Abstract
Knocking out a gene from a genome often causes no phenotypic effect. This phenomenon has been explained in part by the existence of duplicate genes. However, it was found that in mouse knockout data duplicate genes are as essential as singleton genes. Here, we study whether it is also true for the knockout data in Arabidopsis. From the knockout data in Arabidopsis thaliana obtained in our study and in the literature, we find that duplicate genes show a significantly lower proportion of knockout effects than singleton genes. Because the persistence of duplicate genes in evolution tends to be dependent on their phenotypic effect, we compared the ages of duplicate genes whose knockout mutants showed less severe phenotypic effects with those with more severe effects. Interestingly, the latter group of genes tends to be more anciently duplicated than the former group of genes. Moreover, using multiple-gene knockout data, we find that functional compensation by duplicate genes for a more severe phenotypic effect tends to be preserved by natural selection for a longer time than that for a less severe effect. Taken together, we conclude that duplicate genes contribute to genetic robustness mainly by preserving compensation for severe phenotypic effects in A. thaliana.
Keywords: duplicate, Arabidopsis thaliana, phenotypic effect, functional compensation, selection pressure and genetic robustness
Introduction
Knocking out a gene in an organism often causes no phenotypic effect. This phenomenon has been explained in part by the functional compensation of duplicate genes. If duplicate genes play a significant role in functional compensation, the proportion of duplicate genes with knockout phenotypic effects should be lower than the proportion for singleton genes. This is indeed the case in the yeast and in the nematode (Gu et al. 2003; Conant and Wagner 2004) but not in the mouse (Liang and Li 2007; Liao and Zhang 2007). Although recent reports pointed out that the mouse knockout data were biased in both duplication age and function (Su and Gu 2008; Makino et al. 2009), the relationship between phenotypic effect and gene duplication in the mouse appears to be weaker than those in the yeast and the nematode (Hannay et al. 2008). It was proposed that because mouse is more complex than nematode and yeast in terms of the number of different cell types, duplicate genes in mouse tend to undergo functional diversification instead of preserving functional compensation (Zhang 2003; Liao and Zhang 2007). To test this view, we examined the functional compensation by duplicate genes in Arabidopsis thaliana, using single-gene knockout data.
Another question is whether the functional compensation by duplicate genes persists for a long time. Because the loss of a redundant duplicate may cause no serious deleterious effect, a redundant copy may become lost quickly. However, many duplicate genes have been found to have preserved functional redundancy for a long period of evolutionary time (Woollard 2005; Briggs et al. 2006; Tischler et al. 2006; DeLuna et al. 2008; Musso et al. 2008; Roux and Robinson-Rechavi 2008; Vavouri et al. 2008; Kafri et al. 2009). Also, theoretical studies suggest that functional compensation by duplicate genes can be preserved in evolution if it works for unexpected errors of gene function (e.g., null mutations; Nowak et al. 1997; Wagner 2000). Nevertheless, the evolutionary persistence of functional compensation has not been tested in detail in terms of natural selection. Therefore, we examined turnover rates and selection pressures on functional compensation for various phenotypic effects, using single-gene and multiple-gene knockout data.
Materials and Methods
Phenotypic Effects of Single-Gene Knockout
We obtained 5041 insertional mutant lines from our mutant sources (Kuromori et al. 2006) and from the Arabidopsis Biological Resource Center (Columbus, OH). In each insertional mutant line, the tag was inserted into the coding region of a TAIR7 gene. In many cases, a gene had multiple insertional mutant lines, but the phenotypic effect was the same for all lines. Thus, from the 5041 insertional mutant lines, we found 3871 genes that had an insertion(s) of Ds transposon or T-DNA. Among these 3871 genes with insertion, only 253 showed visible phenotypic changes. In addition, we found 1489 genes that had been described in the literature as inducing phenotypic change. In summary, among the 5360 genes included in this study, 1742 showed induced phenotypic changes, whereas 3618 did not (Table S1, Supplementary Material online).
