Abstract
Mutations in either the hereditary hemochromatosis protein, HFE, or transferrin receptor 2, TfR2, result in a similarly severe form of the most common type of iron overload disease called hereditary hemochromatosis. Models of the interactions between HFE, TfR1, and TfR2 imply that these proteins are present in different molar concentrations in the liver, where they control expression of the iron regulatory hormone, hepcidin, in response to body iron loading. The aim of this study was to determine in vivo levels of mRNA by quantitative RT-PCR and concentrations of these proteins by quantitative immunoblotting in human liver tissues. The level of TfR2 mRNA was 21- and 63- fold higher than that of TfR1 and HFE, respectively. Molar concentration of TfR2 protein was the highest and determined to be 1.95 nmoles/g protein in whole cell lysates and 10.89 nmoles/g protein in microsomal membranes. Molar concentration of TfR1 protein was 4.5- and 6.1-fold lower than that of TfR2 in whole cell lysates and membranes, respectively. The level of HFE protein was below 0.53 nmoles/g of total protein. HFE is thus present in substoichiometric concentrations with respect to both TfR1 and TfR2 in human liver tissue. This finding supports a model, in which availability of HFE is limiting for formation of complexes with TfR1 or TfR2.
Keywords: HFE, TfR1, TfR2, hereditary hemochromatosis, human liver
INTRODUCTION
Hereditary hemochromatosis (HH)1 is an autosomal, inherited disorder of iron homeostasis characterized by hepatocellular iron overload that ranges from mild to severe (reviewed in [1; 2; 3]). HH is associated with mutations in at least five genes. On the basis of the gene involved, HH is classified as type 1 (hereditary hemochromatosis, HFE) [4], type 2A (hemojuvelin, HFE2) [5], type 2B (hepcidin, HAMP) [6], type 3 (transferrin receptor 2, TFR2) [7], and type 4 (ferroportin, FPN) [8]. Type 1 is the most common form of HH [4].
HFE is a type I transmembrane protein that belongs to the MHC-I like family of proteins. Like MHC-I proteins, HFE also forms a heterodimer with β2-microglobulin (β2M) [4; 9]. The most common mutation in the HFE protein, C282Y[4], results in destabilization of the α3 domain, which abrogates the interaction between HFE and β2M [4]. As a result, the mutant C282Y-HFE protein has impaired ability to reach the cell surface [4; 10; 11]. The second most common mutation is H63D [4], but the mechanism by which this mutation causes HH is unknown. Interestingly, there is a considerable variation in iron loading in individuals with these two mutations [1; 2]. Such heterogeneity suggests that HFE function depends on the presence of modifiers, which might be proteins that interact with HFE.
The first identified binding partner of HFE was the transferrin receptor 1 (TfR1) [12; 13], a ubiquitous cell surface receptor that binds and internalizes iron-loaded transferrin (holo-Tf). HFE/TfR1 complex dissociates in the presence of holo-Tf because holo-Tf competes with HFE for binding to TfR1 [14; 15; 16]. The discovery that hepcidin, an iron regulatory hormone predominantly expressed in hepatocytes [17], is decreased in both HH type 1 patients [18] and Hfe-/- mice [19; 20] and that HFE is also predominantly expressed in hepatocytes [21] indicated that the primary site of HFE effects on iron homeostasis is the liver. These observations lead to a “hepcidin hypothesis”, in which HFE is an upstream regulator of hepcidin expression (reviewed in [22]). Observations that mice lacking Hfe in the crypt- and villi- enterocytes have no detectable iron loading [23] while mice lacking Hfe in hepatocytes manifest iron overload [24] emphasize the importance of HFE expression in hepatocytes.
Recently, transferrin receptor 2 (TfR2), a homolog of TfR1 that is predominantly expressed in hepatocytes [25], was reported to bind to HFE [26]. Interestingly, the interacting domains of HFE and TfR2 [27] are different from those of HFE and TfR1. First, HFE interacts with TfR2 via its α3 domain, versus with TfR1 via its α1 and α2 domains. Second, the Tf binding site of TfR2 does not overlap with the HFE binding site as it does in TfR1. Thus, in contrast to the HFE/TfR1 complex, the HFE/TfR2 complex does not dissociate, even in the presence of high Tf concentrations [27]. This finding suggests a new model of HFE-dependent regulation of hepcidin expression, in which HFE is released from TfR1 and binds to TfR2, with increasing iron-loaded Tf concentrations. Two recent studies expand these findings. The first work analyzes Hfe and Tfr1 interactions in mice models of HH type 1. Expression of mutant forms of mouse Tfr1 that either prevent or stabilize Hfe/Tfr1 interactions results in an Hfe-dependent induction of hepcidin expression, which occurs only when Hfe is dissociated from Tfr1 [28]. The second study demonstrates that in the presence of holo-Tf, human hepatoma cells that express undetectable HFE but readily detectable TfR1 and TfR2 proteins regulate hepcidin expression only when exogenous HFE is expressed [29]. This study lead to the proposal that TfR1 sequesters HFE from TfR2 under low iron conditions, but under high iron conditions, the increased iron saturation of Tf shifts the balance towards creation of an HFE/TFR2 hepcidin signaling complex [28]. In this process, HFE represents the limiting factor during reorganization of HFE/TfR1 and HFE/TfR2 complexes [29].
In order to better understand the mechanism by which these complexes are formed as well as their response to iron levels, it is important to know the relative amounts of HFE, TfR1, and TfR2 in the liver. Thus, we tested the hypothesis that in human liver, where the HFE-dependent regulation of hepcidin expression occurs, the molar concentration of HFE is similar to or lower than that of TfR1 or TfR2. Both the mRNA and protein levels of HFE, TfR1, and TfR2 in human liver tissues were measured. Our results showed that mRNA and protein levels of TfR2 are significantly higher than TfR1 and HFE levels. The least abundant is the HFE protein, supporting the proposed model of hepcidin regulation in vivo.
