Prochlorococcus marinus strain NATL1A | North Atlantic, 30 | 2,193 | LL, I | 35.0 |
| Prochlorococcus marinus strain NATL2A | North Atlantic, 10 | 1,890 | LL, I | 35.1 |
| Prochlorococcus marinus strain CCMP1375 | Sargasso Sea, 120 | 1,882 | LL, II | 36.4 |
| Prochlorococcus marinus strain CCMP1986 | Mediterranean Sea, 5 | 1,712 | HL, I | 30.8 |
| Prochlorococcus marinus strain MIT 9301 | Sargasso Sea, 90 | 1,907 | HL, II | 31.3 |
| Prochlorococcus marinus strain MIT 9211 | Equatorial Pacific, 83 | 1,855 | LL, III | 39.7 |
| Prochlorococcus marinus strain MIT 9215 | Equatorial Pacific, 0 | 1,983 | HL, II | 31.1 |
| Synechococcus sp. WH8102 | Western Carribean, open ocean strain | 2,517 | Marine A III | 59.4 |
| Synechococcus sp. CC9605 | Off the coast of California | 2,638 | Marine A | 59.2 |
| Synechococcus sp. CC9902 | Coastal seawater (off California) | 2,304 | Marine A | 54.2 |
| Synechococcus sp. CC9311 | Edge of California Current, coastal strain | 2,892 | Marine A | 52.4 |
| Synechococcus sp. RCC307 | Mediterranean Sea, 15 | 2,535 | Marine A | 60.8 |
| Synechococcus sp. WH7803 | Sargasso Sea | 2,533 | Marine A V | 60.2 |
| Synechococcus sp. PCC7002 | Maguyes Island, Puerto Rico | 2,823 | Cluster 3 | 49.6 |
NOTE.—HL, high-light adapted ecotype; LL, low-light adapted ecotype.
Based on results reported in Rocap et al. (2002) and information from NCBI Genomes Web page.
Clades are according to phylogenetic relationships inferred from 233 positions of the 16S–23S rRNA ITS region (Rocap et al. 2002).
Sullivan et al. (2003) found host strain–specific phages infecting Prochlorococcus strains and phages cross-infecting members of different Prochlorococcus ecotypes, as well as both Prochlorococcus and marine Synechococcus. Further studies suggested that recombination within and between Prochlorococcus and Synechococcus may be mediated by phages (Lindell et al. 2004; Sullivan et al. 2005; Zeidner et al. 2005), and some host genes are maintained by phages. In particular, genes encoding unstable components of the photosynthesis machinery are widely spread among cyanophages (Sullivan et al. 2006; Sharon et al. 2007; Sandaa et al. 2008), kept under purifying selection (Zeidner et al. 2005) and expressed during the infection (Lindell et al. 2005). Phycobilisome pigment biosynthesis genes carried by cyanophages were also shown to be transcribed during the infection (Dammeyer et al. 2008). Complete genome sequencing of cyanophages (nine are currently deposited to GenBank) revealed that phage genomes contain not only photosynthesis-related host genes but also other metabolic genes involved in nucleotide metabolism, carbon metabolism, phosphate stress, and lipopolysaccharide biosynthesis (Sullivan et al. 2005; Weigele et al. 2007). These insights into cyanophage genomes suggest that phages might be very important in shaping the genomic content of Prochlorococcus and marine Synechococcus.
When only four genomes were available (Prochlorococcus marinus strains CCMP1375, CCMP1986, and MIT 9313, and marine Synechococcus WH8102), genome-wide analyses involving multiple gene families within several genomes (and utilizing different methodologies) reported that signal recovered from the majority and plurality of genes contradicted the 16S rRNA phylogeny (Zhaxybayeva et al. 2004; Beiko et al. 2005; Zhaxybayeva et al. 2006). Notably, the Prochlorococcus/marine Synechococcus as a group exhibited a large number of genes contradicting this plurality-based phylogenetic signal (Zhaxybayeva et al. 2006), suggesting noncongruent evolutionary histories of individual genes. Such incongruencies have been noted in individual gene analyses as well: for example, a phylogenetic tree reconstructed from ntcA gene also had shown two low-light adapted ecotypes with the largest genomes robustly grouping with Synechococcus sp. (Penno et al. 2006).
More recent studies involving more genomes (Kettler et al. 2007; Dufresne et al. 2008) have concentrated on phylogenetic signal extracted from a concatenation of core genes in the set of genomes (i.e., genes present in all considered genomes). Kettler et al. (2007) mapped patterns of gene gain and loss in Prochlorococcus spp. onto the concatenated gene phylogeny and found that for most genomes, the noncore genes gained by genomes are located in the genomic islands. The analysis of gene families in 11 genomes of marine Synechococcus isolates revealed their complex and mosaic phylogenetic history (Dufresne et al. 2008). Based on bipartition analyses of core genes against the phylogenetic tree reconstructed from the concatenated gene alignment, Dufresne et al. (2008) reported 9.3% of core genes having a history of HGT, and additionally many accessory genes were mapped to genomic islands. Given the conservativeness of bipartition analyses (Zhaxybayeva et al. 2006), this number likely underestimates the overall impact of HGT in this group. In this manuscript, we analyze gene families present in 18 genomes of P. marinus and marine Synechococcus, in an attempt to assess how HGT and vertical inheritance shaped the evolution of these genomes and their adaptation to environmental constraints. We use quartet decomposition (Zhaxybayeva et al. 2006), a more sensitive and robust method in comparison to bipartition analyses, and we include in our analyses gene families not present across all analyzed genomes as well as gene families containing additional homologs in each genome (either in-paralogs or xenologs). Our analyses, based on quartets embedded in gene phylogenies, do not require concatenation of alignments.
Materials and Methods
Genome Data
The 19 genomes used in this study were downloaded from the NCBI's RefSeq database (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/): P. marinus strain MIT 9312, P. marinus strain MIT 9313, P. marinus strain MIT 9303, P. marinus strain MIT 9515, P. marinus strain AS9601, P. marinus strain NATL1A, P. marinus strain NATL2A, P. marinus strain CCMP1375, P. marinus strain CCMP1986, P. marinus strain MIT 9301, P. marinus strain MIT 9211, P. marinus strain MIT 9215, Synechococcus sp. WH8102, Synechococcus sp. CC9605, Synechococcus sp. CC9902, Synechococcus sp. CC9311, Synechococcus sp. RCC307, Synechococcus sp. WH7803, and Synechococcus sp. PCC7002 (see table 1). Provided RefSeq annotations of protein-coding genes were used. Note that Synechococcus sp. PCC7002 genome was added to provide an outgroup.
Detection of Gene Families
All protein-coding open reading frames (ORFs) in each genome were searched against all protein-coding ORFs in every other genome using Protein Basic Local Alignment Search Tool (BLASTP) (Altschul et al. 1997), that is, all pairwise genome comparisons were performed. All matches per query ORF with E value <10−4 were saved. Bit scores of the matches were normalized through dividing the scores by query length. The normalized bit scores were passed through the Markov clustering (MCL) program (by Stijn van Dongen, http://micans.org/mcl/; Enright et al. 2002) with inflation parameter set to 1.1 (in order to obtain large clusters or superfamilies). Each superfamily was broken into gene families using phylogenetic information, as implemented in the BRANCHCLUST program (Poptsova and Gogarten 2007) with parameter MANY = 10. This selection resulted in 1,812 gene families without any paralogs and with members present in at least four genomes, 482 gene families (also present in at least four genomes) with at most eight in-paralogs (i.e., lineage-specific duplications), and 76 families with more than eight in-paralogs. The latter 76 families were not analyzed further (due to their overly complicated evolutionary histories). Families with and without in-paralogs were analyzed separately (see below).
