Abstract
The Bacteroides conjugative transposon CTnDOT encodes an integrase, IntDOT, which is a member of the tyrosine recombinase family. Other members of this group share a strict requirement for sequence identity within the region of strand exchange, called the overlap region. Tyrosine recombinases catalyze recombination by making an initial cleavage, strand exchange and ligation, followed by strand swapping isomerization requiring sequence identity in the overlap region, followed by the second cleavage, strand exchange and ligation. IntDOT is of particular interest because it has been shown to utilize a three-step mechanism: a sequence identity-dependent initial strand exchange that requires two base pairs of complementary DNA at the site of cleavage; a sequence identity-independent strand swapping isomerization, followed by a sequence identity-independent cleavage, strand exchange and ligation. In addition to the sequence identity requirement in the overlap region, Lambda Int interactions with arm-type sites dictate the order of strand exchange regardless of the orientation of the overlap region. Although IntDOT has an arm-binding domain, we show here that the location of sequence identity within the overlap region dictates where the initial cleavage takes place and that IntDOT can recombine substrates containing mismatches in the overlap region so long as a single base of sequence identity exists at the site of initial cleavage.
INTRODUCTION
Bacteroides spp. are Gram-negative anaerobes that colonize the human gastrointestinal tract. They are host to a number of integrated mobile genetic elements that have been implicated in the transfer of antibiotic resistance genes (1–4). These elements are called conjugative transposons, or more recently, integrative and conjugative elements (ICEs) (5). CTnDOT is the best studied Bacteroides ICE. It carries genes encoding tetracycline resistance, tetQ, and erythromycin resistance, ermF (6–8). Excision and integration of CTnDOT are catalyzed by its integrase, IntDOT, which is a member of the large tyrosine recombinase family (9–12). Tyrosine recombinases use a topoisomerase I-type mechanism for recombination (13–15), which involves an initial strand exchange to form a Holliday junction intermediate. In most systems this is followed by a homology-dependent strand swapping isomerization step, and then a second strand exchange to form the product (16). In some cases, recombination stops at the Holliday junction step and the structure is resolved by host processes (17–19). Sequence identity between overlap regions is a requirement for recombination of most tyrosine recombinases. It is assumed that Watson–Crick base pairing must take place after the strand exchange in order for ligation to occur (20).
Site-specific recombinases can be sub-classified into two groups depending on whether or not their recombination reaction is regulated. There are autonomous systems in which the recombinase binds only core-type DNA sites. These enzymes contain core-binding (CB) and catalytic (CAT) domains that interact directly with core-type sites. In the more complex systems the recombinase has an additional N-terminal (N) domain that binds to arm-type sites that flank the core-type sites. These systems require accessory proteins (14,21). There does not appear to be any correlation between the complexity of the system and the regulation of the order of strand exchanges. The order of strand exchange can be influenced by a variety of different factors including additional DNA-binding domains, accessory proteins and DNA structure.
IntDOT contains an N domain and requires accessory proteins for integration and excision, so it was initially assumed that these would influence the order of strand exchange in the integration reaction. In this article, we show that for IntDOT, DNA sequence identity in the overlap region dictates the order and position of strand exchange and that a single base of sequence identity is required for the first strand exchange. Interestingly, unlike most tyrosine recombinases, IntDOT does not require sequence identity for the strand swapping isomerization step or for the second strand exchange. This unique mechanism for strand exchanges may place IntDOT into a new subclass of tyrosine recombinase.
MATERIALS AND METHODS
Preparation of radiolabeled attB substrates
One strand of the attB DNA oligonucleotide (IDT) was 5′-end-labeled with γ-32P-ATP (Perkin-Elmer) using T4 polynucleotide kinase (Fermentas) and free γ-32P removed using G-25 spin columns (Amersham Biosciences). The labeled DNA was mixed with the unlabeled complementary strand at a 1 : 5 molar ratio and annealed in an annealing buffer (0.1 M KCl, 10 mM Tris–HCl pH 8, 5 mM EDTA) by heating to 90°C for 2 min followed by slow cooling to 25°C. The attB DNA substrates containing a nick in either the top or bottom strand were prepared by mixing a labeled intact strand with two unlabeled strands complementary to the labeled strand at ratios of 1 : 5 : 5, respectively. These were annealed as described above. All nicked attB substrates were phosphorylated at the 5′-end of the oligo at the cleavage site. A list of oligonucleotides used in this study is shown in Supplementary Table S1.
In-vitro recombination assay
The attDOT and attB substrates were incubated in a 20 µl reaction volume containing 0.17 µM Escherichia coli IHF, 1 unit of IntDOT, 30 mM Tris–HCl (pH 7.4), 5 mM DTT, 0.1 mg/ml tRNA, 0.07 mg/ml BSA, 2.6% glycerol and 50 mM KCl. The final concentrations of attDOT and attB were 2 nM as determined by OD260. A unit of IntDOT is defined as the minimum amount of IntDOT needed to produce maximum recombination between attDOT and attB (22). The reaction was shown to proceed over 16 h so samples were incubated overnight at 37°C and the reaction quenched with the addition of 5 µl stop solution (30% glycerol, 10% SDS, 0.25% xylene cyanol and 0.25% bromophenol blue). All samples were subjected to electrophoresis on a 1% agarose gel. Gels were dried on a vacuum slab drier, then exposed on phosphorimager screens and the recombination efficiency quantified using a Fujifilm FLA-3000 phosphorimager and Fujifilm Image Gauge software (Macintosh v.3.4).
Restriction digest analysis of the recombinant products
Some experiments were done where products were cleaved by SspI endonuclease. A double volume of the standard recombination assay described above was performed but was terminated by heating for 20 min at 60°C instead of by the addition of stop solution. Magnesium chloride was added to a final concentration of 10 mM. Half the reaction volume was transferred to a fresh microcentrifuge tube and digested with SspI endonuclease (Fermentas). The digest was stopped by heating at 60°C for 20 min and the sample was subjected to electrophoresis on a 1% agarose gel at 100 V for 2 h and analyzed as described above.
Site-directed mutagenesis of attDOT/pUC19
Mutations were made in the 7 bp core overlap region of attDOT using the Stratagene Quikchange Mutagenesis Kit as described by the supplier (Stratagene). Primers carrying the specific mutations are shown in Table 3. The attDOT regions of the plasmid were sequenced to confirm the presence of the desired mutation and to ensure no other mutations were present.
