Abstract
MicroRNAs (miRNAs) are ∼21 nt non-coding RNAs which regulate post-transcriptional gene expression. miRNAs are key regulators of nearly all essential biological processes. Aiming at understanding miRNA’s functions in Euphorbiaceae, a large flowering plant family, we performed a genome-scale systematic study of miRNAs in Euphorbiaceae, by combining computational prediction and experimental analysis to overcome the difficulty of lack of genomes for most Euphorbiaceous species. Specifically, we predicted 85 conserved miRNAs in 23 families in the Castor bean (Ricinus communis), and experimentally verified and characterized 58 (68.2%) of the 85 miRNAs in at least one of four Euphorbiaceous species, the Castor bean, the Cassava (Manihot esculenta), the Rubber tree (Hevea brasiliensis) and the Jatropha (Jatropha curcas) during normal seedling development. To elucidate their function in stress response, we verified and profiled 48 (56.5%) of the 85 miRNAs under cold and drought stresses as well as during the processes of stress recovery. The results revealed some species- and condition-specific miRNA expression patterns. Finally, we predicted 258 miRNA:target partners, and identified the cleavage sites of six out of ten miRNA targets by a modified 5′ RACE. This study produced the first collection of miRNAs and their targets in Euphorbiaceae. Our results revealed wide conservation of many miRNAs and diverse functions in Euphorbiaceous plants during seedling growth and in response to abiotic stresses.
INTRODUCTION
Euphorbiaceae is one of the largest plant families, which consists of more than 7000 species, including monoecious herbs, perennial shrubs and trees. Euphorbiaceus plants are characterized with high photosynthesis and high biomass. They are evolutionally diversified, carry distinct physiologies, and have complex traits adapting to dynamic environmental conditions. There are many agri-economically important species in Euphorbiaceae, such as the Castor bean (Ricinus communis), the Cassava (Manihot esculenta), the Rubber tree (Hevea brasiliensis) and the Jatropha (Jatropha curcas). Castor oil is an important raw material for many industrial products, such as lubricants and paints. The Cassava is the sixth major staple crops in the world and its starchy-enriched root is ideal for bio-ethanol production. The Rubber tree is the most important resource of natural rubber for tires and other products. The Jatropha seeds have high oil content and can be easily processed into bio-diesel.
MicroRNAs (miRNAs) are ∼21 nt non-coding regulatory RNAs which have recently emerged as a major player of post-transcriptional gene regulation. miRNAs are known to mediate a wide variety of biological processes, such as development, organ identity, metabolism and stress response (1–4). In plants, most miRNAs cleave their mRNA targets by perfectly or nearly perfectly matching to the coding regions of the targets (5–7). As in animals, miRNA-mediated translational inhibition has also been recognized in plants (8,9). A large number of miRNAs have so far been identified in various animal and plant species. There are 9539 miRNA entries, distributed in 103 species, in the miRNA Registry Database (miRBase version 13.0, http://microrna.sanger.ac.uk/). However, none of these miRNAs was from Euphorbiaceae, reflecting a disparity between the important values of this plant family and insufficient molecular and genetic studies, including small RNA mediated gene regulation, in Euphorbiaceae.
A substantial number of miRNAs are conserved in different plant species (10). For example, 481 miRNAs in 37 miRNA families have been identified in 79 plant species, and many of them are conserved in lineages from mosses and gymnosperms to monocotyledons and eudicotyledons (11). Moreover, 682 miRNAs have been identified in 155 plant species and 15 miRNA families have been found to be conserved in 11 plant species by searching public databases (12,13). An in-silico search of homologues in public databases can greatly help identify orthologues and paralogues in plants, particularly those in unsequenced species. Such homologous miRNA families typically have conserved and essential regulatory functions across many plants (14).
Individual members of a miRNA family, although they have nearly identical sequences and often share common targets, may have different spatial and temporal expression patterns in response to endogenous cues and environmental stimuli. For example, it has been reported that the expressions of ∼100 miRNAs, which are conserved in fish, chicken, and mouse, were not strictly conserved over time and location (15). It has also been discovered that many miRNAs in different plants, evolved from a common ancestor, can diverge in sequence and expression across time and space (16).
Many plants have evolved to adapt to a harsh environment. Several recent studies have shown that miRNAs in plants play important roles in responding to adverse abiotic stresses. For example, miR395 was shown to be activated to cope with sulfur deficiency (17), miR399 was under close regulation to maintain a balance of phosphorus status in vivo (18), and miR398 was induced to enhance adaptation to oxidative stress (8). However, little is known about the miRNA functions in the remarkable stress adaptation and defense systems in Euphorbiaceae. Study of stress-responsive miRNAs will help increase our understanding of the miRNA function of stress tolerance in Euphorbiaceaes and their specificity in individual Euphorbiaceous plants.
Identification of miRNAs and their targets is challenging and calls for a combination of computational analysis and experimental verification. Several criteria for miRNA identification have been developed, and key criteria include precise excision from stems of hairpin precursors, conservation in evolutionally related species, and Dicer dependence (19). Note that it is more difficult to predict miRNAs in plants than in animals, because plant miRNA precursors are typically longer and have lower conservation, and their secondary structures are often more variable than that in animals. The identification of miRNA targets is essential for understanding miRNA functions, which was often done computationally. The main criterion for target prediction is the sequence complementarity between a miRNA and its targets. Even though computational target prediction has been successful, most de novo computational methods have high false positive rates (20). Therefore, experimental validation must be performed. The most effective method for validating miRNA targets in plants is the modified 5′ RACE to detect miRNA cleavage sites, since the main mode of action of plant miRNAs is target cleavage (21).
Aiming at gaining insight into miRNAs and their important regulatory functions in Euphorbiaceae, we studied miRNAs and their targets in four agri-economically valuable Euphorbiaceous species, the Castor bean (two cultivars), the Cassava (two cultivars), the Rubber tree and the Jatropha, by combining computational prediction and experimental validation and analysis.
MATERIALS AND METHODS
Computational prediction of conserved miRNAs
Mature plant miRNA sequences from miRBase (http://microrna.sanger.ac.uk/sequences/, release 10.1) were aligned to the Castor bean genome (http://Castorbean.tigr.org). We retrieved the flanking genomic sequences around completely matched loci, with different upstream and downstream lengths, to form possible precursors of candidate miRNAs with the RNAfold program (22). We chose those sequences whose folding structures have at least 18 bp in matched regions, one central loop, and folding energy no greater than –18 kcal/mol. The free tails in the secondary structures were then removed. Next, we applied the MiRcheck program (5) to select sequences that have ≤4 mismatches, ≤2 bulged or asymmetrically unpaired nucleotides and ≤2 continuous mismatches in the seed regions. The unique sequences that satisfy these criteria were retained for further analysis.
Our method was able to find 84 of the 97 conserved miRNAs when applied to A. thaliana, with only 2 false positive predictions, indicating that it has a sensitivity of 86.6%.
Computational prediction of candidate targets of miRNAs
We used our recently developed Hitsensor algorithm (Zheng and Zhang, unpublished) to predict miRNA targets in the Castor bean, whose genome was downloaded from http://castorbean.jcvi.org/. Hitsensor searches miRNA complementary sites in coding regions with a modified Smith-Waterman algorithm (23). It scores these sites by giving rewards to key sequence-specific determinants, including seed region, 12–17 nt region, local-AU content around the seed region, and ≤3 mismatches. The parameters of Hitsensor were set such that it was able to predict nearly all experimentally validated miRNA targets with ≤3 mismatches in A. thaliana, which were retrieved from TarBase (http://diana.cslab.ece.ntua.gr/tarbase/).