Quartet Decomposition Analyses
The method of quartet decomposition is described in Zhaxybayeva et al. (2006). In brief, gene families were aligned in ClustalW version 1.83 (Thompson et al. 1994). (In a test run, the alignments were further “cleaned” with the GBLOCKS [Castresana 2000] program. Quartet decomposition results [see below] were not qualitatively affected by this alignment pruning technique [data not shown]. Because removing sites from the alignments does not necessarily produce better phylogenies and results in loss of informative data [Wong et al. 2008], the analyses discussed in this manuscript are based on original ClustalW alignments.) The shape parameter of the gamma distribution for each gene family alignment was calculated in Tree-Puzzle version 5.2 (Schmidt et al. 2002) under the Jones, Taylor, and Thornton (JTT) model (Jones et al. 1992) with among-site rate variation modeled using a gamma distribution approximated by four categories (Yang 1994). One hundred bootstrap samples were generated for each gene family using the SEQBOOT program of the PHYLIP package (Felsenstein 1993), and distance matrices were calculated for each bootstrap sample in Tree-Puzzle version 5.2 using shape parameters estimated for the original alignment (see above). Neighbor-Joining trees were calculated using the NEIGHBOR program from the PHYLIP package (Felsenstein 1993). These phylogenetic analyses were chosen for their speed (calculations of maximum likelihood trees of 100 bootstrap samples per data set were too slow). For each gene family, all embedded quartets were evaluated and results of quartets with at least 80% bootstrap support were summarized in a spectrogram (using scripts from Zhaxybayeva et al. 2006). Quartets containing short internal branches (less than three substitutions over alignment length) or long external branches (10 times longer than internal branch) were excluded from analyses before the summarizing step.
Plurality Tree Calculation and Conflicting Families
Quartet topologies supported by a plurality of gene families were used to reconstruct a supertree using the “matrix representation using parsimony” (MRP) method (Baum 1992; Ragan 1992) as implemented in CLANN version 3.0.2 (Creevey and McInerney 2005). Only quartets that were resolved by at least 30% of gene families that contained it were included. The tree from the resulting MRP matrix was calculated using the PARS program of the PHYLIP package (Felsenstein 1993). Families with at least one quartet with a topology contradicting the plurality tree with more than 80% bootstrap support were identified as “families conflicting with plurality signal.” For gene families with “paralogous” members, each conflict involving an “in-paralog” was counted toward the number of conflicts. Additionally, the MRP matrix was analyzed using the P-distances and NeighborNet method as implemented in SplitsTree 4 (Lapointe et al. 2003; Huson and Bryant 2006).
Assessment of False Positives
In Zhaxybayeva et al. (2006), we used simulations to assess quartet false positives and false negatives and concluded that false positives diminish if quartets that are in general poorly resolved by many gene families (more than 70% of families containing the quartet) are excluded from further analyses. We used the same cutoff (i.e., at least 30% of gene families required to support one of the three quartet topologies with ≥80% bootstrap support) in the present analyses.
Assignment of Functional Categories
Each gene family was searched against the COG database (Tatusov et al. 2003) (August 2005 release obtained from NCBI's FTP site) using BLASTP search with E value cutoff of 10−5. COG category of top-scoring BLASTP hit was assigned as the gene family's functional category.
Rooting of Each Gene Family
Additional homologs from the completely sequenced genomes of Thermosynechococcus elongatus BP-1, Synechocystis sp. PCC6803, and Synechococcus elongatus PCC7942 (all obtained from NCBI RefSeq database) were added to each gene family using BLASTP searches with E value cutoff of 10−20. Each “extended” family was aligned in ClustalW, and trees were obtained using the same methodology as for original gene families (see above). The consensus bipartitions for 100 bootstrap samples were calculated using the CONSENSE program of the PHYLIP package (Felsenstein 1993). The consensus bipartitions were screened for the families with at least two outgroup homologs (genome Synechococcus sp. PCC7002 was considered as a part of the outgroup, if present in a gene family) and where the ingroup formed a monophyletic group with at least 50% bootstrap support. Position of a root per gene family was extracted from the gene families satisfying the above criteria (1,082 of 1,812 gene families without in-paralogs).
Alignment Conservation and Its Impact on Plurality Signal
Proportion of identical sites in a gene family alignment was considered as a proxy for alignment conservation (gaps were treated as missing data). The gene families were divided into five categories of alignment conservation (0–0.2, 0.2–0.4, 0.4–0.6, 0.6–0.8, and 0.8–1.0). For each category, its impact on the plurality topology was assessed.
Assessment of Agreement of Each Gene Family with the Plurality Tree
Each gene family (consisting of n taxa) was given an agreement score, calculated as
the sum being over all quartets in the n-taxon family. The agreement score was normalized using maximum possible score of 100 × C4n. The drawback of this score is that it only shows agreement with plurality and does not distinguish between strong disagreement and poor resolution.
To address the question whether the gene families were on average in agreement with the plurality topology, we performed the following randomizations: for each gene family, taxa assignments on bootstrap trees were reshuffled. The resulting gene families were summarized into plurality topology (see above), and agreement of individual reshuffled gene families were assessed using the score above. The choice of this approach over simulations of trees was made for two reasons: 1) tree shapes of real trees were preserved (and hence no tree shape bias generated) and 2) the bootstrap support values are also preserved (to avoid solving the problem of how to simulate bootstrap values on the simulated tree topologies). Ten randomizations were performed, and the mean and standard deviation of average agreement scores were obtained. The Z score of average real-data agreement score and randomized score (normally distributed) was calculated and its significance assessed.
Genome Divergence as Measured by ANI and AAI
For all protein-coding genes in a genome, we calculated average nucleotide identity (ANI; Konstantinidis and Tiedje 2005a) and amino acid identity (AAI; Konstantinidis and Tiedje 2005b) using modified calculations: 1) if basic local alignment search tool (BLAST) search reported multiple hits per ORF, they were consolidated and 2) identity in the region not reported by BLAST was not considered to be 0% (i.e., the score was normalized by BLAST search match length and not the length of the query). These modifications would err on the side of making ANI and AAI values potentially higher than the values calculated according to Konstantinidis and Tiedje (2005a, 2005b).
Phylogenetic Tree Based on Genome Rearrangements
Genomes (as collection of ORFs in order of their appearance in each genome) were aligned using MUMmer version 3.20 (Kurtz et al. 2004). The MUMmer alignment results were converted into pairwise gene order and strand location information for each pair of genomes (omitting unaligned regions) suitable for estimating INV distance using the GRIMM program (Tesler 2002). The resulting INV distances (number of inversions needed to convert one genome's order into another's) were normalized using the total number of genes in two compared genomes. The tree from the resulting distance matrix was reconstructed using the FITCH program of the PHYLIP package (Felsenstein 1993) with global rearrangements and 10 jumbles.