Identification of Singletons, Duplicates, and Closest Paralog Genes
The amino acid sequences of A. thaliana (TAIR7) and mouse genes (NCBIM37.55) were obtained from TAIR (www.arabidopsis.org) and Ensembl (www.ensembl.org). The longest amino acid sequence in a gene locus was used as the representative protein sequence. Similarity searches of all against all genes were conducted using BlastP (Altschul et al. 1997). A singleton protein was defined as a protein that did not match any other protein in the Blast search with the E value ≤1. To find the closest paralog of a protein, we aligned the protein and all proteins obtained in the Blast search by ClustalW (Thompson et al. 1994) and estimated the amino acid similarity between sequences. The protein with the highest similarity to the one under study was defined as the closest paralog. Following the definition of Gu et al. (2003), a gene is defined as a duplicate if it showed ≥30% similarity and ≥50% alignable region at the protein level with the closest paralog. The synonymous divergence (Ks) and the nonsynonymous divergence (Ka) between the target gene and the closest paralog were estimated by the modified Nei–Gojobori method (Zhang et al. 1998).
Defining Gene Families and Tandem Duplicate Clusters
To define gene families in A. thaliana, all-against-all similarity searches of protein sequences were conducted using BlastP with an E value cutoff of 10−5. Based on the E values, we generated similarity clusters representing gene families with the Markov clustering program (van Dongen 2000). As reported earlier (Hanada et al. 2008), tandem duplicates were defined as genes in any gene pair, T1 and T2, that 1) belong to the same gene family, 2) are located within 100 kb of each other, and 3) are separated by at most ten nonhomologous (not in the same gene family as T1 and T2) genes.
Results and Discussion
Functional Compensation by Duplicate Genes
We examined the phenotypic changes in each of the 3871 knockout mutants and found phenotypic changes in only 253 genes. Furthermore, from the literature we found 1489 genes whose knockout mutants showed phenotypic changes. We then computed the ratio of duplicate and singleton genes with and without phenotypic changes (table 1). Genes with phenotypic changes have a significantly lower proportion of duplicate genes and a higher proportion of singleton genes than genes without phenotypic changes (P = 2.0 × 10−11, χ2 test), indicating that the functional compensation is significantly more common in duplicate genes than in singleton genes in the Arabidopsis genome.
Table 1.
Numbers of Duplicates and Singletons with or without Knockout Effects on Phenotypes
| Phenotypic changes | Number of duplicates (D) | Number of singletons (S) | D/S ratio | P value (Yes vs. No) |
| Yes (either our study or literature) | 1,168 | 98 | 12.0 | 2.0 × 10−11 |
| Yes (our study) | 166 | 19 | 8.7 | 1.6 × 10−5 |
| Yes (literature) | 1,002 | 79 | 12.7 | 4.4 × 10−8 |
| No (our study) | 2,903 | 122 | 23.8 |
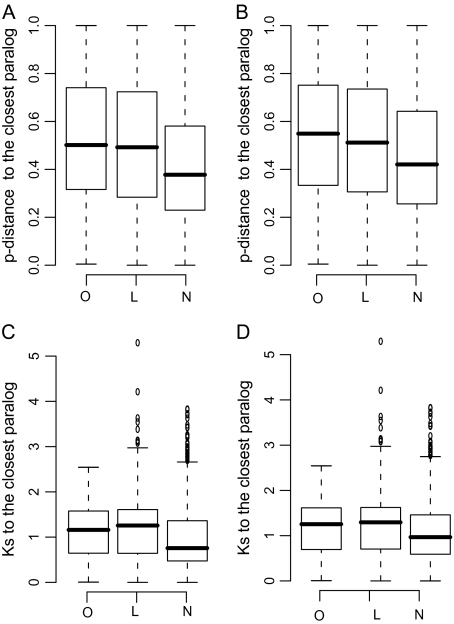
Because the age (sequence divergence) of duplicate genes may influence the chance of functional compensation, we computed the p-distance (proportion of amino acid differences) and the synonymous distance (Ks) between a knockout gene and its closest paralog. Because tandem duplicates may have a higher chance of gene conversion than nontandem duplicates (Gao and Innan 2004), we also did the same analyses using the data set without tandem duplicates. In both data sets, genes without phenotypic changes have, on average, significantly lower p-distance and Ks to the closest paralog than genes with phenotypic change (P < 0.01 in p-distance and P < 0.01 in Ks; fig. 1 and Table S2, Supplementary Material online). These results indicate a higher probability of functional compensation for recently duplicated genes than for anciently duplicated genes. Note that the conclusions drawn from our own data are essentially the same as those drawn from the data collected from the literature (table 1 and fig. 1). Therefore, the pooled data were used in the following analyses.