Plant materials and stress conditions
Developmental seedling stage
Seedlings of two cultivars of the Castor bean (HeLa, whose genome has been sequenced, and Hainan, a variety cultivated in Southern China), two Cassava cultivars (SC124, a cold-prone areas cultivar in China, which exhibit fast leaf falling and fast recovery growth when temperatures dramatically vary, and C4, adapted to geographical high-latitude region of Argentina, can retain leaves and exhibits relatively slow recovery when the temperature drops and returns to normal), and the other two species (the Rubber tree and Jatropha), with apex included, were sampled from 10 days old buds. All plants were grown under the natural conditions (11 h light, 13 h dark and 25°C during the day and 18°C at night).
Stress condition
The above Castor bean HeLa and two Cassava cultivars were subjected to cold/drought control conditions (grown under temperature of 25°C for cold and holding 90% full field water content for drought), stress conditions (14 and 4°C for cold and ∼60–80% water content for drought treatment) and recovery treatment (returning back to 25°C after 5 days cultivation under 14°C for cold, and watered once again after 6 days of irrigation avoidance for drought). The stress experiments on miRNAs are specified in Supplementary Table S1.
Experimental profiling of miRNA expression
Small RNA extraction
Small RNAs (<200 bp) were extracted with the BioTeKe miRNA extraction kit (RP5301, BioTeKe Corporation, China). Young leaves were grinded into powder with liquid nitrogen, and lysis solution and chloroform were added to remove genomic DNA and protein. The suspension was subjected to two rounds of spin-column to remove large RNA and retain small RNA molecules successively. The captured small RNAs were washed twice for purification and dissolved with 65–70°C pre-heated elution buffer. Purified small RNAs, including miRNA, siRNA, tRNA, 5sRNA, etc., were further examined with 12% agarose gel using GoldenView dye.
Poly(A) tail tagging
The extracted small RNAs were polyadenylated by poly(A) polymerase (M0276S, Biolabs) in 20.0 µl volume as follows: 5 µl miRNA (∼2 µg), 5 U Poly(A) Polymerase, 1× buffer, 2 mM ATP and ddH20. The mixture was incubated at 37°C for 1 h. The products were purified with equal volume phenol/chloroform (1:1), and deposited with 0.5 µl of 10 mg/ml mussel glycogen, 1/10 volume 5 M acetate sodium and 4× volume alcohol.
Primer design and first-strand cDNA synthesis
The sequences of the 85 predicted miRNAs, along with their flanking sequences, were classified into 55 groups. For each group, a miRNA-specific primer was designed based on mature miRNA sequences, and extended by 1 to 4 nt on its 5′-end to ensure that the primer had the core sequence for all members of the group. If a miRNA family was divided into different groups; 1–5 nt on the 3′- or 5′-end of the primers were varied to differentiate the groups. The melting temperature (Tm) of all primers was set to be close to that of the anchor primer, which was designed according to the poly (T) adaptor. The 55 miRNA-specific primers, anchor primer and poly (T) adaptor were listed in Supplementary Table S2. Poly(A)-tail tagging and primer extension methods were used to amplify mature miRNAs (24). The amplicons included 21–25 nt miRNA specific primer, 24 continuous A nucleotides and 39 nt adaptors designed for the common reverse primer template, resulting in ∼85 bp target length.
miRNA expression analysis with end-point detection and real-time quantification
RT reactions were conducted as follows: small RNAs with poly(A)-tails were heated to 70°C for 5 min to denature RNAs, incubated under 37°C for 5 min to anneal poly (T) adaptor primers, and then cooled on ice. The remaining reagents (5× reaction buffer, dNTPs, RNase inhibitor, M-MuLV reverse transcriptase, Fermentas K1611) were added and the mixture was incubated at 37°C for 60 min. The reaction mixture was heated to 70°C for 10 min to inactivate reverse transcriptase. The first-strand cDNA products were prepared.
PCRs were performed in reaction volumes (20 µl) as follows: 1× PCR buffer, 200 µM of dNTP, 1 µM miRNA and anchor primers, 2.0 µl diluted first cDNA strand, and 2.5 U Taq DNA polymerase (TaKaRa). Amplification was performed as follows: one step at 94°C for 5 min, 34 cycles at 94°C for 30 s, 57 to 60°C for 30 s, then 72°C for 30 s, and the last step with 72°C for 7 min. A negative control (no template) was included for each primer combination. Target bands were evaluated according to the marker ladder in electrophoresis.
Real-time PCR was performed following a standard SYBR Premix Ex TaqTM kit (TaKaRa) protocol. The reactions (1.0 µl first-stand cDNA product, 1× Universal PCR Master Mix, 1.0 µM miRNA-specific and anchor primers) were incubated in 0.1 ml tubes of Rotor-gene 6000 machine as follows: 95°C for 15 s, then 8 cycles of the touchdown program decreased by 0.5°C each anneal step (starting with 60°C), then followed by 40 cycles of the normal program (95°C for 5 s, 58°C for 15 s and 72°C for 20 s). The procedure ended by a melt-curve ramping from 60 to 95°C, raised by 0.5°C each step.
The target gene and reference gene U6 for each sample were amplified in parallel for 3 replicates. The values of the threshold cycle (CT) were calculated using Rotor-Gene 6000 series software 1.7 (Corbett Robotics, Australia). CT values were converted to relative expression by the △△CT method with the following formula: The relative concentration = 2–△△CT, where △△CT = (△CTsample – △CTcontrol), △CT = CT(miRNA)-CT (U6) in each sample. If the CT value was greater than that of one with no template control (NTC), the miRNA was considered to be not expressed.
Experimental miRNA target validation
RNA ligase-mediated rapid amplification of the 5′cDNA ends (RLM-RACE) GeneRacer Kit (Invitrogen, USA) was used to validate the predicted target, omitting the 5′ phosphates of the truncated mRNA removal and the 5′ cap structure of full-length mRNA removal treatments. Briefly, total RNA was extracted with RNAplant regent (TIANGEN, DP407-01), and the PolyA RNA was isolated using polyAtract mRNA isolation system III (Promega, USA) to eliminate contaminated non-mRNA. Ligation with a 5′ RNA adapter and a reverse transcription were performed next. The resulting cDNA was used as a template for PCR amplification. Two ∼100bp-spaced gene-specific reverse primers (GSP1 and GSP2) were made for each Castor bean target, designed based on the downstream sequence of the miRNA:target binding site at the target. Two GeneRacer 5′ forward primers (included in GeneRacer kit) were used to specifically nest amplify the 3′ cleavage product of the target mRNA. The amplified PCR products were gel purified, cloned into pCR TOPO TA vectors (Invitrogen, USA) and sequenced (Sangon, China). The gene specific primers that we used are provided in Supplementary Table S3.
RESULTS
Candidate conserved miRNAs in four Euphorbiaceous species
Based on the conservation of miRNAs in many plant species and the currently available genome sequence of the Castor bean, we predicted 85 individual conserved miRNAs belonging to 23 families in the Castor bean, Ricinus communis (Table 1). As shown in Table 1, the most abundant are miR169 (14 loci), miR170/171 (9 loci), miR399 (9 loci) and miR156/157 (8 loci). These families also have a large number of loci in A. thaliana, O. sativa and P. trichocarpa (Table 1), indicating their abundance in plants, from monocots to eudicots. The genomic organizations of the 85 individual miRNAs are listed in Supplementary Table S4 and the hairpin structures of 85 pre-miRNAs are provided in Supplementary Figure S1.
Table 1.