Investigation of Potential Impact of Anomalous Gene Trees
According to Degnan and Rosenberg (2006), four-taxon trees containing branches with length below Lcritical = 0.156Ne generations are susceptible to anomalous gene tree (AGT) problem (where Ne = N/2). We attempted to estimate the value of Lcritical for the genomes analyzed in this manuscript, for which we needed data on generation time and estimate of effective population size Ne. In field studies, the observed Prochlorococcus population growth rate is 0.8–1 per day (Partensky et al. 1999), that is, on the order of one division per day. Ne × μ is estimated to be 1.00 for Prochlorococcus populations (Lynch and Conery 2003). Using these estimates, the critical number of substitutions is 0.156 × N × μ = 0.312 × Ne × μ [generations × substitutions/(site × year)]. Corrected for 365 generations per year, we obtained Lcritical = 0.312/365 [substitutions/site] = 0.0008548 [substitutions/site]. Sixty-six gene families were detected to have quartets with internal branch length below Lcritical. However, due to the removal of quartets with short internal branches, all these quartets were already removed from further analyses (see Quartet Decomposition Analyses). If Ne × μ value given above is an overestimate, which might be the case if this estimate was based on interpopulation comparisons, then the value of Lcritical will be even smaller.
Embedded Quartets Scatter Plot Analyses
To test various groupings of examined genomes, we developed quartet-based scores to assess how well a gene family supports a requested grouping of taxa. As a control, a randomized assignment of genomes into two categories was performed as well. For each gene family, we calculated the normalization score as a sum of all embedded quartets in agreement with a data partition multiplied by 100 (analogously to the normalization score of agreement with plurality topology, see above). The agreement with a partition score is a sum of bootstrap values (≥80%) for observed embedded quartets in agreement with a data partition divided by the normalization score (making the score to range between 0 and 1). The disagreement scores are calculated analogously. The resulting scatter plots show how well the data support the selected group and also discriminate poorly resolved gene families from those strongly favoring one or another scenario.
GC Composition and Its Effect on Phylogenetic Reconstruction
For each gene family, average variation of GC content between higher and lower GC content genomes was calculated (see table 1 for genomic GC content information). Correlation between the family's GC content variation and agreement with a data partition by GC content (the score was calculated as difference between agreement and disagreement scores; see Assessment with Plurality Scores) was investigated for the data sets that agree and disagree with plurality topology.
Addition of Phage-Encoded Genes to Gene Families
A total of 932 gene families with no in-paralogs and with conflicts to plurality topology were used in BLASTP searches (with E value cutoff of 10−10) against a database containing nine completely sequenced cyanophage genomes and three additional outgroup genomes (Thermosynechococcus elongatus BP-1, Synechocystis sp. PCC6803, and Synechococcus elongatus PCC7942). Thirty-five gene families with at least one cyanophage homolog satisfied the above criteria and were aligned in ClustalW version 1.83 (Thompson et al. 1994). The phylogenetic trees were reconstructed in PhyML (Guindon and Gascuel 2003) under JTT + G model and with 100 bootstrap samples. The trees were visually examined for cases of conflict that involve phage homologs.
16S rRNA Phylogenetic Tree Reconstruction
The 16S rRNA sequences alignment was retrieved from RDP database version 9.60 (Cole et al. 2007). The phylogenetic tree was reconstructed in the PhyML program version 2.4.5 (Guindon and Gascuel 2003) under Hasegawa–Kishino–Yano (HKY85) model (Hasegawa et al. 1985) with proportion of invariant sites estimated and gamma distribution with four rate categories (with shape parameter of gamma distribution estimated from the data). One hundred nonparametric bootstrap replicates were analyzed.
Results
Detection of Gene Families
A variety of criteria have been used to identify orthologous gene families in a set of completely sequenced genomes (for a recent review of many available methods, see Kuzniar et al. 2008). None of the available methods guarantee that selected families will not contain paralogs (or additionally acquired xenologs), whereas some methods approach this goal by being very strict (with a drawback of making the resulting number of gene families selected for analyses small). We combined BLAST-based selection of clusters using the MCL algorithm with further automated phylogenetic screening of families and paralogs (see Materials and Methods). Using phylogenetic information allows separation of distant paralogs and better identification of gene families (Poptsova and Gogarten 2007). In analyses of 19 genomes (table 1; 18 genomes of Prochlorococcus and marine Synechococcus and 1 outgroup genome of Synechococcus PCC7002), this procedure identified 1,812 families without in-paralogs, 482 families with at most eight in-paralogs (i.e., recent lineage-specific duplications) per gene family, and 76 gene families with many in-paralogs (supplementary fig. 1, Supplementary Material online). (Synechococcus PCC7002 is a marine isolate belonging to cluster 3 of Synechococcus [Herdman et al. 2001] and, according to relationships inferred from 16S rRNA gene depicted in fig. 1C, should be more distantly related to the 18 genomes of the ingroup.) Because there are no generally accepted criteria on how to choose one in-paralog over another, we decided to leave all but 76 families with multiple in-paralogs per genome intact but analyze them separately (as opposed to discarding them altogether, as some studies do; e.g., Swingley et al. 2008). Of the 1,812 families, 962 families without in-paralogs are core genes (i.e., present in 18 genomes of Prochlorococcus and marine Synechococcus) and 831 are also present in the outgroup genome, whereas the remaining gene families are present in at least 4 of 19 genomes. Among 482 gene families with in-paralogs, 193 gene families are core genes. Therefore, 962 + 193 = 1,155 core gene families were identified, which is comparable to 1,273 core gene families identified in an earlier study of 12 Prochlorococcus/marine Synechococcus genomes (Kettler et al. 2007), a subset of genomes in our study. An advantage of using quartet decomposition method (Zhaxybayeva et al. 2006) is that it allows us to combine in a single analysis not only the 1,155 core families but also families present in a smaller number of genomes and therefore include considerably more genomic information.