2D gel electrophoresis
A double volume recombination assay was performed and terminated with stop solution. Two 15 µl samples were subjected to electrophoresis on a 1% agarose gel in 1× TBE for 2 h at 100 mA. One lane was sliced out of the gel and prepared for the second dimension; the other lane was dried and exposed on a phosphorimager screen. The gel slice for the second dimension was soaked in alkaline buffer (50 mM NaOH, 5 mM EDTA) for 1 h before being set in a gel tray with 1% agarose dissolved in alkaline buffer. The gel was electrophoresed in alkaline running buffer (50 mM NaOH, 5 mM EDTA) for 16 h at 30 V, then dried and exposed on a phosphorimager screen.
Detection of covalent DNA/Protein complexes
A double volume recombination assay was performed but was stopped with the addition of 0.1 vol of 10% SDS. A standard SDS/KCl precipitation was performed as described previously (23). The fractions were separated on a 1% agarose gel for 2 h at 100 V.
RESULTS
In this study we used a gel-based in vitro integration assay developed previously (24). The in-vitro integration reaction utilizes two DNA substrates carrying a linear radiolabeled attB site and a supercoiled plasmid containing the attDOT site. The reaction requires IntDOT and E. coli integration host factor (IHF). [The Bacteroides host factor has not yet been identified, but E. coli IHF substitutes (24)]. Because IntDOT-mediated recombination requires an accessory factor, all experiments were performed with IntDOT and E. coli IHF. Recombination between the attDOT and attB sites produces a 3.6 kb linear recombinant that can be separated from unreacted attB DNA on an agarose gel (see ‘Materials and Methods’ section). Several attB sites have been identified (25) but most experiments described here use the attB1 site.
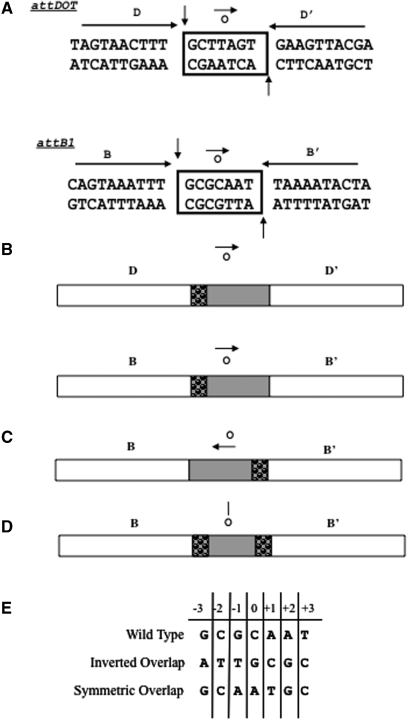
The overlap region defines the sites of cleavage and strand exchange in the top and bottom strands of the attDOT and attB sites (26). It consists of a 5 bp coupling sequence that varies in sequence from site to site, and a conserved GC dinucleotide that is found at the left side of the overlap regions in attDOT and all known naturally occurring attB sites (25) (Figure 1). (The term ‘GC dinucleotide’ is used to describe the complementary 5′ GC/3′ CG sequence at the left end of the overlap region). It has been shown previously that the integration reaction catalyzed by IntDOT proceeds via a three-step mechanism: a pair of top strand exchanges on the left side of the overlap region that are dependent upon sequence identity, followed by a sequence independent isomerization step, and then a pair of bottom strand exchanges on the right side of the overlap region that are independent of sequence identity (26). It appeared that the only sequence identity between the sites that was required for integration was the GC dinucleotide base pairs on the left side of the overlap regions in both attDOT and attB. For example, previous work showed that only the base pairs at the position of the GC dinucleotide need to be identical in both substrates for efficient recombination—attDOT and attB sites where both sites contained CG or AT substituted for the GC dinucleotide were shown to be active (26). Thus, productive recombination occurs when both sites have complementary dinucleotide sequences and the sequence itself is not important.
Figure 1.
(A) Wild-type sequences of the core-binding sites attDOT and attB. The boxed region indicates the overlap region (o). The conserved GC dinucleotide is shown at the left sides of the overlap regions. The remaining bases in the overlap are the coupling sequences that vary from site to site. The small arrow represents orientation of the overlap region relative to flanking arm sites. The longer arrows indicate the imperfect inverted repeats that flank the overlap regions. The D and B sites contain 8 of 10 identical base pairs. The vertical arrows show the sites of cleavage on the top and bottom strands. (B) Simplified schematic diagram of the same core regions. The bubbled area denotes the GC dinucleotide homology between the attDOT and attB1 overlap regions. (C) Schematic diagram of the attB site with an inverted overlap region. The arrow denotes the directionality of the overlap sequence. (D) Schematic diagram of the attB site containing a symmetric overlap region. The line over the ‘o’ represents a line of symmetry within the overlap region. (E) Sequences of the wild type, inverted and symmetric overlap regions.
The overlap regions are flanked by IntDOT core-type binding sites referred to as B and B’ in attB, or D and D’ in attDOT. The D and B sites contain a conserved sequence ending in TTTGC (Figure 1). The first strand exchanges occur after initial cleavage between the T and G in attDOT and attB sites. The second strand exchange occurs by cleavage between the A and C residues on the bottom strand of attDOT and the two A residues on the bottom strand of attB1. Note that ligation of the top strands involves complementary base pairs at the site of ligation, while ligation of the bottom strand occurs in the presence of mismatches in the DNA.
An attB site with an inverted overlap region functions as an efficient recombination substrate
We wanted to distinguish between three possible general models of the mechanism of ordered strand exchange in IntDOT-mediated recombination. The first model proposes that sequences outside the overlap region (the boxed sequences in Figure 1A) play a defining role in determining the order of strand exchange. For example, the arm-type sites could be responsible for determining the order of strand exchange, much like the Lambda Int system (27). A variation of this model is that the core-type sites such as the D and B sites determine the order of strand exchange. If true, the location of the GC dinucleotide within the overlap region should not matter and initial cleavage and strand exchange should always take place at the same site relative to the arm- and core-type sites. The second model proposes that the D and B core-type sites must be contiguous with the GC dinucleotide located in the overlap region so that the entire conserved sequence shared by both sites is responsible for determining the order of strand exchange. If the position of the GC dinucleotide relative to the D and B sites is important, then moving the GC dinucleotide to the opposite end of the overlap region will result in loss of recombination. The third model proposes that only the GC sequence identity within the overlap regions of the recombining att sites is important, and the position (either at the left or right side of the overlap region) of these bases in both attDOT and attB should not affect recombination as long as the sequences are identical in both sites. In addition, initial cleavage and strand exchange will take place at the location of the sequence homology independent of the flanking sequences. In summary, Model 1 predicts that the arm- and/or core-type sites determine the order of strand exchange; Model 2 predicts that the GC dinucleotide must be adjacent to the conserved sequence of the B and D sites and Model 3 predicts that only the GC dinucleotide within the overlap region determines the order of strand exchange.