FIG. 1.—
Sequence divergence in genes with and without phenotypic effects in single-gene knockout. Sequence divergences are in terms of the p-distance (proportion of amino acid differences) and the synonymous distance (Ks) between a knockout gene and the closest paralog. The three data sets (O, L, and N) used are 1) with phenotypic changes observed in our study (O), 2) with phenotypic changes reported in the literatures (L), and 3) no phenotypic changes observed in our study (N). A. The p-distance in the entire data. B. The p-distance in the data without tandem duplicates. C. The Ks in the entire data. D. The Ks in the data without tandem duplicates. The distributions of p-distance and Ks values are shown as box plots with the thick solid horizontal line indicating the median value, the box representing the interquartile range (25–75%), and the dotted lines indicating the first to the 99th percentile. The p-distance and Ks are compared between genes with and without phenotypic effects. Genes with phenotypic effects have a significantly higher p-distance and Ks to the closest paralog than genes without any phenotypic effects (P < 0.01 in p-distance, P < 0.01 in Ks). See Table S2 (Supplementary Material online) for P values.
Persistence of a Phenotype in a Gene Family
Although functional compensation by duplicate genes tends to decrease with the age of duplicate genes, some proteins tend to keep functional redundancy for a long period (Kafri et al. 2008, 2009). It is possible that the rate of decrease may be different for different phenotypic effects. Note, however, that if a gene originally had multiple phenotypic effects, the rate of decrease for functional compensation to a particular phenotype may not be correctly inferred by functional compensation for the phenotype due to the deletion of an extant gene. To address whether a gene has been persistently related to a particular kind of phenotype in evolution or not, we examined the relationship between phenotypic effects and families of genes with phenotypic changes.
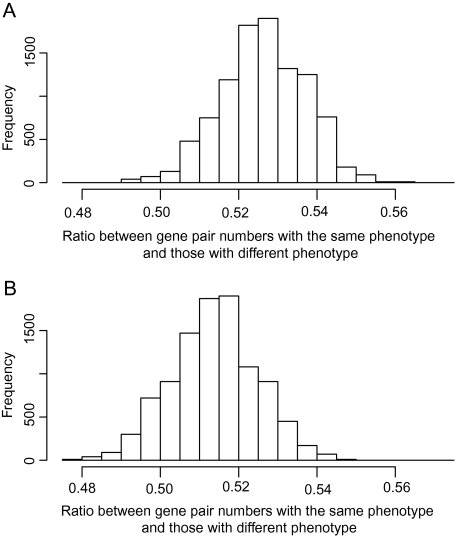
We inferred Arabidopsis gene families by the Markov Clustering algorithm (http://micans.org/mcl/). On the basis of the work of Meinke et al. (2003), we classified phenotypic changes into seed, reproductive, vegetative, and conditional phenotypes (Table S1, Supplementary Material online). Briefly, seed, reproductive, and vegetative phenotypes show abnormally visible changes with developmental stage, whereas the conditional phenotype shows abnormally visible changes only in response to either a biotic or an abiotic treatment. Among the 1742 genes whose knockout mutants showed phenotypic changes, 279, 219, 882, and 362 were classified as genes whose knockout mutants showed changes as the seed, reproductive, vegetative, and conditional phenotypes, respectively. From the 1742 genes, we randomly chose 10,000 pairs of genes either within a gene family or between gene families, and examined the ratio between the number of gene pairs with the same kind of phenotypes (seed, reproductive, vegetative, or conditional) and the number of gene pairs with different kind of phenotypes. We compared the ratios within and between gene families (fig. 2) and found that genes in the same gene family tend to have higher ratios than genes from different families (P < 1 × 10−15, the t-test), indicating that a gene family tends to be persistently maintaining its phenotype.