METHODS
Cell culture
TRVb cells that lack the endogenous transferrin receptor 1 (TFRC) and that do not express detectable HFE and TfR2 (kindly provided by Dr. Timothy McGraw, Cornell University, New York, USA) were grown in F-12 Coon’s Modification, 5% fetal bovine serum (FBS) and 2 mg/ml glucose. TRVb/HFE/β2M cells stably expressing HFE with C-terminal FLAG epitope tag (HFE-FLAG) and β2M [30] were grown in the same medium supplemented with 300 μg/ml hygromycin. HeLa/tTA-HFE-FLAG cells [31] that stably express HFE-FLAG in a tet-off system were grown in DMEM/10 % FBS supplemented with 400 μg/ml G418 (Geneticin, Calbiochem) and 300 ng/ml puromycin, with (dox+) or without (dox−) 1 μg/ml doxycycline. HepG2/tTA HFE cells [29] that stably express HFE-FLAG in a tet-on system were maintained in DMEM/10% FBS supplemented with 2 mg/ml L-glutamate, 5 μg/ml blasticidine, and 400 μg/ml G418. Doxycyline (0.2 μg/ml) was used to induce expression of HFE-FLAG in HepG2/tTA cells. All cell-lines were grown at 37°C with 5% CO2.
Human tissue samples
Human liver tissue samples were obtained from the Oregon Cancer Center Bank by biopsy of normal liver tissues from donors with conditions not related to iron disease. The study was reviewed by the IRB board and considered exempt #6891. The liver specimens were obtained 30-120 minutes after the biopsy. Each specimen was cut into small pieces (~1-2 g) and snap frozen in liquid nitrogen. The frozen tissues were stored at −80°C until the analysis. No information on iron status of the donors was available to us. Therefore, the levels of the liver nonheme iron were determined, where possible, and found normal (100-400 μg/g wet tissue).
Real-time quantitative reverse transcriptase PCR (qRT-PCR)
Eight liver tissues (1787, 2152, 2154, 3273, 7416, 7511, 7696, and 7836) were included in the following analysis. Total RNA prepared using RNAeasy isolation kit (Qiagen, Valencia, CA) was treated with DNAase (Roche Diagnostics) to remove contaminating genomic DNA. RNA (2 μg) was used to synthesize cDNA using Oligo dT primers and Superscript II Reverse Transcriptase (RT) (Invitrogen, Carlsbad, CA). Samples were then analyzed by real-time qRT-PCR using the SYBR green detection system on an ABI PRISM 7900 machine. The primer pairs used for quantitative amplification of GAPDH, HFE, TFRC, and TFR2 cDNAs were previously described [32; 33]. Data were analyzed using the ΔCT (difference in threshold cycles) method [21; 32]. The results for each gene of interest were normalized to the levels of GAPDH.
Quantitative immunoblot analysis
Purified, soluble HFE (sHFE) and TfR2 (sTfR2) proteins were a kind gift from Dr. Pamela Bjorkman (California Institute of Technology, Pasadena, California). Full-length human TfR1 was purified from human placenta as described in [34]. Increasing amounts of purified proteins were separated on 10% SDS-PAGE followed by protein transfer to nitrocellulose. Membranes were probed with primary antibodies (see below) and visualized with fluorescently-labeled Alexa 680 goat anti-rabbit IgG (Molecular Probes, Eugene, OR) or IRDye 800 donkey anti-mouse IgG (Rockland Immunochemicals, Gilbertsville, PA). Membranes were scanned using the Odyssey Infrared Imaging System (Li-Cor, Lincoln, NB). Intensities of individual bands were quantified using Odyssey software. For detection of human TfR1, monoclonal mouse anti-TfR1 3B82A1 [35] at dilution of 1:1,000 was used. HFE and TfR2 were detected with polyclonal rabbit anti-HFE EX1 [10] at dilution of 1:1,000 and anti-TfR2 16637 [33] at dilution of 1:10,000, respectively. Polyclonal rabbit anti-HFE EX1 was a kind gift from Dr. John Feder (Bristol Meyers Squibb). Polyclonal rabbit anti-HFE CT16 (gift from Dr. Robert E. Fleming, Saint Louis University, Saint Louis, Missouri) was used at dilution of 1:1,000. Initially, calibration curves for individual proteins were compared in the absence (−WCL) and presence (+WCL) of whole cell lysates. Here, purified proteins were mixed with whole cell extracts prepared from TRVb cells, which do not express endogenous HFE, TfR1, or TfR2. Addition of extraneous proteins affected the intensities of protein bands for all protein standards (see Supplementary Figure 1 online), presumably by preventing absorption to the plastic tubes. Therefore, whole cell lysates were added to purified protein standards in all analyses described in this work.
Preparation of whole cell lysates and microsomal membranes
Whole cell lysates of cultured cells were prepared by lysing the cells in the ice-cold NET buffer (150 mM NaCl, 5 mM EDTA, 10 mM Tris, pH 7.4) containing 1% Triton X-100 and 1x Complete Mini Protease Inhibitor Mix (Roche Applied Science, Indianapolis, IN). Lysates were clarified by centrifugation at 12,000 × g for 15 minutes. Membranes from HepG2 cells were prepared by homogenization of the cells in the ice-cold homogenization buffer (50 mM NaPi pH 7.2, 250 mM sucrose, 100μM PMSF) followed by centrifugation at 12,000 × g for 10 minutes. Microsomal membranes from the supernatant were pelleted at 120,000 × g for 90 minutes, and resuspended in NET buffer (150 mM NaCl, 5 mM EDTA, 10 mM Tris, pH 7.4) containing 1x Complete Mini Protease Inhibitor Mix. Cell lysates and membranes from liver tissues were prepared as described above, with the exception that the tissue was first crushed in liquid nitrogen in a ceramic mortar, and the crushed tissue was homogenized with 15 strokes in a glass homogenizer. Whole cell lysates and membranes were either immediately used for analyses or snap frozen in liquid nitrogen and stored at −80°C. Protein concentration was determined using the BCA Protein Assay kit (PIERCE, Rockford, IL).