The predicted miRNAs of Castor bean, Ricinus communis (Rco) and the number of individual members in a miRNA family in Arabidopsis thaliana (Ath), Oryza sativa (Osa) and Populus trichocarpa (Ptc)
| miRNA family | Rco | Atha | Osaa | Ptca |
|---|---|---|---|---|
| miR156/157 | 8 | 12 | 12 | 11 |
| miR159/319 | 5 | 6 | 8 | 15 |
| miR160 | 3 | 3 | 6 | 8 |
| miR162 | 1 | 2 | 2 | 3 |
| miR164 | 4 | 3 | 6 | 6 |
| miR166 | 5 | 7 | 7 | 7 |
| miR167 | 3 | 4 | 10 | 8 |
| miR168 | 1 | 2 | 2 | 2 |
| miR169 | 14 | 14 | 17 | 32 |
| miR170/171 | 9 | 4 | 9 | 14 |
| miR172 | 4 | 5 | 4 | 9 |
| miR390 | 2 | 2 | 1 | 4 |
| miR393 | 2 | 2 | 2 | 4 |
| miR394 | 0 | 2 | 1 | 2 |
| miR395 | 5 | 6 | 23 | 10 |
| miR396 | 3 | 2 | 5 | 7 |
| miR397 | 1 | 2 | 2 | 3 |
| miR398 | 2 | 3 | 2 | 3 |
| miR399 | 9 | 6 | 11 | 12 |
| miR403 | 2 | 1 | 0 | 3 |
| miR408 | 1 | 1 | 1 | 1 |
| miR535 | 1 | 0 | 1 | 0 |
| Total | 85 | 89 | 132 | 164 |
aThe numbers of Ath, Osa, Ptc are based on information in miRBase, release 10.1.
miRNAs expression patterns during seedling development
Members of a miRNA family, whose sequences may differ by one or several nucleotides, may possibly possess different regulatory functions. To distinguish individual members of a miRNA family, we designed member-specific primers based on their sequences if such primers can be constructed (see ‘Materials and Methods’ section). On the other hand, if a primer is shared by several members, it is unable to distinguish these members. In the rest of the paper we will mention both individual miRNAs and the number of primers used.
An overview of the experiment validation is listed in Supplementary Table S1. We adopted a two-step strategy to experimentally validate the 85 miRNAs in five Euphorbiaceae seedlings, which included the Castor bean (HeLa and Hainan), the Cassava (SC124), the Rubber tree (73397) and the Jatropha (Landrace).
End-point RT-PCR results showed that 39 primers covering 58 (∼68.2%) miRNAs were able to amplify PCR products in at least one of the five plants (Supplementary Figure S2). The remaining 16 primers, which covered 27 (∼31.8%) of the 85 tested miRNAs, did not produce expected products.
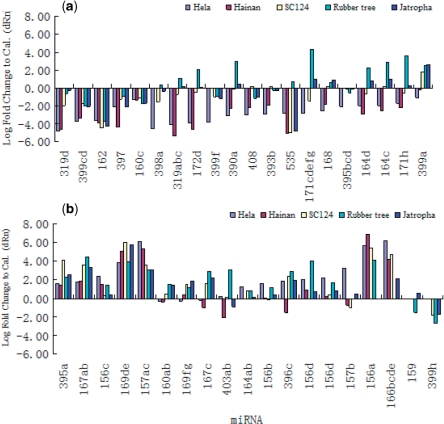
The expressions of the 58 detected miRNAs were further examined using quantitative real-time RT-PCR, in comparison with the expression of the internal control U6. The profiling results are shown in Figure 1. Fourteen of the 58 miRNAs (covered by 10 primers) shared similar expression patterns in the five plants: Eight miRNAs (miR157ac, miR169de, miR156c, miR167ab and miR395a; five primers) were expressed at a higher level and 6 miRNAs (miR160c, miR397, miR162, miR399cd and miR319d; five primers) were expressed at a lower level than U6. The expression of the remaining 44 miRNAs varied substantially in the five plants, and exhibited species-specific expression patterns. In the Castor bean, the lower expressed miRNAs out-numbered the more highly expressed ones (29 versus 14 in HeLa and 20 versus 10 in Hainan) when compared with U6. In the Cassava, about an equal number of miRNAs had lower and higher expression than U6 (23 versus 21), while these numbers were changed to 10 versus 34 in the Rubber tree and 12 versus 32 in the Jatropha. As expected, the expression patterns of these 44 miRNAs were more similar in the two Castor bean varieties than in the other three species analyzed (Figure 1).
Figure 1.
The miRNA expression levels of 58 miRNAs, with respect to their 39 corresponding primers, measured by qRT–PCR in seedlings of the four Euphorbiaceous species. The relative quantity of expression is normalized to the expression of the internal control U6 gene in units of Log Fold Change to Cal. The ‘(a)’ and ‘(b)’ refer to the fold changes of expression that are, respectively, higher and lower than the expression of the internal control U6, and the first five primers of each were uniformly higher-expressed and lower-expressed than U6 in all four species.
miRNAs responding to abiotic stresses and their expression patterns
We followed the same two-step strategy to study the 85 predicted miRNAs under drought and cold stresses in the Castor bean (HeLa) and the two Cassava varieties (SC124 and C4). Fourty eight (56.5%) miRNAs (34 primers) were able to produce distinct PCR bands in either cold or drought conditions or during stress recovery processes (Supplementary Figure S3). The fold changes of the 48 miRNAs—calculated with real-time PCR across two time points separately under the above three stress treatments—varied from –134 to 109 relative to their corresponding control (Supplementary Table S5). In the following, we describe these results in detail.
Expression patterns under cold stress
The expression of the 48 miRNAs in the Castor bean was summarized as follows: 41 (30 primers) were down-regulated and four (2 primers) were up-regulated, while three (2 primers) were not detected, compared to that in normal growth conditions (Supplementary Table S5). The miRNA expression in the Cassava were divided into two time points (after 3 and 24 h of cold treatment), and the classification of expression patterns are shown in Supplemental Table S6: At each time point, the expression of a miRNA can be up-regulated (labeled as ‘+’) or down-regulated (labeled as ‘–’), compared to its expression in the control conditions, or not detected (labeled as ‘n’). Across the two time points, there are 11 combinations of the three expression types (+, – and n). The trends of the fold changes of the 11 combinations could be classified into three main categories, i.e. up-regulation (including ‘down-up’ and ‘non-up’), down-regulation (involving ‘up-down’ and ‘up-non’) from the first time point to the next, and no detection at both time points. Following this classification, Table 2 shows the resulting expression. In SC124, 32 miRNAs belong to the down-regulation class, while only 6 miRNAs fall into the up-regulated class. Interestingly, the result on C4 is reversed: 4 miRNAs in the down-regulated category, and 31 miRNAs in the up-regulated class. The remaining 10 miRNAs (8 primers) in SC124 and 13 miRNAs (9 primers) in C4 were not detected in the two time points.
Table 2.