FIG. 2.—
Relationship between gene family and phenotypic effect. Based on Arabidopsis gene families generated by the Markov Clustering algorithm, 10,000 pairs of genes are randomly chosen within a gene family (A) and between gene families (B). The ratio between the number of gene pairs with the same kind of phenotypes (seed, reproductive, vegetative, or conditional) and the number of gene pairs with different kind of phenotypes is significantly higher between genes in the same gene family than between genes in different gene families (P < 1 × 10−15).
Differential Persistence in Functional Compensation among Phenotypes
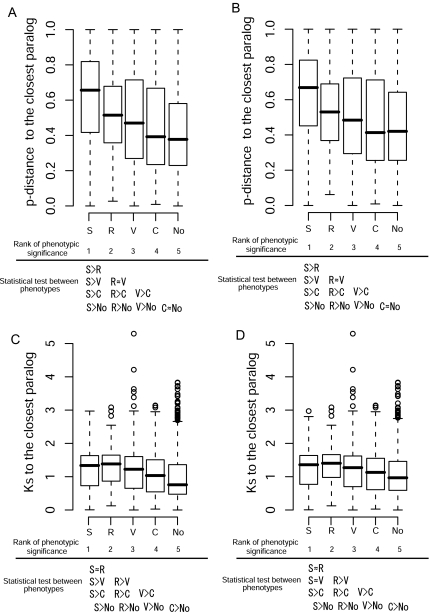
To examine the evolutionary persistence of functional compensation for different phenotypes, we compared the ages of duplicate genes among the four phenotypes in either the entire data set or the data set excluding tandem duplicates (fig. 3). In both data sets, genes whose knockout mutants showed changes in seed, reproductive, vegetative, conditional, and no-phenotypes are ranked in that order of oldest to youngest duplication dates. That is, it is likely that seed, reproductive, vegetative, conditional, and no-phenotypes are ranked in the order of longest to the shortest persistence of functional compensation.
FIG. 3.—
Relationship between sequence divergence and phenotypic effect. A. Relationship between the protein distance (p-distance) between a knockout gene and its closest paralog and phenotypic effect when the entire data were used. B. Relationship between the protein distance (p-distance) between a knockout gene and its closest paralog and phenotypic effect when the data without tandem duplicates were used. C. Relationship between the synonymous distance (Ks) between a knockout gene and its closest paralog and phenotypic effect when the entire data were used. D. Relationship between the synonymous distance (Ks) between a knockout gene and its closest paralog and phenotypic effect when the data without tandem duplicates were used. Phenotypic changes are classified into seed (S), reproductive (R), vegetative (V), conditional (C), or no (No) phenotypes. The distributions of p-distance and Ks values are shown as box plots with the thick solid horizontal line indicating the median value, the box representing the interquartile range (25–75%), and the dotted lines indicating the first to the 99th percentile. It appears that the order of phenotypic effect from the highest to the lowest significance is seed > reproduction > vegetative > conditional > no effect; see text for the ranking. Below each figure, significant differences are shown for each pair of phenotypes by the Wilcoxon test. “X > Y” means that X is significantly greater than Y at P = 0.05 and “X ≈ Y” means that X is not different from Y at the 5% level of significance.