Quantification of TfR1 and TfR2 proteins
Thirteen liver biopsies (1, 2152, 2514, 3237, 5116, 5200, 5207, 5213, 5588, 7416, 7511, 7696, and 7715) were analyzed for TfR1 and TfR2 levels. For quantitative immunoblotting, 5-100 μg of the total whole cell lysate or membrane protein were loaded on 10% SDS-PAGE followed by fluorescent immundetection, as described above. Intensities of individual protein bands were plotted against the standard curves for the purified proteins that were run beside human liver proteins. The number of moles of TfR1 was calculated using a molecular weight of 84,900 for the unglycosylated protein. The number of moles of TfR2 was calculated first using a molecular weight of 74,000 for the soluble protein (sTfR2) and then normalized to a molecular weight of 88,800 for the full-length, glycosylated protein.
Generation of Affi-Gel-10 antibody resin
Mouse monoclonal anti-HFE antibodies 8C10 (kind gift from Dr. Maria DeSousa and Dr. Jorge Pinto, Universidade do Porto, Porto, Portugal) were produced in CELLLine Device (BD Biosciences, Bedford, MA) according to manufacturer’s protocol. Antibodies were then purified on Affigel-Protein A column using MAPS II Kit (BioRad, Hercules, CA), concentrated, and extensively dialyzed against PBS to remove amines. Dialyzed antibodies were coupled to Affigel-10 for 4 hours at 4°C in PBS according to the manufacturer’s instructions and then cross-linked in the presence of 20mM dimethyl pimelimidate [36]. The resulting Affi-Gel-10/8C10 resin was washed in PBS and stored as 10% slurry in PBS supplemented with 0.05% sodium azide and 0.1% Triton X-100. In parallel, reactive groups on an aliquot of the Affi-Gel-10 resin were also blocked, and the resin was stored as described for Affi-Gel-10/8C10. Affi-Gel-10 was used as a control for non-specific binding.
Immunoprecipitation
Whole cell lysates prepared from HeLa/tTA-HFE-FLAG cells (25 μg), HEK293/HFE cells (25 μg), or human liver microsomal membranes solubilized in 1% Triton X-100 (100 μg - 20 mg), were used for immunoprecipitation experiments. Lysates were incubated with 40 μl of either Affi-Gel-10 (negative control) or Affi-Gel-10/8C10 resin for 1 hour at 4°C. Immobilized complexes were pelleted and washed three times with the ice-cold NET buffer containing 1% Triton X-100. Beads were resuspended in 2 × Laemmli buffer, eluted proteins transferred to a new tube, heated at 95°C for 5 minutes, and separated on 10% SDS-PAGE. Immunprecipitated proteins were detected by immunoblotting.
Data analysis
Student paired, two-tailed t test was used to analyze data sets for both mRNA and quantitative immunoblotting analyses.
RESULTS
Levels of HFE, TfR1, and TfR2 mRNAs
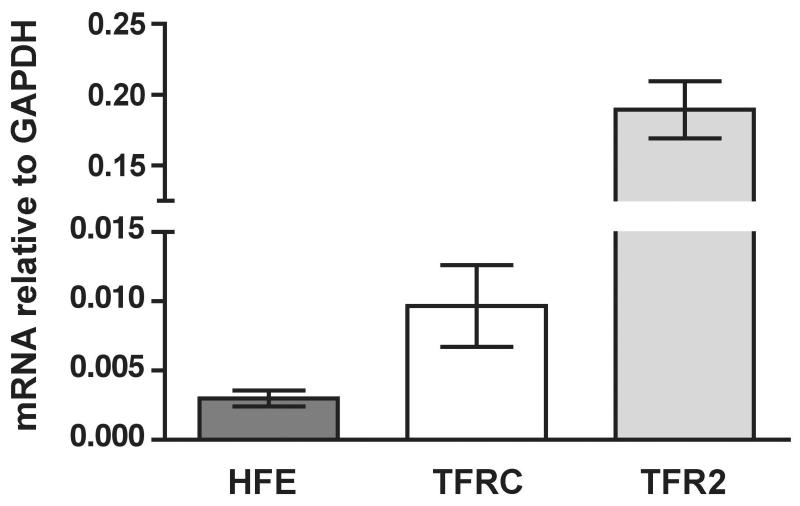
Recently, our group and others have focused on studies of HFE complexes with TfR1 and TfR2 in cultured cells [26; 27] and in mouse models [28] of HH. In this study, we decided to concentrate on analysis of in vivo levels of these individual players because such information is critical for interpretation of biochemical studies performed in hepatic tissues and hepatic cells. First, mRNA levels were analyzed. Total RNA was isolated from eight control liver samples, and the levels of HFE, TFRC, and TFR2 mRNAs were analyzed by qRT PCR (Fig. 1). Individual genes were expressed very differently. TFR2 mRNA was the highest, about 20-fold higher than that of TFRC. HFE mRNA was least abundant, ~ 63-fold and ~ 3-fold lower than TFR2 and TFRC mRNAs, respectively.
Figure 1.