Expression trends of miRNAs in two Cassava varieties across two time-points under stress conditions
| Treatment | Down-regulation form time point 1 to time point 2 |
Up-regulation form time point 1 to time point 2 |
No detection in both time points | ||
|---|---|---|---|---|---|
| Up-Downa | Up-Non | Down-Up | Non-Up | Non-Non | |
| Cold | |||||
| SC124 | |||||
| No. | 22 (13)b | 10 (7) | 4 (4) | 2 (2) | 10 (8) |
| ID | miR156a, c, d, 157ac, 159, 160ab, c, 162, 166a, bcde, 171ab, 395bcde, 396c | miR167c, 319abc, 390b, 397, 398b, 399a, 403ab | miR157b, 168, 395a, 398a | miR156e, 164d | miR164ab, c, 167ab, 399e, f, h, 408, 535 |
| C4 | |||||
| No. | 2 (2) | 2 (2) | 15 (11) | 16 (10) | 13 (9) |
| ID | miR157b, 399h | miR160c, 399a | miR156a, 159, 160ab, 162, 166bcde, 167c, 168, 395a, 396c, 397, 398a | miR156e, c, d, 157ac, 164d, 166a, 171ab, 395bcde, 403ab, 408 | miR164ab, c, 167ab, 319abc, 390b, 398b, 399e, f, 535 |
| Drought | |||||
| SC124 | |||||
| No. | 3 (2) | 1(1) | 14 (9) | 20 (16) | 10 (6) |
| ID | miR160ab, 167c | miR160c | miR156a, 157ac, b, 166a, bcde, 395a, 396c, 398a, 403ab | miR156e, c, d, 159, 162, 164d, 168, 171ab, 390b, 395bcde, 397, 398b, 399e, h, 408, 535 | miR164ab, c, 167ab, 319abc, 399f |
| C4 | |||||
| No. | 1 (1) | 1 (1) | 18 (12) | 13 (8) | 15 (12) |
| ID | miR160c | miR166a | miR156a, 157ac, 159, 160ab, 162, 166bcde, 167c, 168, 396c, 397, 403ab | miR156e, c, d, 164d, 167ab, 171ab, 395a, bcde | miR164ab, c, 319abc, 390b, 398a, b, 399e, f, h, 408, 535 |
| Recovery | |||||
| SC124 | |||||
| No. | 29 (19) | 9 (6) | 1 (1) | 8 (7) | 1 (1) |
| ID | miR156a, c, d, 157ac, b, 159, 160ab, 162, 164d, 166bcde, 167c, 168, 171ab, 390b, 395a, bcde, 396c, 397, 403ab | miR160c, 164ab, 319abc, 398a, 399a, f | miR166a | miR156e, 167ab, 398b, 399e, h, 408, 535 | miR164c |
| C4 | |||||
| No. | 2 (2) | 7 (4) | 20 (14) | 5 (4) | 14 (10) |
| ID | miR156e, 395a | miR160c, 395bcde, 408, 535 | miR156a, d, 157ac, b, 159, 160ab, 162, 166a, bcde, 167c, 168, 396c, 397, 403ab | miR156c, 164d, 171ab, 390b | miR164ab, c, 167ab, 319abc, 398a, b, 399a, e, f, h |
aExpression patterns of miRNAs as specified in Supplementary Table S6.
bNumber in bracket represents the number of primers used, and the number outside of bracket refers to the corresponding miRNA loci of given primers.
The profiling results on SC124 and C4, shown in the first row of Table 3, can facilitate a direct comparison of miRNAs in these two Cassava cultivars under cold stress. Seven miRNAs (seven primers) displayed similar expression patterns across the two time points, including two down-regulated and five up-regulated. However, 26 miRNAs (16 primers) exhibited opposite expression patterns. Moreover, eight miRNAs (six primers) were not detected in either SC124 or C4 across these two time points. The expression patterns of the rest seven miRNAs (five primers) varied dramatically, indicating that although some of these miRNAs were conserved in these two Cassava varieties, the expression or even function of most miRNAs might be cultivar specific, which may have a different impact of chilling injury on these two Cassava varieties.
Table 3.
Comparison of expression patterns between two Cassava varieties under abiotic stresses
| Comparison | Similar expression pattern |
Opposite expression pattern |
No detection |
|||
|---|---|---|---|---|---|---|
| No. | miRNAs | No. | miRNAs | No. | miRNAs | |
| Cold (SC124 and C4) | 7 (7)a | miR156e, 160c, 164d, 168, 395a, 398a, 399a | 26 (16) | miR156a, c, d, 157ac, b, 159, 160ab, 162, 166a, bcde, 167c, 171ab, 395bcde, 396c, 397, 403ab | 8 (6) | miR164ab, c, 167ab, 399e, f, 535 |
| Drought (SC124 and C4) | 27 (18) | miR156a, c, d, e, 157ac, b, 159, 160c, 162, 164d, 166bcde, 168, 171ab, 395a, bcde, 396c, 397, 403ab | 4 (3) | miR160ab, 166a, 167c | 8 (5) | miR164ab, c, 319abc, 399a, f |
| Recovery (SC124 and C4) | 7 (4) | miR160c,166a,395a, bcde | 25 (18) | miR156a, c, d, e, 157ac, b, 159, 160ab, 162, 164d, 166bcde, 167c, 168, 171ab, 390b, 396c, 397, 403ab | 6 (5) | miR164c, 167ab, 398b, 399e, h |
For a specific stress, ‘Similar expression pattern’ means that the expression trend of miRNAs from time point 1 to point 2 is similar between SC124 and C4. ‘Opposite expression pattern’ refers to the expression trend of miRNAs from time point 1 to point 2 being reversed between SC124 and C4. ‘No detection’ is for no detection of expression in the two varieties.
aThe number in bracket represents the number of primers used, and the number outside of the bracket refers to the corresponding miRNA loci of given primers.
Finally, between the Castor bean and the Cassava, the 48 miRNAs behaved even more differently. Among the 48 miRNAs, 47 (33 primers) had different expression patterns, while only miR166a has a lower expression level than U6 in these two species (Supplementary Table S5).
Expression patterns under drought stress
The expression patterns of these 48 miRNAs under drought stress are also shown in Supplementary Table S5. In the Castor bean, the expression patterns under drought were similar to that under cold, i.e. 41 miRNAs (31 primers) were down-regulated, four miRNAs (1 primer) were up-regulated and three miRNAs (2 primers) were not detected.
The 48 miRNAs in the two Cassava varieties were profiled at two time points (after 3 and 6 days of drought treatment). The samples and experiment design were the same as for cold stress (Supplementary Tables S1 and S6). The results are shown in the second row of Table 2: only 4 miRNAs were down-regulated while 34 miRNAs were up-regulated in SC124. The results on C4 were similar: 2 miRNAs were down-regulated and 31 miRNAs were up-regulated. The remaining 10 miRNAs (6 primers) for SC124 and 15 miRNAs (12 primers) for C4 were not detected.
We further compared miRNA expression patterns under the drought conditions in the two Cassava varieties. As shown in Table 3, a considerable number of miRNAs (27 miRNAs, 18 primers) had similar expression patterns. A small portion (four miRNAs, three primers) exhibited opposite expression patterns. In addition, eight miRNAs were not detected at the two time points in either SC124 or C4. The expression patterns of the remaining nine miRNAs (eight primers) varied between SC124 and C4. Even though the miRNAs expression patterns under drought stress were largely different from those under cold stress, a similar conclusion can be drawn for drought, i.e. a substantial portion of the predicted miRNAs shared similar expression patterns, while several miRNAs behaved in a variety specific fashion. The results indicated that the mechanism of drought tolerance, a common property of the Cassava, might be similar in the two Cassava varieties, even though they grow different ecological conditions.
When comparing the Cassava and the Castor bean, the expressions of the 48 miRNAs were similar to those under cold stress: 44 had different expression patterns, and four miRNAs (miR395bcde, 1 primer) expressed at lower level than U6 in these two species (Supplementary Table S5).
Expression patterns during stress recovery
To gain deep insight into environmental adaptation in Euphorbiaceae, we studied miRNA expression during plant recovery from abiotic stresses at two time points (5 and 24 h after cold relief, and 3 and 6 days after drought relief). We used samples mixed with an equal amount of plant materials for cold recovery and drought recovery in each variety, as was done for the control samples. The results are summarized in last columns of Table 2. As shown, 38 miRNAs were down-regulated while only 9 miRNAs were up-regulated in SC124. However, the results were very different in C4: only 9 miRNAs were down-regulated and 25 miRNAs were up-regulated. The remaining 1 miRNA in SC124 and 14 miRNAs (10 primers) in C4 could not be detected during stress recovery.
