Abstract
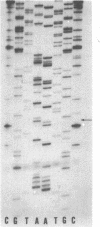
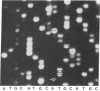
A method for sequencing DNA by using a difluoresceinated primer and laser excitation is described. Dideoxy protocols have been determined that provide sequences for 600 bases starting with base 1 with less than 1% error in a single load. Electrophoresis is at 20 W and the bands are detected 24 cm from the bottom of the loading well with a scanning fluorescence detector. Bands are imaged on a TV screen in two dimensions. The sequences can be read from the TV screen manually or semiautomatically by using a simple software program. The system allows more bases to be read with a lower error rate than any other reported automated sequencing method.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansorge W., Barker R. System for DNA sequencing with resolution of up to 600 base pairs. J Biochem Biophys Methods. 1984 Mar;9(1):33–47. doi: 10.1016/0165-022x(84)90064-2. [DOI] [PubMed] [Google Scholar]
- Ansorge W., Sproat B. S., Stegemann J., Schwager C. A non-radioactive automated method for DNA sequence determination. J Biochem Biophys Methods. 1986 Dec;13(6):315–323. doi: 10.1016/0165-022x(86)90038-2. [DOI] [PubMed] [Google Scholar]
- Ansorge W., Sproat B., Stegemann J., Schwager C., Zenke M. Automated DNA sequencing: ultrasensitive detection of fluorescent bands during electrophoresis. Nucleic Acids Res. 1987 Jun 11;15(11):4593–4602. doi: 10.1093/nar/15.11.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S., Pohl F. M. DNA sequencing with direct blotting electrophoresis. EMBO J. 1984 Dec 1;3(12):2905–2909. doi: 10.1002/j.1460-2075.1984.tb02230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulbecco R. A turning point in cancer research: sequencing the human genome. Science. 1986 Mar 7;231(4742):1055–1056. doi: 10.1126/science.3945817. [DOI] [PubMed] [Google Scholar]
- Garoff H., Ansorge W. Improvements of DNA sequencing gels. Anal Biochem. 1981 Aug;115(2):450–457. doi: 10.1016/0003-2697(81)90031-2. [DOI] [PubMed] [Google Scholar]
- Jablonski E., Moomaw E. W., Tullis R. H., Ruth J. L. Preparation of oligodeoxynucleotide-alkaline phosphatase conjugates and their use as hybridization probes. Nucleic Acids Res. 1986 Aug 11;14(15):6115–6128. doi: 10.1093/nar/14.15.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores J. C. Current approaches to DNA sequencing. Anal Biochem. 1987 May 15;163(1):1–8. doi: 10.1016/0003-2697(87)90084-4. [DOI] [PubMed] [Google Scholar]
- Prentki P., Karch F., Iida S., Meyer J. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene. 1981 Sep;14(4):289–299. doi: 10.1016/0378-1119(81)90161-x. [DOI] [PubMed] [Google Scholar]
- Prober J. M., Trainor G. L., Dam R. J., Hobbs F. W., Robertson C. W., Zagursky R. J., Cocuzza A. J., Jensen M. A., Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987 Oct 16;238(4825):336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. M., Fung S., Hunkapiller M. W., Hunkapiller T. J., Hood L. E. The synthesis of oligonucleotides containing an aliphatic amino group at the 5' terminus: synthesis of fluorescent DNA primers for use in DNA sequence analysis. Nucleic Acids Res. 1985 Apr 11;13(7):2399–2412. doi: 10.1093/nar/13.7.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. M., Sanders J. Z., Kaiser R. J., Hughes P., Dodd C., Connell C. R., Heiner C., Kent S. B., Hood L. E. Fluorescence detection in automated DNA sequence analysis. Nature. 1986 Jun 12;321(6071):674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- Wada A. Automated high-speed DNA sequencing. 1987 Feb 26-Mar 4Nature. 325(6107):771–772. doi: 10.1038/325771a0. [DOI] [PubMed] [Google Scholar]