Quantification of HFE, TFRC, and TFR2 mRNAs in human liver. Bars show the mean value, normalized to GAPDH levels, calculated from at least two independent experiments. Error bars represent standard deviation. Differences between individual pairs were statistically significant with p values 0.0264 for HFE/TFRC, 0.0002 for TFRC/TFR2, and <0.0001 for HFE/TFR2 pair.
Quantitative immunoblot analysis
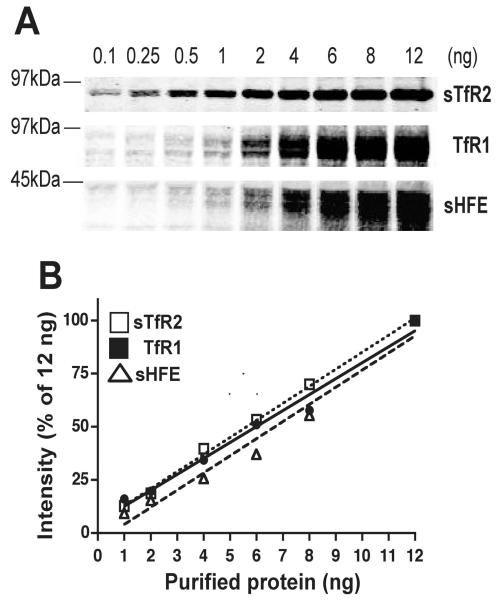
A quantitative immunoblot analysis was developed to determine wheter the protein levels reflected the mRNA levels. Purified proteins were used to determine the sensitivity of available antibodies against HFE, TfR1, and TfR2, as well as the range of protein concentration within which the intensities of signal remained linear. Increasing amounts of individual purified proteins were subjected to SDS-PAGE, followed by immunodetection using appropriate antibodies. The anti-TfR2 antibodies used in this study reproducibly detected 0.1 ng of purified, soluble TfR2 (sTfR2). The anti-TfR1 and anti-HFE antibodies were about 10-20 times less sensitive than the anti-TfR2 antibodies (Fig. 2A). Quantitative analysis of immunoblots revealed that the signal obtained for individual proteins remained linear over the range of protein concentrations measured (Fig. 2B). Protein levels in subsequent experiments were all quantified within the linear range of the assay.
Figure 2.
Quantitative immunoblot analysis of purified sHFE, TfR1, and sTfR2 proteins. (A) Detection limit for individual antibodies. Representative immunoblots are shown for each protein from at least ten independent experiments with similar results. (B) Linearity of quantitative immunoblot analysis. Quantification of individual bands from (A). Correlation coefficients were R2 = 0.9596 for sHFE, R2 = 0.9152 for TfR1, and R2 = 0.9786 for sTfR2.
Stoichiometries of TfR1 and TfR2 proteins
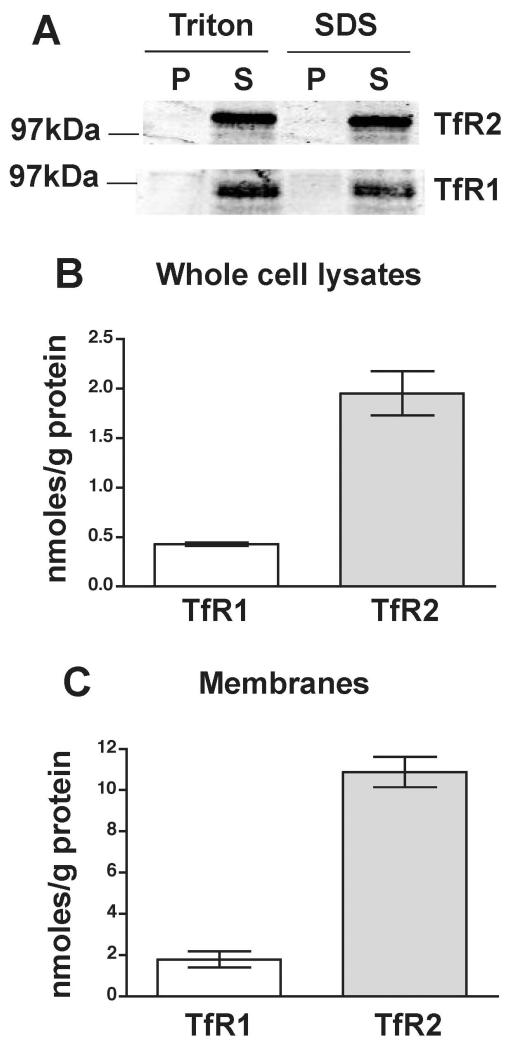
Quantification of TfR1 and TfR2 proteins was done in both whole cell lysates and microsomal membrane preparations. Whole cell lysates were prepared in the presence of Triton X-100 while membranes were prepared in the absence of detergent (see Methods). Triton X-100 extracted both TfR1 and TfR2 with similar efficiency as SDS did in control experiments (Fig. 3A). Whole cell lysates or membranes for each protein were run on SDS-PAGE together with purified standards. TfR1 and TfR2 bands were detected using primary antibodies followed by quantitative analysis of fluorescent signals on immunoblots as described in Methods.
Figure 3.
Molar concentrations of TfR1 and TfR2 proteins in human liver. (A) Extraction of TfR1 and TfR2. Whole cell lysates were prepared in the presence of 1% Triton X-100 alone (Triton) or in the presence of 1% Triton X-100 and 0.3 % SDS (SDS). Similar amounts of total protein (100 μg) were loaded per lane. Representative immunoblots are shown from three independent experiments with similar results. P, pellet; S, supernatant (extracted protein). TfR1 and TfR2 levels in whole cell lysates (B) and microsomal membranes (C). Bars show mean values from at least three independent experiments, each done in duplicates or triplicates for individual liver sample. Error bars show standard deviation. The differences between each TfR1 and TfR2 set were statistically very significant (p < 0.0001).