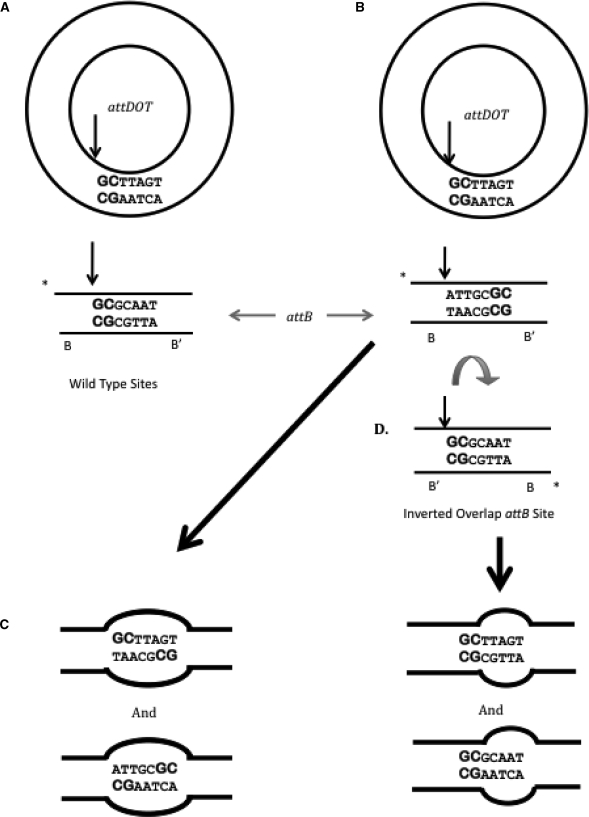
In attempt to distinguish between these three possibilities, we designed an attB1 site with an inverted overlap region (Figure 1B and C). In this substrate, the entire seven base overlap region is rotated 180° so that the GC dinucleotide is relocated to the 5′-end of the bottom strand, contiguous with the B’ site (Figure 2A and B). Synapsis of the wild-type attDOT site and the inverted attB1 site in orientations shown in Figure 2B generates two mismatched base pairs (GC in attDOT and AT in the inverted attB1 site) at the position of the original GC dinucleotide. If the flanking core- or arm-type sites determine the order of strand exchange, then the reaction will proceed with initial cleavage and strand exchange taking place adjacent to the D arm, but recombination will be substantially decreased because a homology-dependent strand exchange cannot take place (Figure 2C). A substrate with the inverted attB site should also not be functional in integrative recombination if the GC dinucleotide must be adjacent to the B site for productive recombination as proposed in the second model.
Figure 2.
Schematic model of how the plasmid containing the attDOT site orients itself relative to the attB to maintain alignment of the GC dinucleotide. (A) Orientation of wild-type attDOT and wild-type attB in the standard integration reaction. The vertical arrow indicates the site of initial cleavage. (B) Possible orientation of wild-type attDOT and the attB site containing an inverted overlap region. (C) Products from the attDOT and attB sites oriented as shown in B if the initial cleavage occurs at the location of the vertical arrow. There is complete heterology within the entire seven base overlap region in both products. (D) Rotation of the inverted overlap attB site by 180° orients the GC dinucleotide in the same position as in the wild-type reaction. The vertical arrow denotes the site of initial cleavage producing the recombinants that both contain the complementary GC dinucleotide on the left side.
We found that the attB1 site with an inverted overlap region recombined nearly as efficiently as the wild-type attB1 with a partner site containing the wild-type attDOT (data not shown). These results suggest a less defining role for the arm-type sites but do not yet distinguish between the other models. As shown in Figure 2D, a 180° rotation of the entire attB1 site would generate an alignment that contained a GC dinucleotide in the correct position in the top strand adjacent to the B’ core site. This could indicate that the position of the GC dinucleotide within the core overlap region dictates the orientation of the att sites in a synaptic complex that undergoes recombination. If correct, neither the core-type sites, nor the arm-type sites control the orientation of sites in a synaptic complex. In addition, since the top strands containing the GC dinucleotides are exchanged first in recombination with wild-type sites (10,26) it is possible that the strands containing the GC dinucleotides in reactions between attDOT and the inverted overlap attB1 site are also exchanged first, supporting the third model. Alternatively, the first strand exchange could be taking place between non-identical base pairs and the location of sequence identity within the overlap region is not as important as the orientation of the B and D sites relative to each other, thereby supporting the first model.