If natural selection affects the persistence of functional compensation, the turnover rate is likely to be slower in genes with functional compensation to more severe phenotypes than genes with functional compensation to less severe phenotypes. We assume that the different phenotypes can be ranked in the order of highest to lowest significance for plant survival. The relationship between phenotype and significance can be explained as follows. Among the four phenotypes studied, an effect on seed phenotype might be most severe because a mutant with an altered seed phenotype may have embryonic defects and may not be able to germinate. Thus, functional compensation by duplicate genes for this phenotype may persist longer than the three other phenotypes. In the case of mutants with changes in reproductive phenotype, these changes largely affect the next generation and seem to reduce survivability. Therefore, the reproductive phenotype is generally severe but less severe for survival than the seed phenotype. For the vegetative phenotype, some mutations in this phenotype do not affect survival, so that this phenotype is likely to be less severe than that of either seed or reproductive phenotype. Changes in conditional phenotype appear only under certain treatments, so the significance of this phenotype is likely to be minimal. No-change phenotype is undoubtedly the least severe for plant survival. We therefore propose the order from the highest to the lowest significance in phenotypes as seed, reproductive, vegetative, and conditional and no-change.
Under this ranking of significance, we tested differences in either p-distance or Ks distance between different pairs of phenotypes by the Wilcoxon test (fig. 3 and Table S2, Supplementary Material online). We found that genes whose knockout mutants showed more severe phenotypes tend to be more anciently duplicated, whereas those whose knockout mutants showed less severe phenotypes are more recently duplicated. Thus, this result supports our expectation that functional compensation to more severe phenotypes tends to persist for a longer time, whereas functional compensation to less severe phenotypes tends to disappear faster. Thus, this relationship between duplication age and phenotypic significance supports the hypothesis that natural selection affects the persistence of functional compensation.
Different Selection Pressures on Functional Compensation among Phenotypes
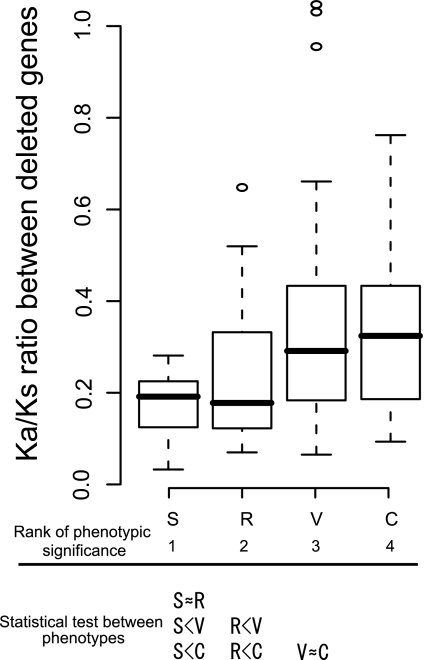
We then tried to evaluate selection pressures using duplicate genes with direct evidence of functional compensation. “Direct evidence” means the observation of phenotypic change only upon knocking out multiple duplicate genes; knocking out a single gene did not induce phenotypic change (Table S1, Supplementary Material online). A total of 163 nontandem duplicate genes that were related to 75 kinds of phenotypic change were classified into 26, 89, 32, and 16 genes whose multiple-knockout mutants showed phenotypic changes in the seed, vegetative, reproductive, and conditional phenotypes, respectively.