The third row of Table 3 presents the results comparing the miRNA expressions during stress recovery in the two Cassava varieties. The number of miRNAs and their expression patterns during stress recovery were similar to those under cold stress. 25 miRNAs (18 primers) showed opposite expression patterns, while a fraction of them (seven miRNAs, four primers) showed similar expression patterns in these two varieties. However, six miRNAs (five primers) were not detected at the two time points in both SC124 and C4. The expression patterns of the rest 10 miRNAs (7 primers) varied in the two Cassava varieties. The result—a large number of miRNAs with different expression patterns—implied that the recovery mechanism might be different in SC124 and in C4.
In summary, the expression patterns of 48 miRNAs detected under the cold stress were relatively similar to those during recovery processes when comparing the two different stress treatments, and the expression trends across the two time points between SC124 and C4 were much more similar to each other in drought treatment.
Common and unique expression patterns in development and stress response
Individual members of a miRNA family or even a single miRNA may have different expression patterns under different physiological or stress conditions. Our study of the 85 candidate miRNAs offered such an opportunity to identify common and distinctive expression patterns of individual miRNAs in the Castor bean and the Cassava under development and different stress conditions. As shown in Table 4, 23 of the 85 miRNAs were expressed exclusively in normal seedling growth condition or expressed in response to abiotic stresses. In particular, six miRNAs (5 primers) were not detected in the seedling development stage, but were expressed under the cold/drought stress conditions. In contrast, 17 miRNAs (10 primers) were expressed in the normal seedling stage but not under the three stresses. Furthermore, 42 miRNAs (29 primers) were expressed during seedling development and under cold and drought stresses. The remaining 20 (23.5%) miRNAs (11 primers) were not detected in development or three stress conditions.
Table 4.
Summary of miRNAs that were expressed in development stage and/or responded to abiotic stresses
| Expression classificatory | miRNAs | Total number |
|---|---|---|
| Seedling stage-only | 156b, 169de, fg, 171cdefg, h, 172d, 319d, 390a, 393b, 399cd | 17 (10)a |
| Abiotic stress-only | 166a,171ab, 390b, 398b, 399e | 6 (5) |
| Both seedling and stress | 156a, c, d, e, 157ac, b, 159, 160ab, c, 162, 164ab, c, d, 166bcde, 167ab, c, 168, 319abc, 395a, bcde, 396c, 397, 398a, 399a, f, h, 403ab,408, 535 | 42 (29) |
| Not detected | 169abc, hij, k, lmn, 171i, 172abc, 393a, 396ab, 399b, g, i | 20 (11) |
aThe number in bracket represents the number of primers, and the number outside of bracket refers to the corresponding miRNA loci of given primers in bracket.
Targets of miRNAs
Candidate miRNA targets
We utilized the currently annotated mRNA genes in the Castor bean (from the CBGD database http://castorbean.jcvi.org) to predict miRNA targets. With our Hitsensor method (see ‘Materials and Methods’ section), we predicted 258 miRNA:target pairs between 63 miRNAs and 83 mRNAs. Some of the predicted targets are listed in Table 5 and the full list was given in Supplementary Table S7.
Table 5.
Some of predicted targets of miRNAs in Castor bean, Ricinus communis, and their conservation in Arabidopsis thaliana (Ath), Oryza sativa (Osa) and Populus trichocarpa (Ptc)
| miR family | Target family | Atha | Osaa | Ptca | Verified targets | Rcob |
|---|---|---|---|---|---|---|
| miR156 | SBP (5) | 11 | 9 | 16 | SPL2/3/4/10 (10,34,48,55) | 5 |
| miR 159/319 | MYB | 8 | 6 | 5 | MYB33, MYB65 (1,30,31) | 3 |
| TCP (1) | 5 | 4 | 7 | TCP2/3/4/10, TCP24 (1) | 0 | |
| miR160 | ARF (5) | 3 | 5 | 9 | ARF10/16/17 (33,48,53) | 4 |
| miR162 | DCL1 (34) | 1 | 1 | 1 | DCL1 (34) | 1 |
| miR164 | NAC (5) | 6 | 6 | 6 | CUC1/2, NAC1, At5g07680, At5g61430 (33,48,55,71) | 2 |
| miR166 | HD-ZIPIII (5) | 5 | 4 | 9 | PHB, PHV, REV, ATHB-8, ATHB-15 (37,45,56,72,73) | DNA-binding protein |
| miR167 | ARF (3,5) | 2 | 4 | 7 | ARF6, ARF8 (34,55) | 0 |
| miR172 | AP2 (3) | 6 | 5 | 6 | AP2, TOE1, TOE2, TOE3 (2,34,48) | 2 |
| miR393 | F-Box (4,44) | 4 | 2 | 5 | TIR1, ABF1/2/3 (44) | 2 |
| miR396 | GRF (44) | 7 | 9 | 9 | GRL1/2/3/7, GRL8, GRL9 (44) | 0 |
| miR397 | Laccase (4,44) | 3 | 15 | 26 | At2g29130, At2g38080, At5g60020 (44) | 7 |
| miR399 | E2-UBC (4) | 1 | 1 | 1 | At2g33770 (55) | 0 |
| Inorganic phosphate transporter (44) | 1 | 4 | 4 | 2 | ||
| miR408 | Laccase (39) | 3 | 2 | 3 | At2g30210 (39,63) | 1 |
aThe numbers of predicted targets in Arabidopsis thaliana (Ath), Oryza sativa (Osa) and Populus trichocarpa (Ptc) are given in review in (74).
bA target is included if it can be targeted by at least one member of a miRNA family.
Table 5 shows 29 miRNA:target pairs of 10 conserved miRNA families in the Castor bean as well as their conservation in A.thaliana, O. sativa and P. trichocarpa. Remarkably, the targets of miR396 and miR167 families were not predicted based on the current Castor bean genome annotation. In addition, miR166 was predicted to target mRNAs different from those targeted by the same miRNAs in A. thaliana, O. sativa, and P. trichocarpa. The predicted targets in the Castor bean were fewer than the counterparts in the other three species mentioned, except the targets of miR162. This might be because of the incomplete annotation of the Castor bean genome. In addition, 229 of the 258 miRNA:target pairs were specific to the Castor bean, indicating these pairs may have functions specific to the Castor bean or Euphorbiaceae.
Stress-related, diverse miRNA cleavage sites
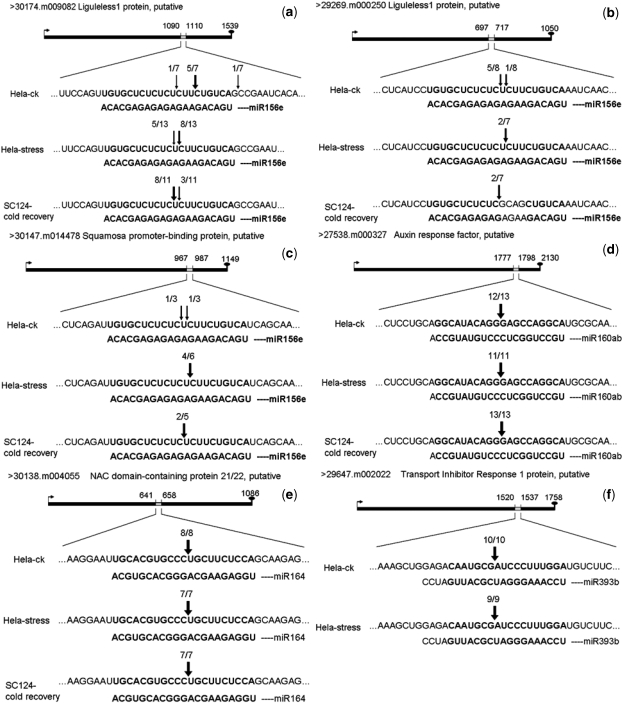
Plant miRNAs are known to regulate their targets preferentially through cleavage of their mRNA targets. To understand miRNA functions and evaluate our miRNA target prediction method, we randomly selected 10 miRNA:target pairs with high Hitsensor scores (see ‘Materials and Methods’ section), and verified them using modified 5′-RACE. Cloning and sequencing the resultant PCR amplicons can verify the target that has undergone miRNA-directed cleavage and determine the nucleotide position at which a cleavage event occurs. Three plant samples were tested for each of the 10 targets. The results are shown in Table 6 and Figure 2.