Whole cell lysates from 13 livers and membranes from seven of these livers were analyzed. The mean value of the TfR2 protein levels was 1.95 nmoles/g protein in whole cell lysates and 10.98 nmoles/g protein in microsomal membranes (Fig. 3B). TfR1 levels were about 4.5-fold lower (0.43nmoles/g protein) in whole cell lysates and about 6.1-fold lower (1.78 nmoles/g protein) in microsomal membranes than those of TfR2 (Fig. 3C). A small discrepancy between the fold differences observed in whole cell lysates and membranes are most likely to higher sensitivity of the immunoblot when membranes, e.g. enriched fraction, are used as the source material. Nevertheless, the findings in both whole cell lysates and membranes show that TfR2 is present in significantly higher molar concentrations in human liver than TfR1. Moreover, both proteins behave as membrane proteins in our assays since they were enriched in the membrane fraction.
Stoichiometric analysis of HFE
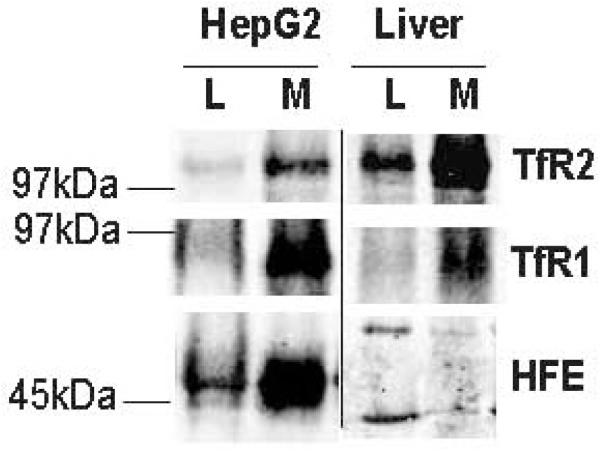
Analysis of the HFE protein levels was similar as those described for TfR1 and TfR2 above (Fig. 3). In HepG2/tTA HFE cells that stably express HFE-FLAG in the presence of doxycyline, HFE was easily detectable and enriched in membrane fraction and thus behaved similarly to TfR1 and TfR2 proteins (Fig. 4, left section). However, the EX1 HFE antibody did not recognize HFE in either whole liver lysates or liver membrane preparations (Fig. 4, right section) despite rigorous optimizations of immunoblot analysis. The EX1 antibody was initially chosen because it had the highest specificity and sensitivity amongst several antibodies tested in HeLa cells stably expressing HFE (data not shown). Rabbit polyclonal anti-HFE CT16 and monoclonal mouse anti-HFE 8C10 showed significantly lower sensitivities than the anti-HFE EX1 antibody against both the purified sHFE and HFE-FLAG expressed in HeLa cells. These two antibodies were also tested for their recognition of a potential HFE band in both human liver whole cell lysates and microsomal membranes; unfortunately, without success (data not shown). Efforts to concentrate HFE by immunoprecipitation also gave negative results (see Supplementary Figure 2 online). The absence of HFE may be caused by the absence of the HFE partner, the β2-microglobulin. However, in all analyzed samples, β2-microglobulin was detectable (see Supplementary Figure 3 online), which excluded the possibility that the HFE protein was undetectable due to the absence of its essential folding partner. The data indicate that the levels of HFE are below the sensitivity of the current immunoblotting procedure, which we estimated to be 1-2 ng of purified sHFE (Fig. 2A), and that the HFE protein is even less abundant than TfR1.
Figure 4.
Detection of HFE protein in human liver. HFE expressed in cell culture behaves as membrane protein. Similar amounts (100 μg) of total protein were loaded per lane. HepG2, HepG2 cells stably expressing HFE-FLAG in the presence of doxycycline; L, whole cell lysate; M, microsomal membranes.
DISCUSSION
In this work, we determined the stoichiometries of TfR1 and TfR2, two key proteins involved in the regulation of hepcidin expression in response to holo-Tf levels. A recent model suggests that TfR1, via its interaction with HFE, controls levels of HFE that are available for interaction with another binding partner such asTfR2 [28; 29]. However, no in vivo data on the amounts of individual candidates existed that would support these suggestions. Therefore, we tested the hypothesis that HFE is limiting in vivo and determined both mRNA and protein levels for each candidate gene in normal human liver tissue. We observed that HFE is the least abundant while TfR2 is the most abundant of the three proteins tested.
TfR2 mRNA levels were about 21-fold and 63-fold higher than those of TfR1 and HFE, respectively. However, the difference in stoichiometries between TfR2 (1.78 nmoles/g in lysates or 10.98 nmoles/g in membranes) and TfR1 (0.43 nmoles/g in lysates or 1.95 nmoles/g in membranes) proteins was not proportional to the mRNA levels. The observed discrepancy between the differences in mRNA and protein levels may reflect the different mechanisms of TfR1 and TfR2 regulation including rate of translation, as well as posttranslational regulation. TfR1 levels are regulated at the posttranscriptional level via iron regulatory proteins (IRPs), which bind to iron responsive elements (IREs) located in the 3′ untranslated portion of TfR1 mRNA [37; 38; 39; 40]. Thus, the steady-state levels of TfR1 mRNA reflect changes in intracellular iron levels. In addition, the half-life of TfR1 protein in cultured cells is relatively long (~23 hrs) [33], which is consistent with relatively low levels of mRNA compared to TfR2. In sharp contrast to TfR1, intracellular iron stores do not affect the stability of TfR2 mRNA. TfR2 is regulated at the level of protein stability in cultured hepatic cells. In this case, addition of holo-Tf to the media increases the half-life of TfR2 by a factor of ~ 2.8 [33; 41]. Moreover, unlike TfR1, TfR2 protein is much less stable, with a half-life of 2-10 hrs in hepatic cell [33; 42]. Therefore, the observed disproportional differences in mRNA and protein levels between TfR1 and TfR2 might merely reflect differential regulation of these two homologous but functionally distinct receptors.