Synaptic complexes containing the attB1 and inverted overlap attB1 sites
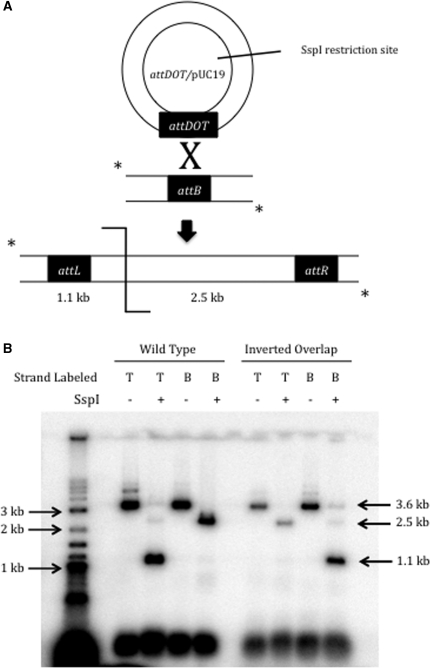
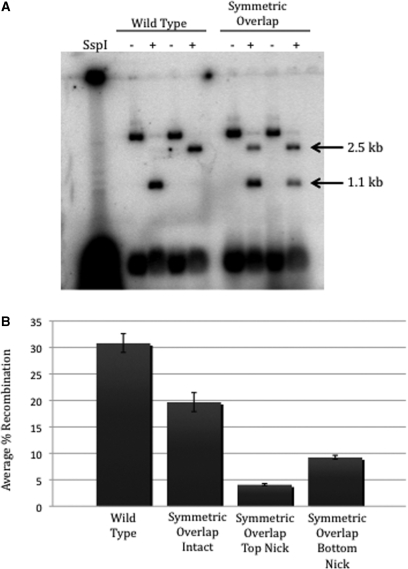
SspI digestion of the linear recombinant product from wild-type att sites produces two fragments of 1.1 and 2.5 kb due to a single asymmetric restriction site within the plasmid containing attDOT (Figure 3A). During synapsis, the attDOT site presumably forms an intasome with four IntDOT monomers and an unknown number of IHF heterodimers bound to the DNA (28,29). There are two potential orientations of the attB1 site relative to the intasome in a synaptic complex. If the second model is correct, then the restriction pattern will be the same as that seen with the wild-type att sites, meaning the intasome will form a productive synapse with the inverted overlap attB site in the same orientation as it does with the wild-type attB site. On the other hand, if the third model is correct then an attB1 site with the inverted overlap will interact with the intasome in the reverse orientation where the GC dinucleotides are exchanged, and the orientation of the integrated attDOT plasmid relative to the ends of the recombinant product will be inverted. When the top strand of a wild-type attB1 is radiolabeled, digestion with SspI of the resulting recombinant yields a 1.1 kb fragment. A 2.5 kb fragment appears if the bottom strand is radiolabeled (Figure 3B). After recombination with the attB1 site containing the inverted overlap region, the opposite fragment pattern appears. With a label on the top strand, the 2.5 kb fragment is produced and the 1.1 kb fragment appears with a bottom strand label. This confirms that the orientation of attDOT relative to the attB sites in the two sites synaptic complexes are different and that the first strand exchange involves exchange of the strands containing the GC dinucleotide. We conclude from this experiment that Model 2 is incorrect.
Figure 3.
(A) Schematic representation of the in-vitro integration assay and the single asymmetric SspI site that produces 1.1 and 2.5 kb fragments from the linear recombinant. (B) Results from an SspI digest of recombinants produced from wild-type attDOT and either wild-type or inverted overlap attB sites. A T for the top strand or a B for the bottom strand indicates the strand that is radiolabeled.
An attB site with an inverted overlap sequence shows a strong bias for bottom strand exchange
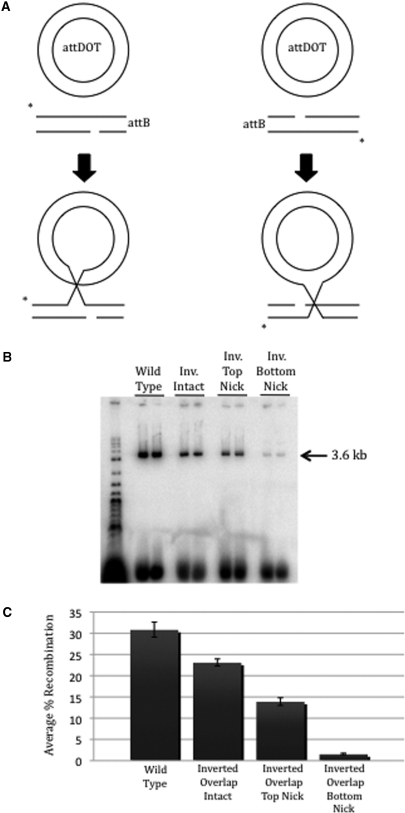
Since the orientation between attDOT and attB during productive synapsis appeared to be changed with the attB with the inverted overlap region, we wanted to determine if the order of strand exchange was also altered. We previously showed that the first strand exchanges between the attDOT and attB1 sites occurred at the top strands at sites adjacent to the GC dinucleotide (26). We used complementary oligonucleotides that introduced a nick at either the top strand or bottom strand cleavage site of attB1. Nicked substrates allow the intact strand to be cleaved and exchanged to form a Holliday junction containing a nick. However a nick at the second cleavage site prevents the exchange of the second strand, thereby trapping a Holliday intermediate. Alternatively, if the nick is at the site of the first strand exchange, no intermediate is formed (Figure 4A). In the integration reaction between wild-type attDOT and wild-type attB1, the top strand is cleaved and exchanged first, so a nick in the top strand inhibits the first step of the reaction and no product is formed. A nick on the bottom strand allows the top strand to be cleaved and exchanged forming the Holliday junction intermediate product that is detectable on an agarose gel. We constructed nicked inverted overlap attB1 sites and tested them with wild-type attDOT. If the bottom strand is cleaved and exchanged first, then the Holliday junction intermediate would be formed in the reaction with the inverted overlap attB1 site containing a nick in the top strand but not from the attB1 site containing a nick in the bottom strand.
Figure 4.
(A) Recombination reactions with attB suicide substrates nicked on either the top or bottom strand. The attB substrates contain a nick at either the top (right) or bottom (left) strand at the cleavage site. Initial strand exchange can occur between the intact strand of attB and the corresponding strand of attDOT, allowing determination of strand exchange order. The strand containing a nick cannot undergo recombination and so traps a Holliday intermediate where the first strand has been exchanged but the nicked strand has not. (B) Integration assay results using wild-type attDOT and either wild-type attB; inverted overlap intact attB; inverted overlap attB containing a nick in the top strand; or inverted overlap attB containing a nick in the bottom strand. Each sample is run in duplicate on the gel. (C) Graph showing the average percent recombination of the intact and nicked attBs over a minimum of four experiments.
The results of the experiments with the inverted overlap attB1 sites are shown in Figure 4B and C. Products were detected from the inverted attB1 site with a nick at the top strand cleavage site, but to a much lesser extent from the inverted attB1 site with a nick at the bottom strand cleavage site. This result indicates that the frequency of first strand exchange between the attDOT top strand and the bottom strand of the inverted attB1 site containing the GC dinucleotide occurs ∼9 times more frequently than the exchange with the top strand.
An attB1 site with a symmetric overlap produces recombinants in both orientations
If the orientation of the integrating attDOT site relative to the attB site is dependent on sequence identity at the position of the first exchange within the overlap region, then presenting an attDOT intasome with a site containing a symmetric overlap region with a GC dinucleotide in both strands should produce recombinants where the attDOT site recombined with the symmetric core in both orientations with equal frequency (Figure 1D). Figure 5A shows the results of experiments where the symmetric overlap attB1 site was labeled on the top or bottom strand. Both the 1.1 and the 2.5 kb products are formed regardless of which strand is labeled (Figure 5). These results suggest that the symmetric overlap attB1 site can synapse with attDOT in both orientations at equal frequencies. These results also support the argument that synapsis occurs at equal frequency with wild-type attB1 in both orientations but only one orientation produces recombinants.