Using the above genes, we examined whether the pressure of natural selection on functional compensation by duplicate genes depends on the phenotype involved. Because selection pressure on genes is commonly estimated by the ratio of the nonsynonymous distance (Ka) to the synonymous distance (Ks), we compared the Ka/Ks ratios of the genes whose knockout mutants showed phenotypic changes in deletion of multiple duplicate genes. Differences in Ka/Ks were tested between phenotypes by the Wilcoxon test (fig. 4 and Table S2, Supplementary Material online). Although the difference in Ka/Ks was not significant between some phenotype pairs, the Ka/Ks ratio increased with decreasing phenotypic significance as a whole, indicating different selection pressures. Indeed, duplicate genes with functional compensation for more severe phenotypes tend to be under strong purifying selection (a low Ka/Ks ratio), and those with functional compensation for less severe phenotypes are subject to relaxed purifying selection (a high Ka/Ks ratio). Because recently (low Ks and p-distance values) and anciently (high Ks and p-distance values) duplicated genes tend to have high and low Ka/Ks ratios, respectively (Lynch and Conery 2000), the differences in Ks and p-distance may mislead the relationship between selection pressures and phenotypes in the case where genes with functional compensation for more severe phenotypes have high Ks and p-distance values. However, genes with functional compensation for more severe phenotypes tend to have lower Ks and P-distance values (Fig. S1, Supplementary Material online). That is, genes with functional compensation for more severe phenotypes have lower Ks values and lower Ka/Ks ratios in comparison with genes with functional compensation for less severe phenotypes (fig. 4 and Fig. S1, Supplementary Material online). Therefore, duplication age unlikely affects our conclusion. These results indicate that there is a stronger tendency for selection pressure to preserve functional compensation to severe phenotype by duplicate genes. Thus, duplicate genes contribute to genetic robustness mainly by preserving compensation of severe phenotypic effects in the A. thaliana genome.
FIG. 4.—
Selection pressures (Ka/Ks ratio) on paralogous genes whose multiple-knockout mutants show phenotypic changes, but single-knockout mutants do not selection pressures on paralogous genes whose multiple-knockout mutants show phenotypic changes in seed (S), vegetative (V), reproductive (R), or conditional (C) phenotypes are inferred by the ratio of the nonsynonymous substitution rate (Ka) to the synonymous substitution rate (Ks). The distributions of Ka/Ks ratios are shown as box plots with the thick solid horizontal line indicating the median value, the box representing the interquartile range (25–75%), and the dotted lines indicating the first to the 99th percentile. Significant differences are shown for each pair of phenotypes by the Wilcoxon test. “X > Y” means that X is significantly greater than Y at P = 0.05 and “X ≈ Y” means that X is not different from Y at the 5% level of significance.
Conclusion
From single-gene knockout data in A. thaliana obtained in our study and from the literature, we found that duplicate genes play a significant role in functional compensation. This conclusion is similar to that in nematode and yeast but different from that in mouse (Gu et al. 2003; Conant and Wagner 2004; Liang and Li 2007; Liao and Zhang 2007). Because mouse is a more complex organism than nematode and yeast, it was proposed that mouse duplicate genes may tend to undergo functional divergence instead of preserving functional compensation (Zhang 2003; Liao and Zhang 2007). However, this view is not supported by our observation in A. thaliana. A possible reason for the difference between mouse and A. thaliana duplicate genes is differences in duplication ages because genes with functional compensation tend to be more recently duplicated (Fig. 1). To examine the duplication age between mouse and A. thaliana, we calculated the p-distance and the Ks between each duplicate gene and its closest paralog in mouse and A. thaliana (Fig. S2, Supplementary Material online). Both p-distance and Ks are, on average, significantly lower in A. thaliana than in mouse (P < 1 × 10−167 in p-distance, P < 1 × 10−120 in Ks) by the Wilcoxon test, indicating that A. thaliana genome has a higher proportion of recently duplicated genes than the mouse genome.
Another interesting finding in this study is that the evolutionary persistence of functional compensation by duplicate genes depends on the phenotype involved. That is, functional compensation by duplicate genes tends to persist for a longer time for a more severe phenotype than a less severe phenotype. We therefore propose that natural selection retains functional compensation to increase genetic robustness, especially by preserving the functional compensation to more significant phenotype. Evidence of functional compensation by duplicate genes was provided by the observation of phenotypic change only upon knocking out multiple duplicate genes. Moreover, we found that the duplicate genes with functional compensation of more severe phenotypes tend to be under strong purifying selection (a low Ka/Ks ratio), and those with functional compensation of less severe phenotypes are subject to relaxed selection (a high Ka/Ks ratio). In conclusion, one reason why many duplicated genes are retained in the Arabidopsis genome is because some or many of them contribute to genetic robustness and are retained by natural selection.