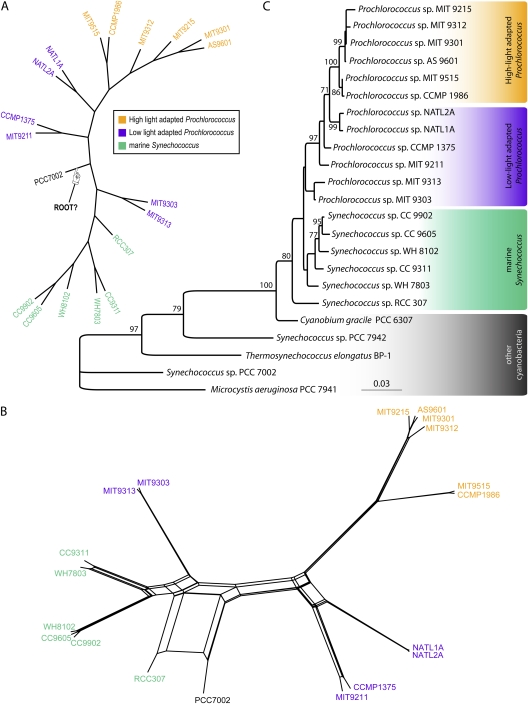
FIG. 1.—
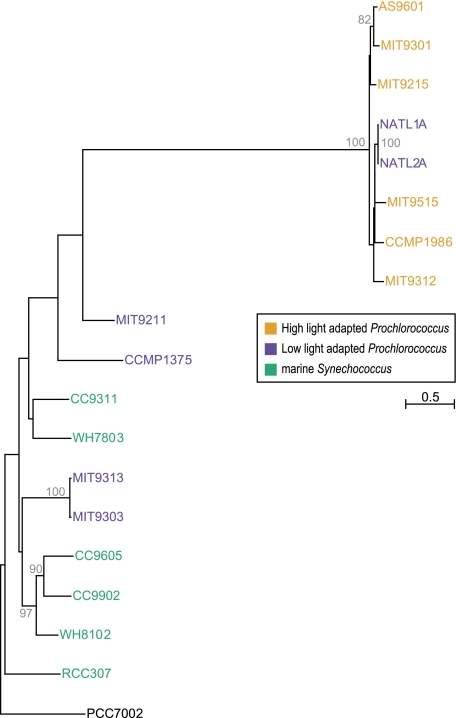
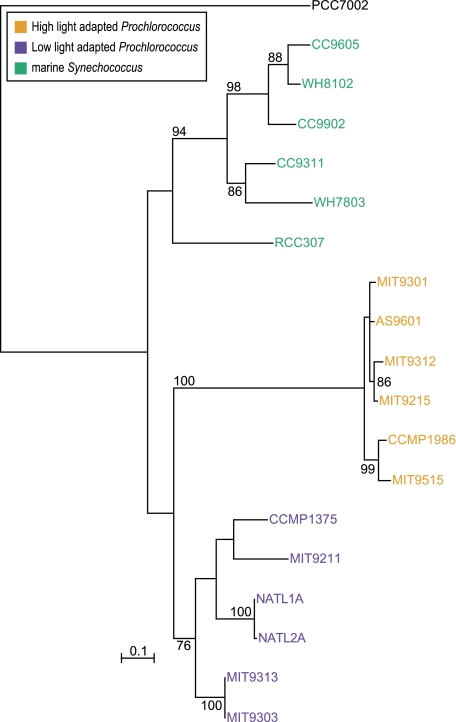
(A) Evolutionary relationships supported by plurality of 1,812 gene families without paralogs. The tree is a supertree reconstructed from embedded quartets supported by the plurality of the gene families (see Materials and Methods). The tree's branch lengths are not scaled with respect to substitutions. The robustness of relationships can be obtained from examining support of individual embedded quartets (see fig. 2). Although a strictly bifurcating tree is observed, it should be noted that this tree topology is strongly contradicted by 932 gene families without in-paralogs and 419 gene families with in-paralogs. Also note the alternative position of the outgroup taxon (Synechococcus sp. PCC7002) in comparison to rRNA tree topology shown in (B). Not all plurality quartets are in agreement with the depicted supertree (NeighborNet reconstruction is shown in supplementary fig. 3, Supplementary Material online). (B) NeighborNet reconstructed from all quartets significantly supported by individual gene families without in-paralogs. In contrast, supplementary figure 3 (Supplementary Material online) is based only on the quartets representing the plurality signal. (C) rRNA tree topology. Bootstrap support values below 70% are not shown. The tree was rooted using “other cyanobacteria” as an outgroup. Low-light adapted Prochlorococcus spp. do not form a monophyletic group (as noted earlier; Coleman and Chisholm 2007). Whereas 16S rRNA analysis does not provide enough resolution to obtain good bootstrap support for all branches, aphylogenetic tree based on 16S–23S rRNA ITS region has the same topology with better resolution (Rocap et al. 2002).
Phylogenetic Signal of 1,812 Gene Families without Paralogs
The spectrogram shown in figure 2 indicates that a large number of families (932, i.e., 51%) conflict with the plurality (fig. 1A) with a bootstrap support value of at least 80%. The gene families in conflict with the plurality phylogenetic signal span all functional categories (supplementary fig. 2, Supplementary Material online). The phylogenetic network inferred from the significantly supported embedded quartets in all analyzed gene families is shown in figure 1B (see also supplementary fig. 3, Supplementary Material online). The relationships among the 18 genomes are mostly in agreement with the phylogeny reported earlier for repeatedly concatenated randomly selected core gene sets of 100 (Kettler et al. 2007). The exception is the uncertainty around the location of the outgroup genome, Synechococcus sp. PCC7002, as indicated by unresolved splits in the NeighborNet (fig. 1B). This uncertainty is at the root of disagreement between 16S rRNA phylogeny (fig. 1C) and earlier genome-wide analyses (Zhaxybayeva et al. 2004, 2006; Beiko et al. 2005).
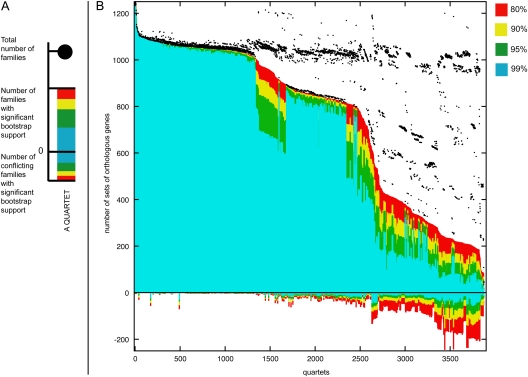
FIG. 2.—
Quartet decomposition analysis of 19 Prochlorococcus and marine Synechococcus genomes (cf., table 1). (A) A single component of quartet decomposition analysis (for more details on methodology, see Zhaxybayeva et al. 2006). Each embedded quartet is represented by a vertical bar and a black dot. The black dot indicates how many data sets contain this embedded quartet. The vertical bar shows the number of data sets having the topology of the quartet that is supported by a plurality of gene families (value above zero) and the number of data sets having one of the other two quartet topologies (value below zero). The bar is color coded with respect to bootstrap support. (B) The quartet spectrum of 1,812 gene families. Columns are sorted according to the number of supporting data sets with at least 80% bootstrap support. Quartets with a very short internal branch or very long external branches as well as those resolved by less than 30% of gene families were excluded from the analyses to minimize artifacts of phylogenetic reconstruction. Quartets above the x axis are combined into a plurality signal (see fig. 1A). Quartets below the x axis are embedded into 932 unique gene families. Note that for every quartet, at least one gene family is in conflict with the quartet topology supported by the plurality.
Rooting
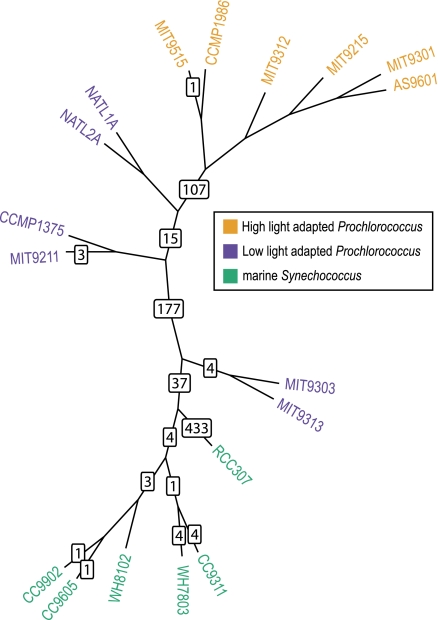
The uncertain position of Synechococcus sp. PCC7002 prompted us to consider the possibility that frequent gene sharing extends beyond the Prochlorococcus/marine Synechococcus group. Therefore, we added homologs from other closely related cyanobacteria (with completely sequenced genomes; see Materials and Methods) and asked where the root is located. Additional requirements for presence of homologs in at least two genomes of the outgroup, for monophyly of the ingroup and for at least 50% bootstrap support for the branch separating the ingroup from the outgroup, resulted in only 830 core (i.e., present in all 18 genomes of ingroup but not required to be present in Synechococcus sp. PCC7002) gene families being useful for rooting analyses. No unique location of the root emerged from this analysis (see fig. 3 and supplementary table 3, Supplementary Material online). This was not an unexpected result: each gene has a different phylogenetic history (e.g., see simulations in Zhaxybayeva and Gogarten 2004), and hence rooting of organismal phylogenies based on individual molecular phylogenetic trees is a somewhat arbitrary procedure. However, 433 gene families (52%) placed the root in the branch leading to Synechococcus sp. RCC307, which agrees with 16S rRNA topology (fig. 1C). The second largest number of genes, 177 (21%), placed the root on the branch where Synechococcus sp. PCC7002 is located in the plurality topology (fig. 1A). The variation in inferred position of the root supports the hypothesis that Synechococcus sp. PCC7002 participates in gene exchanges with the Prochlorococcus/marine Synechococcus group.