Table 6.
The results of experimental validation of miRNA–target interactions in three samples
| miRNAs | Target gene ID | Target gene function | Cleavage sitea |
||
|---|---|---|---|---|---|
| HeLa-ck | HeLa-stress (cold+drought) | SC124-cold recovery | |||
| 160ab | 27538.m000327 | Auxin response factor | 10th 12/13) | 10th (11/11) | 10th (13/13) |
| 393b | 29647.m002022 | Transport inhibitor response1 protein | 11th(10/10) | 11th(9/9) | |
| 164 | 30138.m004055 | NAC domain-containing protein 21/22 | 10th(8/8) | 10th(7/7) | 10th (7/7) |
| 156e | 30147.m014478 | Squamosa promoter -binding protein | 9th(1/3), 10th(1/3) | 9th(4/6) | 10th (2/5) |
| 156e | 30174.m009082 | LIGULELESS1 protein | −1st(1/7), 6th(5/7), 9th(1/7) | 9th(8/13), 10th(5/13) | 9th (3/11), 10th (8/11) |
| 156e | 29269.m000250 | LIGULELESS1 protein, putative | 9th(5/8), 10th(1/8) | 9th(2/7) | 10th (2/7) |
| 160ab | 27494.m000045 | Auxin response factor | |||
| 393b | 30131.m006863 | Transport inhibitor response1 protein | |||
| 171e | 30174.m008828 | Hypothetical protein | |||
| 399a | 30026.m001455 | Conserved hypothetical protein | |||
aThe detailed miRNA–guided cleavage of target mRNAs, the number of cleavage position is counted based on the mature miRNA sequence from the 5′- to 3′-end, the numerator and denominator of fraction to indicate the number of individuals and the whole clones which were cloned to vector and achieve the sequence.
The three-sample conditions listed as following:
HeLa-cold/drought ck: a mixture of equal quantity of HeLa-cold ck and HeLa-drought ck.
SC124-cold recovery: SC124 returning back to 25°C after 5 days cultivation under 14°C.
HeLa-stress: a mixture of equal quantity of HeLa-cold and HeLa-drought.
Figure 2.
The identification of miRNA-guided cleavage products of target genes in Euphorbiaceous plants. The cleavage sites of six selected targets in four miRNA as identified by 5′-RACE analysis. For each miRNA, the target sequence is shown on the top and the miRNA sequence on the bottom. The numbers indicate the fraction of cloned PCR products when PCR were terminated at different positions. (a) The cleavage site of 30174.m009082 by miR156e. (b) The cleavage site of 29269.m000250 by miR156e. (c) The cleavage site of 30147.m014478 by miR156e. (d) The cleavage site of 27538.m000327 by miR160ab. (e) The cleavage site of 30138.m004055 by miR164. (f) The cleavage site of 29647.m002022 by miR393b.
Among the 10 targets tested, the cleavage sites on six genes were detected in both the control and the stressed Castor bean, and the cleavage sites of five genes were found in the stressed Cassava. A close examination of these cleavage sites showed that two targets in miR160ab:27538.m000327 and miR164:30138.m004055 were cut at the canonical 10th position in HeLa-ck, HeLa-stress and SC124-cold recovery. Another target (miR393b:29647.m002022) was cleaved at the 11th position under the two conditions of the Castor bean, but not in the Cassava. The remaining three genes had different cleavage sites in the three samples. miR156e sliced its target 30147.m014478 at the 9th position in the Castor bean HeLa-stress and at the 10th position in the Cassava SC124-cold recovery. These two cleavage sites existed in unstressed Castor bean (HeLa-ck). The cleavage of two other targets of miR156e (30174.m009082 and 29269.m000250) was quite different. Gene 30174.m009082 had two cleavage sites (at the 9th and the 10th) in both the Castor bean HeLa-stress and the Cassava SC124-cold recovery samples. The difference lies in the percentages of these two cleavage sites: the 9th cleavage site was more frequent than the 10th site (8/11 versus 3/11) in the Castor bean HeLa-stress while the 9th cleavage site was less frequent than the 10th site in the Cassava SC124-cold recovery (3/11 versus 8/11). However, the cleavage of this gene in HeLa-ck had the common cleavage site at the 9th position and another two noncanonical cleavage sites, one at the 6th position and the other right before the miRNA binding site. Gene 29269.m000250 was cleaved by miR156e at the 9th and the 10th positions in HeLa-ck, but it was uniquely cleaved under stress conditions, i.e. exclusively at the 9th position in the Castor bean HeLa-stress and exclusively at the 10th position in the Cassava SC124-cold recovery. In short, the targets of miR156e were prone to be cut at the 10th position in the Cassava, while preferred to be cleaved at the 9th position in the stressed Castor bean but with variable cleavage positions in the unstressed Castor bean. In summary, there was a substantial difference on the cleavage sites between stressed and unstressed samples of the same plant, as well as between the two species.
The cleaved products of four targets failed to be detected in our experiment. Gene 27494.m000045, another target of miR160ab, was not cleaved. This may be because it was not expressed in the tested conditions since miR160ab were detected in the same conditions and cleaved one of the targets, 27538.m000327 (see above). The same reasoning can be applied to 30131.m006863 and 30026.m001455, since their targeting miRNAs (miR393b and miR399a, respectively) were detected in both developmental stage and stress conditions. On the other hand, miR171e was detected in normal development. No cleavage site on 30174.m008828 may be due to no expression of 30174.m008828 in HeLa-ck and no co-expression of miR171e and 30174.m008828 in other two stressed samples. Besides the co-expression of miRNA and targets, no detection of the four targets may reflect false positive predictions, or indicate that miRNAs may function through translation inhibition.
DISCUSSION
Our results showed that a substantial number of miRNAs which were previously identified and characterized in well studied model plants are conserved in four agri-economically important Euphorbiaceous plants, the Castor bean (Ricinus communis), the Cassava (Manihot esculenta), the Rubber tree (Hevea brasiliensis) and the Jatropha (Jatropha curcas L). Despite broad conservation across the four Euphorbiaceous species, these miRNAs also exhibited diverse expression patterns in development and abiotic stress responses. Our results also revealed that various target cleavage sites exist and different cleavage sites seemed to have a propensity to correlate with developmental and abiotic stress conditions, providing information of condition-specific cleavage. All our results formed the first collection of miRNAs and targets in Euphorbiaceae, which will be deposited to miRBase.
Relationship between miRNA and its target in Euphorbiaceous species
It is known that miRNAs can have a widespread effect on their targets (25). It has even been estimated that one third of mRNAs in the human genome are regulated by miRNAs (26). We predicted 258 miRNA:target pairs for 63 miRNAs in the Castor bean, giving an average miRNA to target ratio of 4.1 (258/63), which is greater than the ratios of 2.5 for A. thaliana and 3.87 for O. sativa but less than that of 6.45 for P. tricocharpa (27).