Figure 5.
(A) SspI restriction digest of recombinants produced from either wild-type attB1 or attB containing a symmetric overlap sequence. (B) Graph comparing the average percent recombination of wild-type attB1 and an attB site containing a symmetric overlap region. All attB sites were combined with a wild-type attDOT and averaged over a minimum of four experiments.
These results are confirmed by experiments done using attB sites with a symmetric overlap region containing a nick at either the top or bottom strand cleavage site. Products were detected regardless of which strand contained the nick, indicating that synapsis and cleavage takes place with the substrates in either orientation (Figure 5B). The higher amount of product detected with the attB site containing the nick at the bottom strand cleavage site may suggest a slight preference for a particular orientation of the D and B sites relative to each other in the synaptic complex.
Other known attB sites produce recombinants similar to attB1
There are six Bacteroides attB sites that have been identified and sequenced (25). All six sites contain a conserved sequence on the left side, ending in TTTGC (Table S2). We wanted to determine whether other attB sites behave similarly to attB1 sites containing different overlap and B’ sequences. Accordingly, we constructed attB2-6 sites using complementary oligonucleotides and tested them in the in vitro integration assay with a wild-type attDOT. We also constructed attB3 and attB5 sites with inverted overlap regions to confirm that the location of the GC dinucleotide within the overlap region dictates the orientation of the integrating attDOT. The attB3 and attB5 sites were chosen for experiments because their sequences are the least similar to the attDOT sequence. The results of the in-vitro integration assay show that all six attB sites behave in a similar fashion and have relatively similar recombination efficiencies, with attB4 being the least efficient of the attB sites (Supplementary Table S2). The site of initial cleavage takes place between the T and G, making the conserved GC dinucleotide part of the overlap region. The recombinants produced from attB3 and attB5 sites containing an inverted overlap region show the same restriction pattern as recombinants produced from the attB1 site with an inverted overlap region (data not shown). We conclude from these results that the known attB sites appear to act similarly with respect to the mechanism for ordered strand exchange.
The overlap region dictates orientation of integrating attDOT
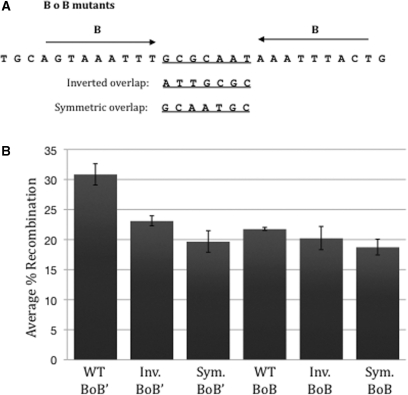
If the B core-type site of attB plays a role in orienting the integrative intasome for productive recombination, then having the B site flanking both sides of the overlap region should also induce integration in both orientations. On the other hand, if sequence identity in the overlap region is the key, then recombination will depend on the location of sequence identity in the overlap region. To further investigate the role of the conserved B region, an attB1 site where both core sites contain B sequences were constructed using complementary oligonucleotides. The core overlap regions were varied to be wild type, symmetric or inverted (Figure 6A). Analysis of BoB derived recombinant products containing a wild type, inverted or symmetric overlap region shows similar frequencies as attB1 derived recombinants with the same overlap regions (Figure 6B). Taken together, the data strongly suggest that the core overlap region is responsible for determining how the integrating attDOT is oriented relative to the attB during synapsis that leads to recombination. It also determines which strand is cleaved and exchanged first.
Figure 6.
(A) Sequence of BoB attB mutants with each core overlap sequence. The B region of the core is normally only on the 5′ side. In the BoB mutant, it is present on both sides. In this study, they are combined with wild-type attDOT containing both the D and D’ core-type sites. (B) Comparison of the average percent recombination of wild-type attDOT combined with wild-type (WT BoB’), inverted overlap (Inv. BoB’), or symmetric overlap attB1 (Sym. BoB’). And wild-type attDOT combined with BoB attB sites containing wild-type (WT BoB), inverted overlap (Inv. BoB) or symmetric overlap (Sym. BoB) regions. The average recombination was taken over a minimum of four experiments.
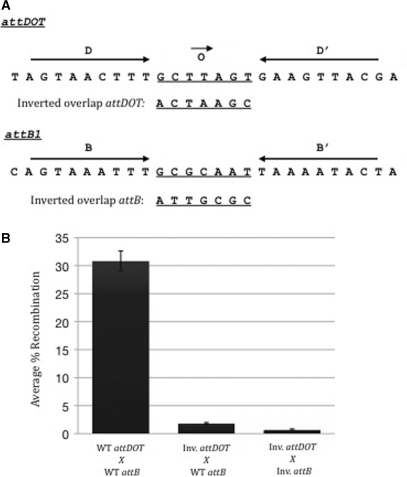
An attDOT site with an inverted overlap recombines with attB1 in the opposite orientation compared to wild-type attDOT
If the hypothesis that the orientation of the overlap region within an attB1 site dictates the order of strand exchange is correct, then a substrate containing an inverted overlap attDOT site should integrate in the opposite orientation as the wild-type attDOT when recombined with either wild-type or inverted overlap attBs. An attDOT derivative containing an inverted overlap region between the D and D’ sites was produced via site-directed mutagenesis to test this proposition (Figure 7). The results are consistent with this model. The restriction pattern from the wild-type attDOT × wild-type attB product showed a 1.1 kb product when the top strand was labeled and a 2.5 kb product when the bottom strand was labeled and the opposite pattern when the attB1 site contained the inverted overlap region (Figure 3). The restriction pattern from the inverted overlap attDOT × wild-type attB1 product showed the predicted pattern but recombination levels were depressed nearly 15-fold (Figure 7B). Thus it is likely that the identity of the core sites of attDOT plays a role in the efficiency of the integration reaction.
Figure 7.
(A) Sequences of attDOT and attB1 with wild-type or inverted overlap regions. (B) Integration assay results from combinations of wild-type (WT) and inverted overlap (Inv.) attDOT and attBs. The average recombination was taken over a minimum of four experiments.