FIG. 3.—
Locations of root as inferred from the individual gene families. The backbone tree is the plurality tree depicted in figure 1A, with the PCC7002 genome removed. Numbers on the branches indicate how many gene families support position of the root on that branch (see Materials and Methods on details of finding the root location). Additionally, 35 gene families supported positioning the root on 14 branches that are in conflict with the plurality tree topology and hence cannot be depicted (see supplementary table 3, Supplementary Material online).
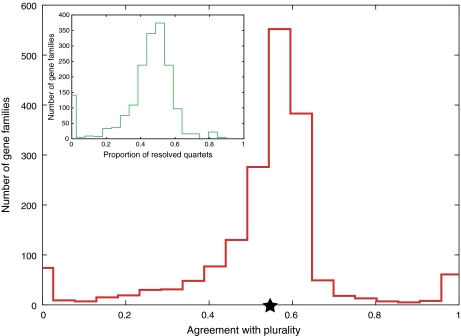
FIG. 4.—
Agreement of individual gene families with plurality tree. For details on score calculation, see Materials and Methods. Average agreement score was 0.54 and is indicated by a star. The main graph shows the agreement based on all embedded quartets (regardless of their resolution). The inset shows the distribution of proportions of unresolved quartets per gene family. On average, 45% of quartets per family were unresolved, explaining why agreement scores are somewhat low. However, the agreement with plurality topology is significantly better than random, for which average agreement score was 0.294 ± 0.003.
Robustness of the Plurality Signal
We tried to identify if the observed topological discrepancy is due to differences in evolutionary histories of the plurality of genes in the genomes and 16S rRNA (and presumed organismal history) or to artifacts in inferring the phylogenetic signal, such as 1) noncore (accessory) genes having different evolutionary histories from core genes, and if the former are abundant, affecting the overall signal; 2) poor quality of automatically generated alignments; and 3) the inferred compound signal not reflecting individual gene histories. Here, we show the robustness of the plurality tree to these potential artifacts.
First, the topology extracted from the analyses of only 962 core genes is identical to the one shown in figure 1 (data not shown). Therefore, noncore genes (while forming almost a half of the analyzed gene families) do not notably bias the resulting plurality signal. Second, we divided the gene families into five groups reflecting alignment conservation, as captured in the proportion of identical sites of an alignment (supplementary fig. 4, Supplementary Material online), and analyzed these five groups separately. Qualitatively, the results (supplementary figs. 5 and 6, Supplementary Material online) are not affected by degree of alignment conservation. Third, we investigated if individual gene families are in agreement with the plurality quartet topologies on average. Using the developed agreement-scoring scheme, we found that on average a gene family agrees with a plurality signal significantly better (average score of a real gene family is 0.5432 and average score of randomized families is 0.2939 ± 0.0028; Z score = 87.92) than a random tree agrees with a plurality (see fig. 3). The individual gene family agreement score is rather low (see Materials and Methods on details what this score means and how it is calculated), which is due to a large proportion of branches with low bootstrap support per gene tree (data not shown) and not due to a number of significant disagreements. Therefore, we conclude that the individual gene families collectively do contain a signal reflected in the plurality topology.
False Positives/Biases/Potential Artifacts
A more difficult question to address is the reliability of the individual gene tree reconstructions, which form the basis of embedded quartet analyses. In our previous analyses (Zhaxybayeva et al. 2006), we performed sequence simulations to derive an empirical cutoff for overall quartet resolution in order to minimize the amount of false positives. Notably, that resulted in an increase of the number of false negatives (i.e., legitimate HGT cases that were thrown away). In this manuscript, we utilized the same approach: gene families that poorly resolve relationship of a quartet were excluded from further analyses.
Recently, a systematic error termed AGT was described (Degnan and Rosenberg 2006). Briefly, if the true organismal tree is of a certain topological conformation (an asymmetric tree) and contains short branches, there is a chance that most frequently observed topologies (of gene trees) differ from the true (organismal) tree. Degnan and Rosenberg (2006) provided a critical branch length below which gene trees are susceptible to the AGT problem. We calculated an approximate critical branch value for Prochlorococcus (see Materials and Methods) and evaluated if gene trees used in our analyses contained branches below the critical value. Indeed, 54 gene families without in-paralogs and 12 gene families with in-paralogs contained at least one such branch. However, in the performed quartet decomposition analyses we had already screened out short branches from the trees, and all branches below critical length were already excluded from the analyses. Thus, the AGT artifact should not have contributed to inferred plurality signal.
GC bias has been shown to carry over to amino acid composition of encoded proteins, producing amino acid bias (due to skewed codon usage, which was demonstrated for Prochlorococcus spp.; Dufresne et al. 2005). The recovered plurality signal (fig. 1A) supports division of the tree into two groups: lower versus higher GC content genomes, hinting at possible hidden GC bias artifact in phylogenetic reconstruction of individual gene families. Notably, the 16S rRNA gene trees also group organisms with lower GC content together (high-light adapted ecotypes). The 16S–23S rRNA ITS region was noted to have skewed GC content as well (Rocap et al. 2002). To test for this potential artifact, we used an alternative measure of phylogenetic distance: the number of rearrangements required to convert gene order in one genome into the order of another (Tesler 2002). Although dot plots for many genome pairs suggested lack of overall synteny (data not shown), the localized synteny was retained (at least 700 genes were alignable for a genome pair within Prochlorococcus and marine Synechococcus group), allowing us to obtain an estimate of the number of required pairwise genome rearrangements (ranging from 14 to 276 rearrangements within the Prochlorococcus and marine Synechococcus group). The resulting tree topology (supplementary fig. 7, Supplementary Material online) is similar to the one shown in figure 1A, in terms of supporting the bipartition that divides the genomes by GC content. However, we noted that the rearrangement tree topology also supports a bipartition dividing the tree into small versus large genomes (INV distance measure is sensitive to the genomic size). Because we also observe a strong correlation between GC content and genome size (supplementary fig. 8, Supplementary Material online), these two measures do not appear independent. Kettler et al. (2007) made a tree topology based on gene content. Although they also did not see differences between gene-based and gain/loss-based topologies, the distance calculated from presence/absence of genes would also be sensitive to genome size, falling into the same category as rearrangement tree we reconstructed. However, in a complementary exploration of GC bias, we investigated if gene families, divided into those with the greatest or the least range of GC content within the family, show equal support for GC bipartition. Results (fig. 5) reveal only very weak correlation (r2 = 0.1232) between GC bias within each gene family and its support of GC bipartition, suggesting that GC bias is not an artifact driving the observed GC-based bipartition.