Supplementary Material
Supplementary materials Table S1, Table S2, Figure S1 and Figure S2 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Supplementary Material
Acknowledgments
We thank Kenneth H Wolfe, Takashi Makino, and Nishiyama Tomoaki for discussions. We also thank TAIR (http://www.arabidopsis.org/) for giving us gene sequence data. This work was supported by the RIKEN Plant Science Center and by National Institutes of Health grant (GM30998 to W.H.L.).
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS. Unequal genetic redundancies in Arabidopsis—a neglected phenomenon? Trends Plant Sci. 2006;11:492–498. doi: 10.1016/j.tplants.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wagner A. Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc Biol Sci. 2004;271:89–96. doi: 10.1098/rspb.2003.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuna A, et al. Exposing the fitness contribution of duplicated genes. Nat Genet. 2008;40:676–681. doi: 10.1038/ng.123. [DOI] [PubMed] [Google Scholar]
- Gao LZ, Innan H. Very low gene duplication rate in the yeast genome. Science. 2004;306:1367–1370. doi: 10.1126/science.1102033. [DOI] [PubMed] [Google Scholar]
- Gu Z, et al. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008;148:993–1003. doi: 10.1104/pp.108.122457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannay K, Marcotte EM, Vogel C. Buffering by gene duplicates: an analysis of molecular correlates and evolutionary conservation. BMC Genomics. 2008;9:609. doi: 10.1186/1471-2164-9-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Dahan O, Levy J, Pilpel Y. Preferential protection of protein interaction network hubs in yeast: evolved functionality of genetic redundancy. Proc Natl Acad Sci USA. 2008;105:1243–1248. doi: 10.1073/pnas.0711043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Springer M, Pilpel Y. Genetic redundancy: new tricks for old genes. Cell. 2009;136:389–392. doi: 10.1016/j.cell.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Kuromori T, et al. A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 2006;47:640–651. doi: 10.1111/j.1365-313X.2006.02808.x. [DOI] [PubMed] [Google Scholar]
- Liang H, Li WH. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet. 2007;23:375–378. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23:378–381. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Makino T, Hokamp K, McLysaght A. The complex relationship of gene duplication and essentiality. Trends Genet. 2009;25:152–155. doi: 10.1016/j.tig.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Meinke DW, et al. A sequence-based map of Arabidopsis genes with mutant phenotypes. Plant Physiol. 2003;131:409–418. doi: 10.1104/pp.014134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso G, et al. The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res. 2008;18:1092–1099. doi: 10.1101/gr.076174.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- Roux J, Robinson-Rechavi M. Developmental constraints on vertebrate genome evolution. PLoS Genet. 2008;4:e1000311. doi: 10.1371/journal.pgen.1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Gu X. Predicting the proportion of essential genes in mouse duplicates based on biased mouse knockout genes. J Mol Evol. 2008;67:705–709. doi: 10.1007/s00239-008-9170-9. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler J, Lehner B, Chen N, Fraser AG. Combinatorial RNA interference in Caenorhabditis elegans reveals that redundancy between gene duplicates can be maintained for more than 80 million years of evolution. Genome Biol. 2006;7:R69. doi: 10.1186/gb-2006-7-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen S. Graph clustering by flow simulation [PhD thesis] [Utrecht (The Netherlands)]: University of Utrecht. 2000 [Google Scholar]
- Vavouri T, Semple JI, Lehner B. Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet. 2008;24:485–488. doi: 10.1016/j.tig.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Wagner A. The role of population size, pleiotropy and fitness effects of mutations in the evolution of overlapping gene functions. Genetics. 2000;154:1389–1401. doi: 10.1093/genetics/154.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard A. The C. elegans Research Community, WormBook. 2005. Gene duplications and genetic redundancy in C. elegans. WormBook, ed; pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.