A single base sequence identity adjacent to the site of cleavage and ligation is sufficient for recombination
We established above that the location of the GC dinucleotide in the overlap region determines the order of strand exchange and provides enough sequence identity for recombination to occur. We next asked if a single base pair of sequence identity between both att sites is sufficient for recombination and whether that single base pair needs to be at the site of cleavage. Presenting a wild-type attDOT site with an attB site containing a CC in place of the GC dinucleotide should decrease recombination due to a lack of sequence identity between the bases at either cleavage site. As expected, recombination was abolished (data not shown). This indicates that sequence identity is required at the base adjacent to cleavage in the overlap region and that the presence of the C of the GC dinucleotide is not sufficient for recombination to occur.
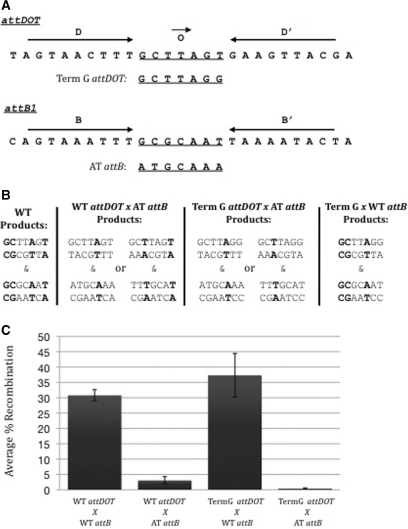
We then constructed an attDOT mutant containing a terminal G substitution in the overlap region (TermG attDOT), making the overlap sequence GCTTAGG (Figure 8). We combined this attDOT site with both wild-type attB and an attB site containing the overlap sequence ATGCAAA (AT attB). Not surprisingly, the attDOT containing the terminal G substitution recombined well with the wild-type attB site since both sites share a G at the cleavage site (Figure 8B and C). Also as expected, the TermG attDOT site recombined almost negligibly with the AT attB site (<0.5%). The wild-type attDOT site however, was able to recombine with the AT attB site, albeit poorly (∼3%). This is explained by the fact that the two sites share a single base of sequence identity at the cleavage sites—the T at the 3' end of the attDOT overlap region and the T at both ends of the overlap region in an inverted AT attB site (Figure 8B). Taken together, these data suggest that only one base of sequence identity is sufficient for low levels of recombination, but that two bases of sequence identity are required for maximum recombination. The sequence identity must be located at the site of cleavage and ligation.
Figure 8.
(A) Sequence comparison of wild-type attDOT and an attDOT site with a terminal G mutation in the overlap region, and wild-type attB with an attB site containing substitutions at both ends of the overlap region. (B) Sequences of products formed from the recombination of wild-type or TermG attDOT with either wild-type or AT attB. The second pair of sequences in each combination is the products from an inverted attB site. Complementary base pairs are in bold. (C) Integration assay results from combinations of wild-type or TermG attDOT sites with either wild-type or AT attB sites. The average recombination was taken over a minimum of four experiments.
DISCUSSION
There are two main subclasses of tyrosine recombinases. The first is the autonomous class that only contains two domains—CB and CAT domains. These recombinases recognize core-type DNA sites and do not require accessory proteins. The second subclass is characterized by the presence of an N-terminal domain that binds to arm-type DNA sites, and a requirement for additional proteins to determine directionality.
Some tyrosine recombinases also have a preferred order of strand exchanges while others do not (30). There does not appear to be a connection between complexity of the recombination systems and the mechanism of regulating the order of strand exchanges. For example, Flp is an autonomous recombinase that does not have a defined order of strand exchange—it will cleave and exchange both strands with equal frequency (31). Cre recombinase also falls under the autonomous class, yet when it binds to its loxP substrate it induces an asymmetric bend that determines which strand will be exchanged first (30,32–34). The Xer system involves two tyrosine recombinases, XerC and XerD, that work in tandem to ensure chromosome and plasmid segregation during cell division. XerC/D can work as autonomous or regulated recombinases depending upon the substrate. For example, the chromosomal dif site only consists of CB sites for XerC and XerD, while the psi site on the plasmid pSC101 requires accessory proteins bound to additional DNA sites. XerC makes the initial cleavage at psi sites resulting in ordered strand exchange, while XerD can initiate cleavage at dif sites (19,35–38).
Lambda Int is a factor-assisted recombinase that contains three domains. It requires accessory proteins to form a protein–DNA complex called an intasome that catalyzes excision and integration (28,39). The arm-type DNA sites contain both Int and accessory protein-binding sites, and are arranged asymmetrically around the core-type sites where cleavage and strand exchange takes place (40). Kitts and Nash used inverted attP sites to determine whether the core or arm-type sites dictate the order of strand exchange. They found that regardless of the orientation of the core, initial cleavage takes place at the site of the overlap region closest to the P arm. Their results established that the order of strand exchange is determined by the asymmetry of the arm-type sites (41). Interestingly, when the N-terminal arm-binding domain of Lambda Int is attached to Cre, the chimera becomes dependent on IHF, Xis and Fis to regulate directionality in the same fashion as Lambda Int (42). This suggests that the factor-assisted recombinases are particularly reliant upon the contribution of the accessory proteins and the spatial arrangement of the additional DNA sites around the core to determine the mechanism and order of strand exchange.
Like Lambda Int, IntDOT possesses an arm-binding N-terminal domain. The attDOT site contains five IntDOT-binding sites arranged asymmetrically around the core site, and intasome formation requires additional accessory proteins (25,43). Despite the asymmetric arrangement of the arm-type sites relative to the core sites, the order of strand exchange appears to be predominantly dictated by the core sites. In a reaction with wild-type sites, IntDOT preferentially cleaves and exchanges the top strand first. The first strand exchange requires a sequence identity between the recombining att sites, but the second strand exchange is independent of sequence identity. It has also been shown that the identity of the base pairs at the site of cleavage is not important, so long as they are identical between the two sites (26). We have shown here that the location of sequence identity within the overlap region determines the order of strand exchange in IntDOT-catalyzed integration, and that the arm-type sites are less important than in other systems.
Tyrosine recombinases catalyze recombination by performing a pair of strand exchanges at opposite ends of the overlap region. Most tyrosine recombinases have an absolute requirement for sequence identity within the overlap region of recombining substrates, and often the initial strand exchange is dependent upon sequence identity between recombining sites. It has been suggested that Watson–Crick base pairing must take place between the partner strands after strand swapping in order for the ligation reaction to occur (16). If no base pairing can occur, then the ligation step is not likely to take place. This idea is supported by work done on Flp showing that the ligation step is the sequence identity dependent step (44). Likewise, mutations in the overlap region of loxP nearly abolishes recombination between wild-type and mutant loxP sites (45).