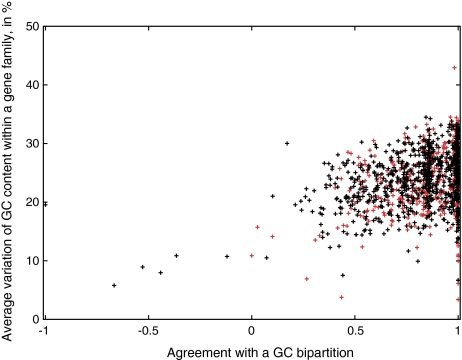
FIG. 5.—
Correlation between GC content observed within each gene family and their agreement with GC bipartition. The genomes were divided into two groups of higher and lower GC genomes (see table 1). A total of 1,271 gene families without paralogs contained at least two genomes of lower and higher GC content and were used in the analyses. GC variation within each family was calculated as average GC variation between each pair of lower–higher GC genes. Agreement with GC bipartition was calculated as described in Materials and Methods. Red crosses refer to all gene families, whereas black crosses indicate gene families that conflict plurality topology. Under the assumption that the observed plurality topology relationships are influenced by GC bias, one would expect a strong correlation. In contrast, the observed trend line of y = 8.1929x + 17.065 is only weakly supported by data (r2 = 0.1232).
Phylogenetic Signal from 482 Gene Families with In-paralogs
So far, only indisputably orthologous families were analyzed, that is, the families did not have any additional homologs (either in-paralogs or xenologs) intermingled within the gene families. However, families with in-paralogs might contain both support for the plurality tree and conflicts with it. Usually these families are excluded from genome-wide analyses that assess HGT due to uncertainty of paralogy versus xenology. Quartet decomposition, on the other hand, makes it easy to compare all possible “alternative” (i.e., containing one or the other in-paralog) embedded quartets with those of plurality tree. Due to the relatively recent divergence of genomes in this group, most gene duplications should result in in-paralogs grouping together and therefore should not produce conflicts with embedded quartets of plurality signal. Quartet decomposition analyses shown in supplementary figure 9 (Supplementary Material online) reveal that 419 (87%) gene families conflict with the plurality topology at 80% bootstrap cutoff, which suggests that most of additional homologs are not the result of recent within-lineage duplications (e.g., see supplementary fig. 10, Supplementary Material online). However, whether the conflicts are due to more ancient paralogy or HGT remains unsolved by these analyses.
Support for Ecotypes/Geography and Other Potential Groupings
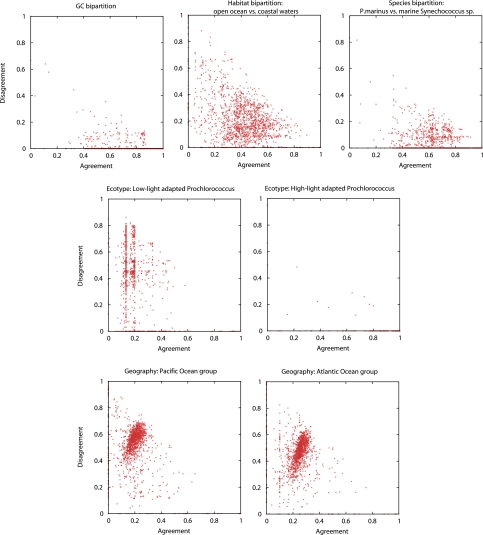
Quartet decomposition allows partitioning of the data according to some particular scenario and retrieval of gene families that support or conflict it. We examined several such scenarios. To evaluate support of a scenario, we introduce scatter plots (fig. 6), in which the gene families (represented by individual data points on the plot) strongly supporting the tested scenarios are situated close to the x axis and away from the origin, strongly conflicting gene families are found near the y axis and away from the origin, and gene families near the origin are unresolved with respect to the tested scenario. The expected distribution of gene families on the x-y plane in a random division of genomes into two groups is shown in supplementary figure 11 (Supplementary Material online).
FIG. 6.—
Scatter plots of agreement of individual gene families with selected data partitions (see graph titles). Each gene family is represented by a red dot. The position of the dot within an xy coordinate system depends on how many embedded quartets within gene family agree with the data partition (x value) and how many disagree (y value). Gene families with poor phylogenetic signal will be close to (0,0). The scatter plots observed for random data partitions are shown in supplementary figure 11 (Supplementary Material online).
In the first scenario, we divided the genomes into three groups, based on “ecotype”: high-light adapted Prochlorococcus versus low-light adapted Prochlorococcus versus Synechococcus sp. (fig. 6). This could help to delineate genes that are ecologically relevant. High-light adapted Prochlorococcus (a grouping supported by the plurality topology) was overwhelmingly supported by the majority of the gene families (1,128). However, 16 gene families (among which are three core gene families) showed disagreement with score of 0.7 or above (this score cutoff was used throughout the analyses presented in this section; see supplementary table 4, Supplementary Material online). For example, a gene from the transcription and translation (J) functional category, 16S rRNA pseudouridylate synthase (fig. 7), supports two low-light strains (NATL1A and NATL2A) grouping within the high-light adapted clade; a hydrolase belonging to the metallo beta lactamase superfamily (supplementary fig. 12, Supplementary Material online) and an aromatic ring hydrolase (supplementary fig. 13, Supplementary Material online) involve the same two low-light strains (NATL1A and NATL2A) grouping within the clade of high-light adapted Prochlorococcus. Almost all (987) gene families disagreed with low-light adapted Prochlorococcus as a group. However, a handful of genes widely represented in 18 analyzed genomes (including three core families) strongly supported the grouping (supplementary table 5, Supplementary Material online). Among the latter gene families are adenosine triphosphate (ATP) synthase delta subunit (fig. 8) and ferredoxin (supplementary fig. 14, Supplementary Material online). Support for two Synechococcus spp. grouping together and separate from one low-light and one high-light Prochlorococcus was found in 366 families but strongly conflicted by 451 gene families, among which are 19 ribosomal proteins (included in 64 gene families involved in transcription and translation; supplementary table 6, Supplementary Material online). Many of the latter conflicts are due to the two largest P. marinus genomes (strains MIT 9313 and MIT 9303) grouping within Synechococcus spp. (for robust examples, see supplementary figs. 15 and 16, Supplementary Material online). The latter relationship is also visible on the tree based on number of rearrangements (supplementary fig. 7, Supplementary Material online). Conflicts observed for the 19 ribosomal proteins do not all correspond to the same evolutionary scenario, and the locations of these genes in the genomes are neither all adjacent nor fully preserved in Prochlorococcus/marine Synechococcus group, possibly due to many rearrangements (Dufresne et al. 2008). Therefore, many of the conflicts within ribosomal proteins probably represent independent transfer events.
FIG. 7.—
Phylogeny of 16S rRNA pseudouridylate synthase. In this example, two low-light adapted Prochlorococcus strains group within the clade of high-light adapted strains with 100% bootstrap support. Synechococcus sp. PCC7002 is used as an outgroup. The tree was reconstructed in PhyML under JTT + G model with 100 bootstrap replicates. Bootstrap values below 70% are not shown.
FIG. 8.—
Phylogeny of delta subunit of ATP synthase. In this example, low-light adapted Prochlorococcus strains form a monophyletic group. This would be an ideal scenario for an ecotype model: low-light adapted Prochlorococcus, high-light adapted Prochlorococcus, and marine Synechococcus form separate clades. There is also a conflict with plurality topology within the high-light adapted Prochlorococcus and marine Synechococcus clades. The tree was reconstructed in the PhyML program under JTT + G model with 100 bootstrap replicates. Bootstrap values below 70% are not shown.