Some well conserved miRNA:target pairs previously detected in several species were also found in Euphorbiaceae. Members of the miR156 family are known to target SBP protein in A. thaliana, O. sativa, and P. trichocarpa (28,29). As expected, the miR156 family was also predicted to target a putative SBP protein in the Castor bean. Six other miRNA:target pairs were found to be conserved in the Castor bean: (i) miR159/319:MYB transcription factors (30,31); (ii) miR160:pARF (putative Auxin Response Factor) (32,33); (iii) miR162:pDCL1 (putative Dicer1) (34,35); (iv) miR164:pNAC (putative NAC domain containing protein 21/22) (36,37); (v) miR397:pLaccase (putative Laccase) (4,38); (vi) miR408:Laccase (39). A commonality of these miRNA:target pairs is that some members of each of these miRNA families were expressed in both seedling development and stress conditions. Consequently, it is viable to infer that some members of the conserved targets play roles in the Castor bean seedling development and stress response. Other three targets of broadly expressed miRNAs were not predicted to target genes of similar functions in the current Castor bean database: miR159/319:TCP (Teosinte Branched/Cycloide/PCF) transcription factors (40), miR167:ARFs (Auxin Response Factors) (41,42), miR396:GRF (Growth-Regulating-Factor) transcription factors (43,44). Moreover, miR166:HD-ZIPIII transcription factors for embryo patterning and vascular development in A. thaliana and Medicago truncatula (45,46), were predicted to target DNA binding proteins in the Castor bean.
Some miRNAs which were detected uniquely in seedling development in our experiment, were also predicted to target similar genes, for example, miR172:pAP2 (putative Floral Homeotic protein APETALA2), and miR393:pTIR1 (putative Transport Inhibitor Response 1). The first pair regulates floral organ identity (47), vegetative-to-reproductive transition (48), and leaf development (49) in Arabidopsis and rice. The second pair is related to stress activities and root development in Arabidopsis (44,50). This indicates that some members of these families have retained similar functions in the Castor bean. Finally, miR399, which targets inorganic phosphate transporter and functions in phosphate-starvation response in Arabidopsis (18,51,52), was found to target 2 similar genes in the Castor bean. Nine members of the miR399 family were observed in all four expression classes (Table 4). The diverse expression patterns of miR399 variants indicate extensive functional specification of this miRNA family in the Castor bean.
It has been observed that the targets of ‘non-conserved’ miRNAs have a wider range of functions than the targets of ‘conserved’ miRNAs (14). In our study, although there were 29 ‘conserved’ miRNA:target pairs, the functions of the remaining 229 miRNA:target pairs may be conceivably more diverse, therefore, may have functions specific to the Castor bean.
Many miRNAs function in plant developmental processes
From our results, fourteen miRNAs showed similar expression patterns in the four Euphorbiaceous species. However, many miRNA families exhibited diverse expression characteristics in varieties of a species, or across Euphorbiaceous plants. Such different expression of miRNAs may play an important role in shaping developmental and physiological changes across species.
The 23 predicted miRNA families were all detected during seedling development in four Euphorbiaceous species (Supplementary Table S8), including 20 miRNA families that are known to be related to development, and 3 novel families (miR157, miR395 and miR399) to be involved in development in Euphorbiaceae. The development-related functions of the 20 miRNA families were analyzed previously: The Auxin signaling related miRNAs (miR160, miR164 and miR167 families) play critical roles in plant leaf and floral morphology as well as root and shoot formation (33,41,53). miR164 functions in organ boundary formation/shoot apical meristem (SAM) initiation (54,55). miR165/166 regulate organ polarity (56,57). miR172 and miR159 play roles in floral organ identity and reproductive development (58,59). miR172, miR156 and miR159 influence vegetative-to-reproductive transition (30,47,60). miR319 is related to crinkly leaves (1), and miR162, miR168 and miR403 function in small RNA metabolism (34,51,61). Recently, 8 additional families (miR169, miR171, miR393, miR396, miR397, miR398, miR408 and miR535) and three development related miRNAs (miR160, miR167 and miR172) were confirmed in rice seed development by MPSS sequencing (49). All of these 20 miRNA families were also detected in seedling development of Euphorbiaceous plants. Three families (miR157, miR395 and miR399), which were not reported before, were expressed in the seedling development of Euphorbiaceous plants that we studied, and might be involved in developmental and metabolic processes specific to this family.
Many miRNAs are induced by abiotic stresses
miRNAs could be up- or down-regulated by stress as documented previously, some of which are included in Supplementary Table S8 miR395 and miR399 were up-regulated during sulphate and phosphate deficiency in Arabidopsis (17). Similarly, all five members of the miR395 family and four members of the miR399 family (miR399aefg) were detected in response to cold and/or drought stimuli in our study, suggesting conserved stress regulation of miR395 and miR399 in the Castor bean and the Cassava. miR398, which was down-regulated in abiotic and biotic stress in Arabidopsis (8,62), is stress responsive in the Castor bean and the Cassava. Of 13 tension or compression stress related miRNA families in P. trichocarpa (63,64), 8 of them (miR156, miR162, miR164, miR408, miR159, miR160, miR172 and miR168) could be detected in the stressed Euphorbiaceous plants. miR319 and miR397, which respond to dehydration, salinity and cold stresses and treatment of phytohormone abscisic acid (4), overlap with the stress-related miRNAs that we tested. Of 11 UV-B radiation induced families (65), 9 (miR156/157, miR159/319, miR160, miR166, miR167, miR169, miR171, miR398 and miR401) were detected under stress conditions in the Castor bean and the Cassava. Moreover, four cold-inducible miRNAs were identified in Arabidopsis (66), three of which (miR166, miR169 and miR396) responded to our tested stresses. miR169g was induced by drought, and miR169g and miR169n responded to high salinity (67,68). In our study, however, none of the 14 members of the miR169 family (6 primers) was detected in the stress conditions of the Castor bean or the Cassava.
In summary, of the 23 miRNA families in Euphorbiaceous plants that we studied: (i) 18 of them showed stress responsiveness; (ii) three families (miR169, miR172 and miR393), which were shown to be stress responsive (64,67–69), were not detected; and (iii) two families (miR403 and miR535), which were not reported before, were regulated under stress conditions. All of these data implied that most conserved miRNAs in Euphorbiaceae shared common functions with their counterparts in other species. However, some miRNAs showed regulatory roles specific to Euphorbiaceous plants. The stress-related miRNAs that we detected in Euphorbiaceous plants may offer excellent opportunities for studying how Euphorbiaceous plants adapt to their harsh, dynamic and resource-lacking environments.
Variability of miRNA cleavage sites
Plant miRNAs exert gene silencing mainly by slicing its target transcripts. The cleavage sites for 6 out of 10 predicted targets were verified. Even though most plant miRNAs cleave their targets preferentially at positions around the 10th nucleotide, some may slice their targets at other locations. Sequencing of the 5′-RACE products of six miRNA targets showed that most cleavage sites were mapped to miRNA complementary sequences, as were observed on other miRNA targets (63). It is interesting to highlight an exceptional cleavage site on 30174.m009082, a target of miR156e, which was mapped to the first nucleotide the 5′-end upstream of the miRNA:target binding site. Similar cleavage sites outside of miR:target basepairing regions have been reported in (70).
It is important to mention the variability of cleavage sites of a target in different conditions. Even in evolutionarily closely-related species as the Castor bean and the Cassava, the cleavage sites of different targets or even a target of a miRNA (e.g. miR156e) exhibited a variable characteristic across different species or even different conditions in the same plant. Distinct cleavage sites may suggest diverse functions in different species. We may even hypothesize that multiple miRNA cleavage sites allow species- and condition-specific fine-tuning of gene regulation in response to endogenous cues and/or exogenous stimuli.