HFE mRNA levels were about one third of those determined for TfR1. Our data on the abundance of HFE protein molecules in human liver tissues are indirect; nevertheless, they suggest that the levels of HFE protein are much lower than those of TfR1 (below 2 nmoles/g of total protein). This suggestion comes from experiments in which we tried to increase sensitivity of the quantitative immunoblots analysis. Membrane preparations resulted in 5-10 fold enrichment of TfR1 and TfR2 in both HepG2 cells and human liver. Similar fold of enrichment was observed for HFE expressed in HepG2 cells, but no HFE-specific band was detected in human liver. The detection limit of anti-HFE EX1 antibody is about 2 ng of sHFE. A band with similar intensity would correspond to 0.53 nmoles of HFE per gram of total protein, if detected in 100 μg of solubilized membranes. In immunoprecipitation experiments, microgram to milligram amounts of solubilized membrane preparations were used. We therefore conclude that the levels of HFE in human liver are below 0.53 nmoles/g of total protein. These calculations on the amount of HFE in human liver samples argue for the presence of substoichiometric amounts of HFE molecules in vivo with respect to TfR1 and support the prediction that TfR1 can bind all available HFE molecules and release them for interaction with TfR2 when concentrations of holo-Tf increase above a critical level [28]. These ideas are predicated on similar binding affinities of HFE for TfR1 and TfR2, which are presently unknown.
Changes in the composition of protein complexes in response to different stimuli represent one of the mechanisms by which cellular processes are tuned. Recent studies suggest dynamic interactions between HFE and TfR1, as well as between HFE and TfR2 [26; 27; 28]. These models were based on the systems in which either a single or more than one protein was exogenously expressed, and thus the actual in vivo stoichiometries have not been taken into consideration. The finding that the amount of HFE in human liver is significantly lower than that of TfR1 or TfR2 indicates that HFE may be a limiting factor during the rearrangement of HFE/TfR1 and HFE/TfR2/Tf protein complexes in vivo.
Supplementary Material
Supplementary Figure 1. Effect of extraneous proteins on intensity of the immunobands. Representative immunoblots are shown for each protein from at least three independent experiments with similar results. WCL, whole cell lysate prepared from TRVb cells.
Supplementary Figure 2. Immunoprecipitation of HFE. Whole cell lysates from HeLa/tTA-HFE-FLAG (lane 1), HEK293/HFE (lane 2), and solubilized human liver membranes (lane 3). Immunoprecipitation reaction with Affi-Gel-10/8C10 beads for HeLa/tTA-HFE-FLAG (lane 4), HEK293/HFE (lane 5), solubilized human liver membranes (lane 6), and HEK293 cells not expressing HFE (lane 8). Affi-Gel-10 beads in the presence of solubilized human liver membranes (lane 7) served as negative control of immunoprecipitation. Similar results were obtained when lysates from HeLa/tTA-HFE-FLAG and HEK293/HFE cells were incubated with Affi-Gel-10 beads. Similar amounts of total whole cell lysate (25 μg) or human liver membrane protein (750 μg) were used for immunoprecipitation of HFE. Addition of C-terminal FLAG tag affected mobility of HFE protein (compare lanes 1 and 4 with lanes 2 and 5, respectively). Rabbit polyclonal anti-HFE-EX1 antibody was used for immunodetection. A representative immunoblot from two independent experiments, with similar results, is shown.
Supplementary Figure 3. Detection of β2-microglobulin in human liver. Similar amounts (200 μg) of total membrane protein were loaded per lane and separated on 17% SDS-PAGE. Mouse monoclonal anti-β2M (Immunotech, Marseille, France) was used for immunodetection at concentration 0.2 μg/ml. C, whole cell lysate from TRVb cells stably expressing human HFE-FLAG (50 μg). A representative immunoblot from three independent experiments, with similar results, is shown.
AKNOWLEDGEMENTS
We thank to Kristin Diez-Sauter, Tul-Dim Cing, and Dara Partovi for technical assistance. We are also grateful to Katarina Luciakova, Juxing Chen, Junwei Gao, Julia Maxson and Kristina Nicholson for critical reading of the manuscript. This work was supported by National Institutes of Health Grants DK072166 and DK54488 (to C.A.E.) and in part by Medical Research Foundation of Oregon ACEBD0082 (to M.C.).
Footnotes
- β2M
- β2-microglobulin
- FPN
- ferroportin
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HAMP
- hepcidin
- HEK293/HFE
- HEK293 cells stably expressing non-tagged, full-length human HFE
- HeLa/tTA-HFE-FLAG
- HeLa cells stably expressing human HFE with C-terminal FLAG epitope from the tet-off promoter
- HepG2/tTA-HFE-FLAG
- HepG2 cells stably expressing human HFE with C-terminal FLAG epitope from the tet-on promoter; hereditary hemochromatosis protein
- sHFE
- soluble HFE
- HH
- hereditary hemochromatosis
- HJV
- hemojuvelin
- Tf
- transferrin
- TFRC
- transferrin receptor 1 gene
- TfR1
- transferrin receptor 1 protein
- TfR2
- transferrin receptor 2
- sTfR2
- soluble TfR2
- IREs
- iron responsive elements
- IRPs
- iron responsive proteins.
In this article, we have followed the numbering system of HFE, which starts at the first amino acid translated [43].