The second scenario considered a division by genome nucleotide composition: higher versus lower GC content (which also coincides with division by smaller vs. larger number of ORFs per genome; see table 1). A total of 960 gene families show strong support for division of the genomes into two groups according to the GC content (this bipartition is also embedded into plurality topology). Of those, 139 gene families are in disagreement with this bipartition.
In a data partition by named genus (Prochlorococcus vs. Synechococcus), it was no surprise to see larger number of conflicts (495 gene families), given that this data partition is in conflict with plurality signal. This demonstrates large gene flow occurring between these two genera. This division is similar to the Synechococcus spp. as a group scenario discussed above. The reason that the Prochlorococcus versus Synechococcus scenario has a larger number of conflicting families (495 vs. 451) is because this bipartition did not have the additional requirement of the two Prochlorococcus to be one low-light and one high-light adapted.
Four of 19 examined genomes were isolated from coastal waters, and hence we asked if any genes support such grouping (vs. genomes from “open ocean” habitat). Only 37 gene families supported this grouping, most of which are present only in few genomes. Hence, their signal could be due to insufficient taxonomic sampling.
In a division by geography (Atlantic vs. Pacific vs. other [Mediterranean and Arabian Seas] strain isolation locations), the scatter plots are not very different from those obtained by chance (compare to supplementary fig. 11, Supplementary Material online). Therefore, we do not find any evidence for a biogeographical pattern, as suggested in Zwirglmaier et al. (2008). However, this could be solely due to very scarce sampling of our data set. A more extensive data set will be needed to properly test such division.
Gene Families with Homologs in Sequenced Cyanophages and Their Phylogenies
Viruses have been shown to influence the evolutionary histories of some genes (photosynthesis genes in particular) in the Prochlorococcus/marine Synechococcus group (Zeidner et al. 2005; Lindell et al. 2007). Perhaps, the exchange of other genes also can be achieved through viral intermediates. Thirty-five gene families with conflict to plurality were identified to contain at least one homolog among genes in nine sequenced cyanophage genomes (see supplementary table 7, Supplementary Material online). In most cases, the phylogenetic trees had shown very poor resolution (perhaps due to within-gene recombination) or very high level of divergence between cyanobacterial and cyanophage homologs. In several selected examples (which show sufficient bootstrap support values, see supplementary figs. 17 and 18, Supplementary Material online), phage homologs clearly group within the Prochlorococcus/marine Synechococcus group, and it is possible that phage-mediated HGT plays a role in the observed branching patterns.
Among the “host” genes found in sequenced cyanophage genomes was a phosphate-inducible pstS gene (Sullivan et al. 2005). This gene was found to be present in multiple copies in many genomes of the Prochlorococcus/marine Synechococcus group (Martiny et al. 2006) and therefore is part of the gene families with in-paralogs. Reconstructed phylogenetic tree of cyanobacterial and cyanophage genes (supplementary fig. 19, Supplementary Material online) show that cyanophage homologs group within Prochlorococcus spp. and might be responsible to the observed numerous conflicts with the plurality topology.
Discussion
Does the Plurality Signal Reflect Organismal Evolution?
Because Prochlorococcus is assumed to derive from a phycobilisome-containing ancestor (Ting et al. 2002), it was puzzling (and unexpected) to see phycobilisome-containing marine Synechococcus grouping within Prochlorococcus spp. based on cumulative phylogenetic signal (as noticed earlier in Beiko et al. 2005; Zhaxybayeva et al. 2006). Analyses presented here revealed uncertainty at the node in question. The emerging most plausible explanation is that plurality of genes does not reflect the organismal evolution of these genomes but rather reflects “highways of gene sharing” (Beiko et al. 2005). In this light, for example, it makes sense that larger low-light adapted Prochlorococcus spp. genomes are placed closer to marine Synechococcus if we assume that they acquired many of their genes from outside the Prochlorococcus clade and especially from marine Synechococcus. To complicate the evolutionary histories of Prochlorococcus spp. even more, the members of the genus Prochlorococcus experience gene transfer from outside of Prochlorococcus/marine Synechococcus clade, even outside of the cyanobacteria, as can be exemplified by threonyl-tRNA synthetase, which was acquired by Prochlorococcus from gammaproteobacteria (Zhaxybayeva et al. 2006; Luque et al. 2008). Such cases of HGT will produce conflicting signals in our analyses, but we would not be able to trace their source.
Embedded Quartet Scatter Plots as a Tool for Establishing Correlations between Gene Content and Ecological Variables
We have here introduced a new method to correlate various environmental or geographical factors with phylogenetic information from the genomes. This method allows identification of genes whose evolutionary history correlates with selected factors and not necessarily with their phylogeny. For example, we found that the majority of genes support a high-light adapted Prochlorococcus spp. as a group (and light is considered one of the important factors that determine this group; Martiny et al. 2009), whereas the low-light adapted group is held together only by a handful of shared genes, and there is significant gene flow between Synechococcus and Prochlorococcus spp. The limited number of genomes available for analyses did not allow a thorough investigation of other factors that may have contributed to the evolution of these organisms. Once more genomes will become available from known, as well as new (Martiny et al. 2009) groups of Prochlorococcus and Synechococcus, the scatter plots might become a useful way to assess which parts of genomes are responsible for observed ecophysiology.
Scenarios of Prochlorococcus/Marine Synechococcus Evolution
As noted in other earlier analyses, gene gain and loss play a significant role in the evolution of these genera (Coleman et al. 2006; Kettler et al. 2007; Dufresne et al. 2008). In this manuscript, we focused on evolutionary histories of genes shared by these genera. The inferred network-like phylogenetic signal supports the following scenario of Prochlorococcus evolution: since divergence from a Synechococcus-like ancestor, a process that created the many synapomorphies that characterize the genus Prochlorococcus, low-light adapted strains of Prochlorococcus (and in particular the two largest genomes, MIT 9303 and MIT 9313) experience frequent introgression, resulting in genomes that become more “Synechococcus-like” but still maintain genes for their ecological niche (i.e., low-light open ocean environment). Most exchanges between low-light adapted strains and marine Synechococcus are not very recent because we would expect GC content distribution of all genes in these two genomes to be bimodal (more ancestral, higher GC content genes and recently acquired, lower GC content genes). However, the distribution is clearly unimodal (data not shown).
Introgressions, such as the one described above, frequently occur during speciation: they have been observed in Galapagos finches (Grant BR and Grant PR 2008) and recently have been reported for two Campylobacter species that show signs of “despeciation” (Sheppard et al. 2008). The frequent gene exchange may eventually lead to a phylogenetic signal reflecting gene sharing and not organismal histories (Gogarten et al. 2002), a process that in analogy to despeciation (Sheppard et al. 2008) could be labeled as “degeneration.”
Supplementary Material
Supplementary figures 1–19 and tables 1–7 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Funding
Canadian Institutes of Health Research (Postdoctoral Fellowship to O.Z., MOP-4467 to W.F.D.); National Science Foundation Assembling the Tree of Life (DEB 0830024 to J.P.G. and R.T.P.); National Aeronautics and Space Administration Exobiology (NNX08AQ10G and NNX07AK15G to J.P.G.); University of Connecticut Research Foundation (to R.T.P.).
Supplementary Material
Acknowledgments
We thank Dr Edward Susko for suggestions on some statistical analyses, Dr Maria Poptsova for discussions on best ways to identify gene families, and Dr Douglas Campbell for critically reading the manuscript.