Sites containing heterology between the overlap regions are very poor recombination substrates (46). For example, Lambda ‘site affinity’ mutants containing mutations within the overlap region of attP or attB reduce recombination, but when the same mutations are made in both att sites, recombination is restored. Mutations made directly to the left of the cleavage sites, outside of the overlap region, had no effect on recombination (47). It has also been shown that sequence identity in the left side of the overlap region is strongly conserved in Lambda secondary attachment sites, supporting a critical role for sequence identity between recombining substrates (48). Perhaps the sequence identity on the left side promotes initial strand exchange to form the Holliday junction intermediate. IntDOT can resolve the HJ into products due to its tolerance of heterology whereas the Lambda HJ requires host processes to resolve it into products in the presence of heterology.
Studies done on IntDOT using synthetic ligation substrates containing p-nitrophenol at the site of cleavage and ligation have shown that IntDOT can perform the ligation reaction in the presence of heterology (26). The same study also used bridging phosphorothioate substrates to show that cleavage can also take place in the presence of a heteroduplex in the overlap region (26). These results make the role of sequence identity in IntDOT-mediated recombination unclear and raise the question as to why sequence identity is so important within the DOT overlap region for one strand exchange while heterology is tolerated extremely well for the second strand exchange.
We assume that IntDOT, like other tyrosine recombinases, forms an intasome complex with attDOT during integration and that the intasome synapses with a naked attB site (28). Synapsis of the intasome with attB can occur in two orientations: the intasome can synapse with the attB site from the ‘top’, or from the ‘bottom’. Theoretically, cleavage and initial strand exchange could take place in either orientation, but the reaction does not proceed unless Watson–Crick base pairing can take place between the newly exchanged strands. If a complementary base is not available, the reaction is reversed. The attDOT and attB sites are formed, and the attB site is released. If a complementary base is available in the partner site, the ligation reactions occur and first strand exchange is executed. A second round of cleavage and ligation reactions occur at the other border of the overlap region, which do not require Watson–Crick base pairs at the ligation sites and recombinants are formed. This is consistent with the results of our symmetric overlap recombination experiments. Because the conserved GC dinucleotide is present on both the top and bottom strands of attB at the cleavage sites, the attDOT/intasome utilizes both sites of sequence identity with equal frequency.
Warren et al. (42) suggest that two evolutionary steps would be required for an autonomous recombinase like Cre to evolve into the more factor-assisted recombinase like Lambda Int. These steps are the addition of another DNA-binding domain and a ‘detuning’ of the recombinase, which involves a decrease in the affinity for particular DNA sites and/or reduced protein–protein interactions. IntDOT appears to be an evolutionary intermediate that has gained the second DNA-binding domain but has not yet lost its relatively high affinity for the core-type DNA sites. Combined with a relaxed requirement for sequence identity within the overlap region, these characteristics suggest that IntDOT may be a member of a new subclass of tyrosine recombinase that could potentially include the integrases of other conjugative transposons such as Tn916 and Tn1545 that recombine att sites containing non-identical overlap regions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institutes of Health [GM 28717].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Abigail Salyers, Nadja Shoemaker and Margaret McCurdy for helpful discussions, and Scott Janus for technical assistance.
REFERENCES
- 1.Macy JM, Probst I. The biology of gastrointestinal bacteroides. Ann. Rev. Microbiol. 1979;33:561–594. doi: 10.1146/annurev.mi.33.100179.003021. [DOI] [PubMed] [Google Scholar]
- 2.Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 2001;67:561–568. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whittle G, Shoemaker NB, Salyers AA. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell Mol. Life Sci. 2002;59:2044–2054. doi: 10.1007/s000180200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song B, Shoemaker NB, Gardner JF, Salyers AA. Integration site selection by the Bacteroides conjugative transposon CTnBST. J. Bacteriol. 2007;189:6594–6601. doi: 10.1128/JB.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vincent Burrus GP, Bernard D, Guèdon G. Conjugative transposons: the tip of the iceberg. Mol. Microbio. 2002;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 6.Shoemaker NB, Barber RD, Salyers AA. Cloning and characterization of a Bacteroides conjugal tetracycline-erythromycin resistance element by using a shuttle cosmid vector. J. Bacteriol. 1989;171:1294–1302. doi: 10.1128/jb.171.3.1294-1302.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedzyk LA, Shoemaker NB, Young KE, Salyers AA. Insertion and excision of Bacteroides conjugative chromosomal elements. J. Bacteriol. 1992;174:166–172. doi: 10.1128/jb.174.1.166-172.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoemaker NB, Salyers AA. A cryptic 65-kilobase-pair transposonlike element isolated from Bacteroides uniformis has homology with Bacteroides conjugal tetracycline resistance elements. J. Bacteriol. 1990;172:1694–1702. doi: 10.1128/jb.172.4.1694-1702.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esposito D, Scocca JJ. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 1997;25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malanowska K, Salyers AA, Gardner JF. Characterization of a conjugative transposon integrase, IntDOT. Mol. Microbiol. 2006;60:1228–1240. doi: 10.1111/j.1365-2958.2006.05164.x. [DOI] [PubMed] [Google Scholar]
- 11.Nunes-Duby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajeev L, Malanowska K, Gardner JF. Challenging a paradigm: the role of DNA homology in tyrosine recombinase reactions. Microbio. Mol. Bio. Rev. 2009;73:300–309. doi: 10.1128/MMBR.00038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng C, Kussie P, Pavletich N, Shuman S. Conservation of Structure and Mechanism between Eukaryotic Topoisomerase I and Site-Specific Recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 14.Azaro M, Landy A. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: ASM Press; 2002. pp. 119–148. [Google Scholar]