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Cullen LM, Anderson GJ, Ramm GA, et al. Genetics of hemochromatosis. Annu Rev Med. 1999;50:87–98. doi: 10.1146/annurev.med.50.1.87. [DOI] [PubMed] [Google Scholar]
- [2].Bothwell TH, MacPhail AP. Hereditary hemochromatosis: etiologic, pathologic, and clinical aspects. Sem Hematol. 1998;35:55–71. [PubMed] [Google Scholar]
- [3].Batts KP. Iron overload syndromes and the liver. Mod Pathol. 2007;20(Suppl 1):S31–9. doi: 10.1038/modpathol.3800715. [DOI] [PubMed] [Google Scholar]
- [4].Feder JN, Gnirke A, Thomas W, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- [5].Papanikolaou G, Samuels ME, Ludwig EH, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- [6].Roetto A, Papanikolaou G, Politou M, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–2. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- [7].Camaschella C, Roetto A, Cali A, et al. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25:14–5. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- [8].Montosi G, Donovan A, Totaro A, et al. Autosomal-dominant hemochromatosis is associated with a mutation in the ferroportin (SLC11A3) gene. J Clin Invest. 2001;108:619–23. doi: 10.1172/JCI13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wilson IA, Bjorkman PJ. Unusual MHC-like molecules: CD1, Fc receptor, the hemochromatosis gene product, and viral homologs. Curr Opin Immunol. 1998;10:67–73. doi: 10.1016/s0952-7915(98)80034-4. [DOI] [PubMed] [Google Scholar]
- [10].Feder JN, Tsuchihashi Z, Irrinki A, et al. The hemochromatosis founder mutation in HLA-H disrupts β2-microglobulin interaction and cell surface expression. J Cell Biol. 1997;272:14025–14028. doi: 10.1074/jbc.272.22.14025. [DOI] [PubMed] [Google Scholar]
- [11].Waheed A, Parkkila S, Zhou XY, et al. Hereditary hemochromatosis: effects of C282Y and H63D mutations on association with beta2-microglobulin, intracellular processing, and cell surface expression of the HFE protein in COS-7 cells. Proc Natl Acad Sci USA. 1997;94:12384–9. doi: 10.1073/pnas.94.23.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Feder JN, Penny DM, Irrinki A, et al. The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci USA. 1998;95:1472–7. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Parkkila S, Waheed A, Britton RS, et al. Association of the transferrin receptor in human placenta with HFE, the protein defective in hereditary hemochromatosis. Proc Natl Acad Sci USA. 1997;94:13198–202. doi: 10.1073/pnas.94.24.13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lebron JA, West AP, Jr., Bjorkman PJ. The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. J Mol Biol. 1999;294:239–45. doi: 10.1006/jmbi.1999.3252. [DOI] [PubMed] [Google Scholar]
- [15].Lebron JA, Bjorkman PJ. The transferrin receptor binding site on HFE, the class I MHC-related protein mutated in hereditary hemochromatosis. J Mol Biol. 1999;289:1109–18. doi: 10.1006/jmbi.1999.2842. [DOI] [PubMed] [Google Scholar]
- [16].Bennett MJ, Lebron JA, Bjorkman PJ. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature. 2000;403:46–53. doi: 10.1038/47417. [DOI] [PubMed] [Google Scholar]
- [17].Ganz T. Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood. 2003;102:783–8. doi: 10.1182/blood-2003-03-0672. [DOI] [PubMed] [Google Scholar]
- [18].Bridle KR, Frazer DM, Wilkins SJ, et al. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361:669–73. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- [19].Muckenthaler M, Roy CN, Custodio AO, et al. Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat Genet. 2003;34:102–7. doi: 10.1038/ng1152. [DOI] [PubMed] [Google Scholar]
- [20].Nicolas G, Viatte L, Lou DQ, et al. Constitutive hepcidin expression prevents iron overload in a mouse model of hemochromatosis. Nat Genet. 2003;34:97–101. doi: 10.1038/ng1150. [DOI] [PubMed] [Google Scholar]
- [21].Zhang AS, Xiong S, Tsukamoto H, Enns CA. Localization of iron metabolism-related mRNAs in rat liver indicate that HFE is expressed predominantly in hepatocytes. Blood. 2004;103:1509–14. doi: 10.1182/blood-2003-07-2378. [DOI] [PubMed] [Google Scholar]
- [22].Fleming RE, Britton RS. Iron Imports. VI. HFE and regulation of intestinal iron absorption. Am J Physiol Gastrointest Liver Physiol. 2006;290:G590–4. doi: 10.1152/ajpgi.00486.2005. [DOI] [PubMed] [Google Scholar]
- [23].Spasic M. Vujic, Kiss J, Herrmann T, et al. Physiologic systemic iron metabolism in mice deficient for duodenal Hfe. Blood. 2007;109:4511–7. doi: 10.1182/blood-2006-07-036186. [DOI] [PubMed] [Google Scholar]
- [24].Spasic M. Vujic, Kiss J, Herrmann T, et al. Hfe acts in hepatocytes to prevent hemochromatosis. Cell Metab. 2008;7:173–8. doi: 10.1016/j.cmet.2007.11.014. [DOI] [PubMed] [Google Scholar]
- [25].Fleming RE, Migas MC, Holden CC, et al. Transferrin receptor 2: continued expression in mouse liver in the face of iron overload and in hereditary hemochromatosis. Proc Natl Acad Sci USA. 2000;97:2214–9. doi: 10.1073/pnas.040548097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281:28494–8. doi: 10.1074/jbc.C600197200. [DOI] [PubMed] [Google Scholar]
- [27].Chen J, Chloupkova M, Gao J, et al. HFE modulates transferrin receptor 2 levels in hepatoma cells via interactions that differ from transferrin receptor 1-HFE interactions. J Biol Chem. 2007;282:36862–70. doi: 10.1074/jbc.M706720200. [DOI] [PubMed] [Google Scholar]