References
- Ahlgren NA, Rocap G, Chisholm SW. Measurement of Prochlorococcus ecotypes using real-time polymerase chain reaction reveals different abundances of genotypes with similar light physiologies. Environ Microbiol. 2006;8:441–454. doi: 10.1111/j.1462-2920.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41:3–10. [Google Scholar]
- Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chisholm SW, et al. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature. 1988;334:340–343. [Google Scholar]
- Cole JR, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Chisholm SW. Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol. 2007;15:398–407. doi: 10.1016/j.tim.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Coleman ML, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- Creevey CJ, McInerney JO. Clann: investigating phylogenetic information through supertree analyses. Bioinformatics. 2005;21:390–392. doi: 10.1093/bioinformatics/bti020. [DOI] [PubMed] [Google Scholar]
- Dammeyer T, Bagby SC, Sullivan MB, Chisholm SW, Frankenberg-Dinkel N. Efficient phage-mediated pigment biosynthesis in oceanic cyanobacteria. Curr Biol. 2008;18:442–448. doi: 10.1016/j.cub.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006;2:e68. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, et al. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 2008;9:R90. doi: 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package) Seattle (WA): Department of Genetics, University of Washington; 1993. Distributed by the author. [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Grant BR, Grant PR. Fission and fusion of Darwin's finches populations. Philos Trans R Soc Lond B Biol Sci. 2008;363:2821–2829. doi: 10.1098/rstb.2008.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Herdman M, Castenholz RW, Waterbury JB, Rippka R. Form-genus XIII. Synechococcus. In: Garrity GM, editor. Bergey's manual of systematic bacteriology. 2nd ed. Vol. 1. New York (NY): Springer; 2001. pp. 508–512. [Google Scholar]
- Hu J, Blanchard JL. Environmental sequence data from the Sargasso Sea reveal that the characteristics of genome reduction in Prochlorococcus are not a harbinger for an escalation in genetic drift. Mol Biol Evol. 2009;26:5–13. doi: 10.1093/molbev/msn217. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kettler GC, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005a;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005b;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzniar A, van Ham RC, Pongor S, Leunissen JA. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24:539–551. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Lapointe F-J, Wilkinson M, Bryant D. Matrix representations with parsimony or with distances: two sides of the same coin? Syst Biol. 2003;52:865–868. [PubMed] [Google Scholar]
- Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- Lindell D, et al. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci USA. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell D, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–86. doi: 10.1038/nature06130. [DOI] [PubMed] [Google Scholar]
- Luque I, Riera-Alberola ML, Andujar A, Ochoa de Alda JAG. Intraphylum diversity and complex evolution of cyanobacterial aminoacyl-tRNA synthetases. Mol Biol Evol. 2008;25:2369–2389. doi: 10.1093/molbev/msn197. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Coleman ML, Chisholm SW. Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc Natl Acad Sci USA. 2006;103:12552–12557. doi: 10.1073/pnas.0601301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Tai AP, Veneziano D, Primeau F, Chisholm SW. Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol. 2009;11:823–832. doi: 10.1111/j.1462-2920.2008.01803.x. [DOI] [PubMed] [Google Scholar]
- Moore LR, Chisholm SW. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol Oceanogr. 1999;44:628–638. [Google Scholar]
- Palenik B, Haselkorn R. Multiple evolutionary origins of prochlorophytes, the chlorophyll b-containing prokaryotes. Nature. 1992;355:265–267. doi: 10.1038/355265a0. [DOI] [PubMed] [Google Scholar]
- Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penno S, Lindell D, Post AF. Diversity of Synechococcus and Prochlorococcus populations determined from DNA sequences of the N-regulatory gene ntcA. Environ Microbiol. 2006;8:1200–1211. doi: 10.1111/j.1462-2920.2006.01010.x. [DOI] [PubMed] [Google Scholar]
- Poptsova MS, Gogarten JP. BranchClust: a phylogenetic algorithm for selecting gene families. BMC Bioinformatics. 2007;8:120. doi: 10.1186/1471-2105-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan MA. Phylogenetic inference based on matrix representation of trees. Mol Phylogenet Evol. 1992;1:53–58. doi: 10.1016/1055-7903(92)90035-f. [DOI] [PubMed] [Google Scholar]
- Rocap G, Distel DL, Waterbury JB, Chisholm SW. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocap G, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Sandaa RA, Clokie M, Mann NH. Photosynthetic genes in viral populations with a large genomic size range from Norwegian coastal waters. FEMS Microbiol Ecol. 2008;63:2–11. doi: 10.1111/j.1574-6941.2007.00400.x. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Sharon I, et al. Viral photosynthetic reaction center genes and transcripts in the marine environment. ISME J. 2007;1:492–501. doi: 10.1038/ismej.2007.67. [DOI] [PubMed] [Google Scholar]
- Sheppard SK, McCarthy ND, Falush D, Maiden MC. Convergence of Campylobacter species: implications for bacterial evolution. Science. 2008;320:237–239. doi: 10.1126/science.1155532. [DOI] [PubMed] [Google Scholar]
- Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, et al. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 2006;4:e234. doi: 10.1371/journal.pbio.0040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424:1047–1051. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- Swingley WD, Blankenship RE, Raymond J. Integrating Markov clustering and molecular phylogenetics to reconstruct the cyanobacterial species tree from conserved protein families. Mol Biol Evol. 2008;25:643–654. doi: 10.1093/molbev/msn034. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesler G. GRIMM: genome rearrangements web server. Bioinformatics. 2002;18:492–493. doi: 10.1093/bioinformatics/18.3.492. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CS, Rocap G, King J, Chisholm SW. Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 2002;10:134–142. doi: 10.1016/s0966-842x(02)02319-3. [DOI] [PubMed] [Google Scholar]
- Urbach E, Robertson DL, Chisholm SW. Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature. 1992;355:267–270. doi: 10.1038/355267a0. [DOI] [PubMed] [Google Scholar]
- Waterbury JB, Watson SW, Guillard RRL, Brand LE. Widespread occurrence of a unicellular, marine, planktonic, cyanobacterium. Nature. 1979;277:293–294. [Google Scholar]
- Weigele PR, et al. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ Microbiol. 2007;9:1675–1695. doi: 10.1111/j.1462-2920.2007.01285.x. [DOI] [PubMed] [Google Scholar]
- Wong KM, Suchard MA, Huelsenbeck JP. Alignment uncertainty and genomic analysis. Science. 2008;319:473–476. doi: 10.1126/science.1151532. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Zeidner G, et al. Potential photosynthesis gene recombination between Prochlorococcus and Synechococcus via viral intermediates. Environ Microbiol. 2005;7:1505–1513. doi: 10.1111/j.1462-2920.2005.00833.x. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Gogarten JP. Cladogenesis, coalescence and the evolution of the three domains of life. Trends Genet. 2004;20:182–187. doi: 10.1016/j.tig.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Gogarten JP, Charlebois RL, Doolittle WF, Papke RT. Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events. Genome Res. 2006;16:1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O, Gogarten JP, Doolittle WF. A hyperconserved protein in Prochlorococcus and marine Synechococcus. FEMS Microbiol Lett. 2007;274:30–34. doi: 10.1111/j.1574-6968.2007.00806.x. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Lapierre P, Gogarten JP. Genome mosaicism and organismal lineages. Trends Genet. 2004;20:254–260. doi: 10.1016/j.tig.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Zwirglmaier K, et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol. 2008;10:147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.