While plant miRNAs regulate their targets primarily by degrading the transcripts, some of them may function at times by repressing translation (8,48,60). As no cleavage site was detected for four target genes in our study, we cannot rule out possible translational repression on these target genes. Due to the complex miRNA-mediated mRNA regulation, a combination of miRNA cleavage site analysis, co-expression profile assay and protein profiling is needed to determine the overall mode of action of miRNA gene regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chinese National Basic Research and 35 Development Program (grants 2006CB101701-6 and 2010CB126602); an earmarked fund for China Modern Agro-Industry Technology Research System (nycytx-17). United States National Science Foundation grants (IIS-0535257 and DBI-0743797). Funding for open access charge: Chinese National Basic Research and Development Program (grants 2006CB101701-6 and 2010CB126602).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Pablo Rabinowicz for providing seeds of the Castor bean HeLa and Dr Peng Zhang for providing stemcut of the Cassava C4 variety.
REFERENCES
- 1.Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 2.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 6.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Dugas DV, Bartel B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol. Biol. 2008;67:403–417. doi: 10.1007/s11103-008-9329-1. [DOI] [PubMed] [Google Scholar]
- 9.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 10.Arteaga-Vazquez M, Caballero-Perez J, Vielle-Calzada JP. A family of microRNAs present in plants and animals. Plant Cell. 2006;18:3355–3369. doi: 10.1105/tpc.106.044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang BH, Pan XP, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 12.Sunkar R, Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008;8:37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibanez-Ventoso C, Vora M, Driscoll M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE. 2008;3:e2818. doi: 10.1371/journal.pone.0002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willmann MR, Poethig RS. Conservation and evolution of miRNA regulatory programs in plant development. Curr. Opin. Plant Biol. 2007;10:503–511. doi: 10.1016/j.pbi.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RHA. Differences in vertebrate microRNA expression. Proc. Natl Acad. Sci. USA. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha M, Pang MX, Agarwal V, Chen ZJ. Interspecies regulation of microRNAs and their targets. BBA-Gene Regul. Mech. 2008;1779:735–742. doi: 10.1016/j.bbagrm.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiou TJ. The role of microRNAs in sensing nutrient stress. Plant Cell Environ. 2007;30:323–332. doi: 10.1111/j.1365-3040.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 18.Doerner P. Phosphate starvation signaling: a threesome controls systemic P-i homeostasis. Curr. Opin. Plant Biol. 2008;11:536–540. doi: 10.1016/j.pbi.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen XM, Green PJ, et al. Criteria for annotation of plant microRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat. Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 22.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 24.Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 25.Farh KKH, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 26.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Li AL, Mao L. Evolution of plant microRNA gene families. Cell Res. 2007;17:212–218. doi: 10.1038/sj.cr.7310113. [DOI] [PubMed] [Google Scholar]
- 28.Guo AY, Zhu QH, Gu XC, Ge S, Yang J, Luo JC. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418:1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Riese M, Hohmann S, Saedler H, Munster T, Huijser P. Comparative analysis of the SBP-box gene families in P-patens and seed plants. Gene. 2007;401:28–37. doi: 10.1016/j.gene.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 30.Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- 31.Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are MicroRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 33.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 35.Hirsch J, Lefort V, Vankersschaver M, Boualem A, Lucas A, Thermes C, d'A;ubenton-Carafa Y, Crespi M. Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol. 2006;140:1192–1204. doi: 10.1104/pp.105.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development. 2009;136:823–832. doi: 10.1242/dev.031625. [DOI] [PubMed] [Google Scholar]
- 37.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 38.Abdel-Ghany SE, Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in arabidopsis. J. Biol. Chem. 2008;283:15932–15945. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of MicroRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Huson D, et al. Sequence and expression differences underlie functional specialization of Arabidopsis MicroRNAs miR159 and miR319. Dev. Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 41.Yang JH, Han SJ, Yoon EK, Lee WS. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006;34:1892–1899. doi: 10.1093/nar/gkl118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu MF, Tian Q, Reed JW. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- 43.Liu DM, Song Y, Chen ZX, Yu DQ. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plantarum. 2009;136:223–236. doi: 10.1111/j.1399-3054.2009.01229.x. [DOI] [PubMed] [Google Scholar]
- 44.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 45.Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 46.Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 2008;54:876–887. doi: 10.1111/j.1365-313X.2008.03448.x. [DOI] [PubMed] [Google Scholar]
- 47.Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl Acad. Sci. USA. 2005;102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue LJ, Zhang JJ, Xue HW. Characterization and expression profiles of miRNAs in rice seeds. Nucleic Acids Res. 2009;37:916–930. doi: 10.1093/nar/gkn998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pruss GJ, Nester EW, Vance V. Infiltration with Agrobacterium tumefaciens induces host defense and development-dependent responses in the infiltrated zone. Mol. Plant Microbe In. 2008;21:1528–1538. doi: 10.1094/MPMI-21-12-1528. [DOI] [PubMed] [Google Scholar]
- 51.Allen E, Xie ZX, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 53.Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peaucelle A, Morin H, Traas J, Laufs P. Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development. 2007;134:1045–1050. doi: 10.1242/dev.02774. [DOI] [PubMed] [Google Scholar]
- 56.Williams L, Grigg SP, Xie MT, Christensen S, Fletcher JC. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development. 2005;132:3657–3668. doi: 10.1242/dev.01942. [DOI] [PubMed] [Google Scholar]
- 57.Zhou GK, Kubo M, Zhong RQ, Demura T, Ye ZH. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell. Physiol. 2007;48:391–404. doi: 10.1093/pcp/pcm008. [DOI] [PubMed] [Google Scholar]
- 58.Zhao L, Kim YJ, Dinh TT, Chen XM. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 2007;51:840–849. doi: 10.1111/j.1365-313X.2007.03181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen RS, Li JY, Stahle MI, Dubroue A, Gluber F, Millar AA. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl Acad. Sci. USA. 2007;104:16371–16376. doi: 10.1073/pnas.0707653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007;49:683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- 61.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jagadeeswaran G, Saini A, Sunkar R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta. 2009;229:1009–1014. doi: 10.1007/s00425-009-0889-3. [DOI] [PubMed] [Google Scholar]
- 63.Lu SF, Sun YH, Shi R, Clark C, Li LG, Chiang VL. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu SF, Sun YH, Chiang VL. Stress-responsive microRNAs in Populus. Plant J. 2008;55:131–151. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- 65.Zhou XF, Wang GD, Zhang WX. UV-B responsive microRNA genes in Arabidopsis thaliana. Mol. Syst. Biol. 2007;3:103. doi: 10.1038/msb4100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou XF, Wang GD, Sutoh K, Zhu JK, Zhang WX. Identification of cold-inducible microRNAs in plants by transcriptome analysis. BBA-Gene Regul. Mech. 2008;1779:780–788. doi: 10.1016/j.bbagrm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 67.Zhao BT, Liang RQ, Ge LF, Li W, Xiao HS, Lin HX, Ruan KC, Jin YX. Identification of drought-induced microRNAs in rice. Biochem. Bioph. Res. Co. 2007;354:585–590. doi: 10.1016/j.bbrc.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 68.Zhao BT, Ge LF, Liang RQ, Li W, Ruan KC, Lin HX, Jin YX. Members of miR-169 amily are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 2009;10:29. doi: 10.1186/1471-2199-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sunkar R, Chinnusamy V, Zhu JH, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 70.Moxon S, Jing RC, Szittya G, Schwach F, Pilcher RLR, Moulton V, Dalmay T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18:1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;131:4311–4322. doi: 10.1242/dev.01320. [DOI] [PubMed] [Google Scholar]
- 72.Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, Kim JA, Lee M, Lee Y, Narry Kim V, et al. microRNA directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005;42:84–94. doi: 10.1111/j.1365-313X.2005.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.