- [28].Schmidt PJ, Toran PT, Giannetti AM, et al. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7:205–14. doi: 10.1016/j.cmet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gao J, Chen J, Kramer M, et al. Interaction of the hereditary hemochromatosis protein HFE with transferrin receptor 2 is required for transferrin-induced hepcidin expression. Cell Metab. 2009;9:217–27. doi: 10.1016/j.cmet.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Carlson H, Zhang AS, Fleming WH, Enns CA. The hereditary hemochromatosis protein, HFE, lowers intracellular iron levels independently of transferrin receptor 1 in TRVb cells. Blood. 2005;105:2564–70. doi: 10.1182/blood-2004-03-1204. [DOI] [PubMed] [Google Scholar]
- [31].Gross CN, Irrinki A, Feder JN, Enns CA. Co-trafficking of HFE, a nonclassical major histocompatibility complex class I protein, with the transferrin receptor implies a role in intracellular iron regulation. J Biol Chem. 1998;273:22068–74. doi: 10.1074/jbc.273.34.22068. [DOI] [PubMed] [Google Scholar]
- [32].Davies PS, Enns CA. Expression of the Hereditary Hemochromatosis Protein HFE Increases Ferritin Levels by Inhibiting Iron Export in HT29 Cells. J Biol Chem. 2004;279:25085–92. doi: 10.1074/jbc.M400537200. [DOI] [PubMed] [Google Scholar]
- [33].Johnson MB, Enns CA. Diferric transferrin regulates transferrin receptor 2 protein stability. Blood. 2004;104:4287–93. doi: 10.1182/blood-2004-06-2477. [DOI] [PubMed] [Google Scholar]
- [34].Williams AM, Enns CA. A mutated transferrin receptor lacking asparagine-linked glycosylation sites shows reduced functionality and an association with binding immunoglobulin protein. J Biol Chem. 1991;266:17648–17654. [PubMed] [Google Scholar]
- [35].Vogt T, Blackwell A, Giannetti A, et al. Heterotypic interactions between transferrin receptor and transferrin receptor 2. Blood. 2002;101:2008–14. doi: 10.1182/blood-2002-09-2742. [DOI] [PubMed] [Google Scholar]
- [36].Schneider C, Newman RA, Sutherland DR, et al. A one-step purification of membrane proteins using a high efficiency immunomatrix. J Biol Chem. 1982;257:10766–10769. [PubMed] [Google Scholar]
- [37].Casey JL, Hentze MW, Koeller DM, et al. Iron-responsive elements: regulatory RNA sequences that control mRNA levels and translation. Science. 1988;240:924–928. doi: 10.1126/science.2452485. [DOI] [PubMed] [Google Scholar]
- [38].Mullner EW, Neupert B, Kuhn LC. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell. 1989;58:373–382. doi: 10.1016/0092-8674(89)90851-9. [DOI] [PubMed] [Google Scholar]
- [39].Mullner EW, Kuhn LC. A stem-loop in the 3′ untranslated region mediates iron dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988;53:815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- [40].Owen D, Kuhn LC. Noncoding 3′ sequences of the transferrin receptor gene are required for mRNA regulation by iron. EMBO J. 1987;6:1287–1293. doi: 10.1002/j.1460-2075.1987.tb02366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Robb A, Wessling-Resnick M. Regulation of transferrin receptor 2 protein levels by transferrin. Blood. 2004;104:4294–4299. doi: 10.1182/blood-2004-06-2481. [DOI] [PubMed] [Google Scholar]
- [42].Johnson MB, Chen J, Murchison N, et al. Transferrin receptor 2: evidence for ligand-induced stabilization and redirection to a recycling pathway. Mol Biol Cell. 2007;18:743–54. doi: 10.1091/mbc.E06-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lebron JA, Bennett MJ, Vaughn DE, et al. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–23. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Effect of extraneous proteins on intensity of the immunobands. Representative immunoblots are shown for each protein from at least three independent experiments with similar results. WCL, whole cell lysate prepared from TRVb cells.
Supplementary Figure 2. Immunoprecipitation of HFE. Whole cell lysates from HeLa/tTA-HFE-FLAG (lane 1), HEK293/HFE (lane 2), and solubilized human liver membranes (lane 3). Immunoprecipitation reaction with Affi-Gel-10/8C10 beads for HeLa/tTA-HFE-FLAG (lane 4), HEK293/HFE (lane 5), solubilized human liver membranes (lane 6), and HEK293 cells not expressing HFE (lane 8). Affi-Gel-10 beads in the presence of solubilized human liver membranes (lane 7) served as negative control of immunoprecipitation. Similar results were obtained when lysates from HeLa/tTA-HFE-FLAG and HEK293/HFE cells were incubated with Affi-Gel-10 beads. Similar amounts of total whole cell lysate (25 μg) or human liver membrane protein (750 μg) were used for immunoprecipitation of HFE. Addition of C-terminal FLAG tag affected mobility of HFE protein (compare lanes 1 and 4 with lanes 2 and 5, respectively). Rabbit polyclonal anti-HFE-EX1 antibody was used for immunodetection. A representative immunoblot from two independent experiments, with similar results, is shown.
Supplementary Figure 3. Detection of β2-microglobulin in human liver. Similar amounts (200 μg) of total membrane protein were loaded per lane and separated on 17% SDS-PAGE. Mouse monoclonal anti-β2M (Immunotech, Marseille, France) was used for immunodetection at concentration 0.2 μg/ml. C, whole cell lysate from TRVb cells stably expressing human HFE-FLAG (50 μg). A representative immunoblot from three independent experiments, with similar results, is shown.