- 15.Craig NL, Nash HA. The mechanism of phage lambda site-specific recombination: site- specific breakage of DNA by Int topoisomerase. Cell. 1983;35:795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- 16.Nunes-Düby SE, Azaro MA, Landy A. Swapping DNA strands and sensing homology without branch migration in lambda site-specific recombination. Curr. Biol. 1995;5:139–148. doi: 10.1016/s0960-9822(95)00035-2. [DOI] [PubMed] [Google Scholar]
- 17.McCulloch R, Coggins LW, Colloms SD, Sherratt DJ. Xer-mediated site-specific recombination at cer generates Holliday junctions in vivo. EMBO J. 1994;13:1844–1855. doi: 10.1002/j.1460-2075.1994.tb06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blakely G, Colloms S, May G, Burke M, Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991;3:789–798. [PubMed] [Google Scholar]
- 19.Colloms SD, McCulloch R, Grant K, Neilson L, Sherratt DJ. Xer-mediated site-specific recombination in vitro. EMBO J. 1996;15:1172–1181. [PMC free article] [PubMed] [Google Scholar]
- 20.Nunes-Düby SE, Yu D, Landy A. Sensing homology at the strand-swapping step in lambda excisive recombination. J. Mol. Biol. 1997;272:493–508. doi: 10.1006/jmbi.1997.1260. [DOI] [PubMed] [Google Scholar]
- 21.Van Duyne GD. A Structural View of Tyrosine Recombinase Site-specific Recombination. Washington, DC: ASM Press; 2002. [Google Scholar]
- 22.Nash HA. Purification and properties of the bacteriophage lambda Int protein. Methods Enzymol. 1983;100:210–216. doi: 10.1016/0076-6879(83)00057-9. [DOI] [PubMed] [Google Scholar]
- 23.Trask DK, DiDonato JA, Muller MT. Rapid detection and isolation of covalent DNA/protein complexes: application to topoisomerase I and II. EMBO J. 1984;3:671–676. doi: 10.1002/j.1460-2075.1984.tb01865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Q, Wesslund N, Shoemaker NB, Salyers AA, Gardner JF. Development of an in vitro integration assay for the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 2002;184:4829–4837. doi: 10.1128/JB.184.17.4829-4837.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Q, Paszkiet BJ, Shoemaker NB, Gardner JF, Salyers AA. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 2000;182:4035–4043. doi: 10.1128/jb.182.14.4035-4043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malanowska K, Yoneji S, Salyers AA, Gardner JF. CTnDOT integrase performs ordered homology-dependent and homology-independent strand exchanges. Nucleic Acids Res. 2007;35:5861–5873. doi: 10.1093/nar/gkm637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitts PA, Nash HA Laboratory of Molecular Biology, N.I.o.M.H.B.M.D. Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J. Mol. Biol. 1988;204:95–107. doi: 10.1016/0022-2836(88)90602-x. [DOI] [PubMed] [Google Scholar]
- 28.Richet E, Abcarian P, Nash HA. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein–DNA complex. Cell. 1988;52:9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- 29.Hsu PL, Ross W, Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980;285:85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee L, Sadowski PD. Progress in Nucleic Acid Research and Molecular Biology. Vol. 80. New York: Academic Press; 2005. pp. 1–42. [DOI] [PubMed] [Google Scholar]
- 31.Amin AA, Beatty LG, Sadowski PD. Synaptic intermediates promoted by the FLP recombinase. J. Mol. Biol. 1990;214:55–72. doi: 10.1016/0022-2836(90)90146-D. [DOI] [PubMed] [Google Scholar]
- 32.Guo F, Gopaul DN, Van Duyne GD. Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proc. Natl Acad. Sci. USA. 1999;96:7143–7148. doi: 10.1073/pnas.96.13.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin SS, Pulido E, Chu VC, Lechner TS, Baldwin EP. The order of strand exchanges in Cre-LoxP recombination and its basis suggested by the crystal structure of a Cre-LoxP Holliday junction complex. J. Mol. Biol. 2002;319:107–127. doi: 10.1016/S0022-2836(02)00246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee L, Sadowski PD. Sequence of the loxP site determines the order of strand exchange by the Cre recombinase. J. Mol. Biol. 2003;326:397–412. doi: 10.1016/s0022-2836(02)01429-8. [DOI] [PubMed] [Google Scholar]
- 35.Hallet B, Arciszewska LK, Sherratt DJ. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: an enzymatic switch in site-specific recombination. Mol. Cell. 1999;4:949–959. doi: 10.1016/s1097-2765(00)80224-5. [DOI] [PubMed] [Google Scholar]
- 36.Arciszewska LK, Sherratt DJ. Xer site-specific recombination in vitro. EMBO J. 1995;14:2112–2120. doi: 10.1002/j.1460-2075.1995.tb07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 38.Recchia GD, Aroyo M, Wolf D, Blakely G, Sherratt DJ. FtsK-dependent and -independent pathways of Xer site-specific recombination. EMBO J. 1999;18:5724–5734. doi: 10.1093/emboj/18.20.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patsey RL, Bruist MF. Characterization of the interaction between the lambda intasome and attB. J. Mol. Biol. 1995;252:47–58. doi: 10.1006/jmbi.1995.0474. [DOI] [PubMed] [Google Scholar]
- 40.Ross W, Landy A. Bacteriophage lambda Int protein recognizes two classes of sequence in the phage att site: characterization of arm-type sites. Proc. Natl Acad. Sci. USA. 1982;79:7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitts PA, Nash HA. Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J. Mol. Biol. 1988;204:95–107. doi: 10.1016/0022-2836(88)90602-x. [DOI] [PubMed] [Google Scholar]
- 42.Warren D, Laxmikanthan G, Landy A. A chimeric Cre recombinase with regulated directionality. Proc. Natl Acad. Sci. USA. 2008;105:18278–18283. doi: 10.1073/pnas.0809949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dichiara JM, Mattis AN, Gardner JF. IntDOT interactions with core- and arm-type sites of the conjugative transposon CTnDOT. J. Bacteriol. 2007;189:2692–2701. doi: 10.1128/JB.01796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J, Jayaram M. Role of partner homology in DNA recombination. J. Biol. Chem. 1995;270:4042–4052. doi: 10.1074/jbc.270.8.4042. [DOI] [PubMed] [Google Scholar]
- 45.Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in PI site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weisberg RA, Enquist LW, Foeller C, Landy A. Role for DNA homology in site-specific recombination. The isolation and characterization of a site affinity mutant of coliphage lambda. J. Mol. Biol. 1983;170:319–342. doi: 10.1016/s0022-2836(83)80151-x. [DOI] [PubMed] [Google Scholar]
- 47.Bauer CE, Gardner JF, Gumport RI. Extent of sequence homology required for bacteriophage lambda site-specific recombination. J. Mol. Biol. 1985;181:187–197. doi: 10.1016/0022-2836(85)90084-1. [DOI] [PubMed] [Google Scholar]
- 48.Ross W, Landy A. Patterns of lambda Int recognition in the regions of strand exchange. Cell. 1983;33:261–272. doi: 10.1016/0092-8674(83)90355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.