Abstract
We present an improved MR brain image registration algorithm, called TPS-HAMMER, which is based on the concepts of attribute vectors and hierarchical landmark selection scheme proposed in the highly successful HAMMER registration algorithm. We demonstrate that TPS-HAMMER algorithm yields better registration accuracy, robustness, and speed over HAMMER owing to: 1) the employment of soft correspondence matching, and 2) the utilization of thin-plate splines (TPS) for sparse-to-dense deformation field generation. These two aspects can be integrated into a unified framework, to refine the registration iteratively by alternating between soft correspondence matching and dense deformation field estimation. Compared with HAMMER, TPS-HAMMER affords several advantages: 1) Unlike the Gaussian propagation mechanism employed in HAMMER, which can be slow and often leaves unreached blotches in the deformation field, the deformation interpolation in the non-landmark points can be obtained immediately with TPS in our algorithm; 2) The smoothness of deformation field is preserved due to the nice properties of TPS; 3) Possible misalignments can be alleviated by allowing the matching of the landmarks with a number of possible candidate points and enforcing more exact matches in the final stages of the registration. Extensive experiments have been conducted, using the original HAMMER as a comparison baseline, to validate the merits of TPS-HAMMER. The results show that TPS-HAMMER yields significant improvement in both accuracy and speed, indicating high applicability for the clinical scenario.
Keywords: Deformable registration, Thin-plate splines, Soft correspondence, HAMMER
1. Introduction
Image registration is a critical prerequisite for clinical analysis of longitudinal and cross-sectional medical data. Many registration algorithms have therefore been proposed, and they can be classified as intensity-based or feature-based [1–6]. HAMMER [2, 7] is one of the feature-based non-rigid registration algorithms, which has been successfully applied to many clinical applications. For example, HAMMER has been employed in the study of brain morphometry of XXY males (Klinefelter’s syndrome) [8]. Tissue density maps, generated using HAMMER by aligning individual brain images onto a template, were proposed in Davatzikos et al. [9] as morphological signatures for atrophic region detection and morphological classification [10]. In the study of the hypercortisolism in alcohol dependence and its relationship to hippocampal volume loss [11, 12], HAMMER was used to quantitatively measure hippocampal volume loss due to chronic heavy drinking. HAMMER has also been extended to measure temporal morphological changes [13].
HAMMER takes as input the white matter (WM), gray matter (GM), ventricle (VN), and cerebrospinal fluid (CSF) labeled tissue maps. Geometric moment invariants (GMIs) of each map are computed as morphological signatures for correspondence matching. To avoid local minimal, a small set of driving voxels, which are usually located at the ventricular boundaries, sulcal roots, and gyral crowns, is selected for initial correspondence matching. For non-driving-voxels, their deformations are estimated by interpolation from the deformations of nearby driving voxels. In the end of each iteration, isotropic Gaussian smoothing is employed for deformation field regularization.
HAMMER is able to establish reasonable anatomical correspondences between template and subject brain images. However, it has several limitations: 1) Gaussian smoothing is not sufficient to ensure the smoothness and continuity of deformation field, and may sometimes result in deformations which are over aggressive, especially in the areas with rich edges; 2) deformations are unable to efficiently propagate to the points far away from the driving voxels through dissemination in small neighborhood, thus preventing the registration to converge in short time; 3) since false correspondence is always possible, some mechanism is necessary to alleviate the misalignment from ambiguities during the registration. HAMMER, which is based on ‘hard’ correspondence detection, is sometimes susceptible to such pitfall.
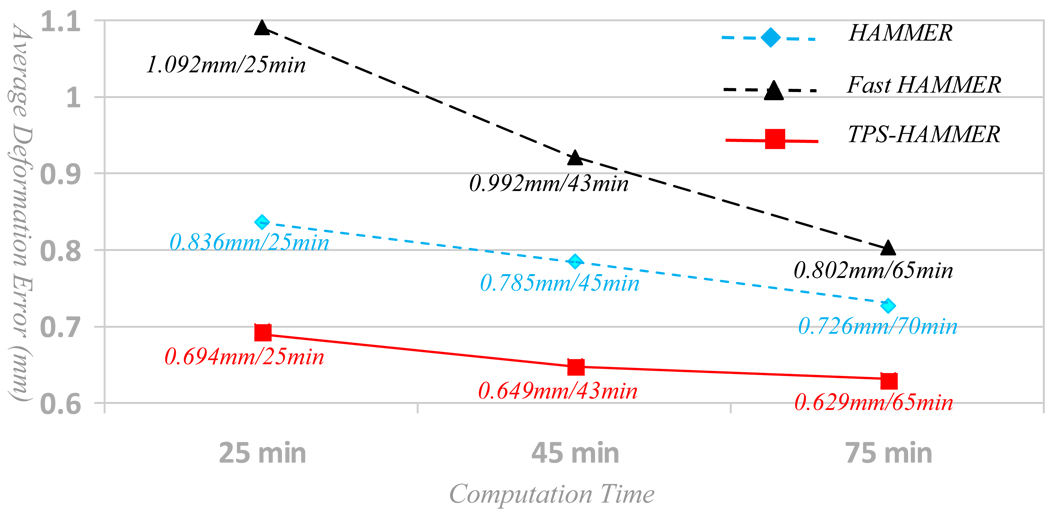
To remedy these limitations, we reformulate HAMMER in the form of a soft correspondence matching [14, 15] and thin-plate splines based deformation interpolation [16], which can be optimized by alternating between the establishment of sparse correspondences and the estimation of dense deformation field. The GMI-based attribute vector and the driving voxel selection scheme in HAMMER will be adopted in this paper. A small set of distinctive driving voxels will be selected for both template and subject images in the initial registration stage. Since the correspondences of these driving voxels can be determined more reliably, it is reasonable to allow them to drive the deformation, rather than the less distinctive voxels. During correspondence matching, robustness is achieved by 1) employing soft assignment where multiple correspondences are allowed, instead of solving for exact one-to-one correspondence in the beginning of registration; 2) considering not only the voxelwise similarity but also subvolume similarity. It is worth noting that soft correspondence assignment has been explored to achieve fast registration by Shen [15], however, the estimation of dense deformation is still implemented using a Gaussian propagation mechanism. Under the same computation time (25 min), its registration accuracy falls much behind ours, as confirmed by our experimental results below (Fig. 6).
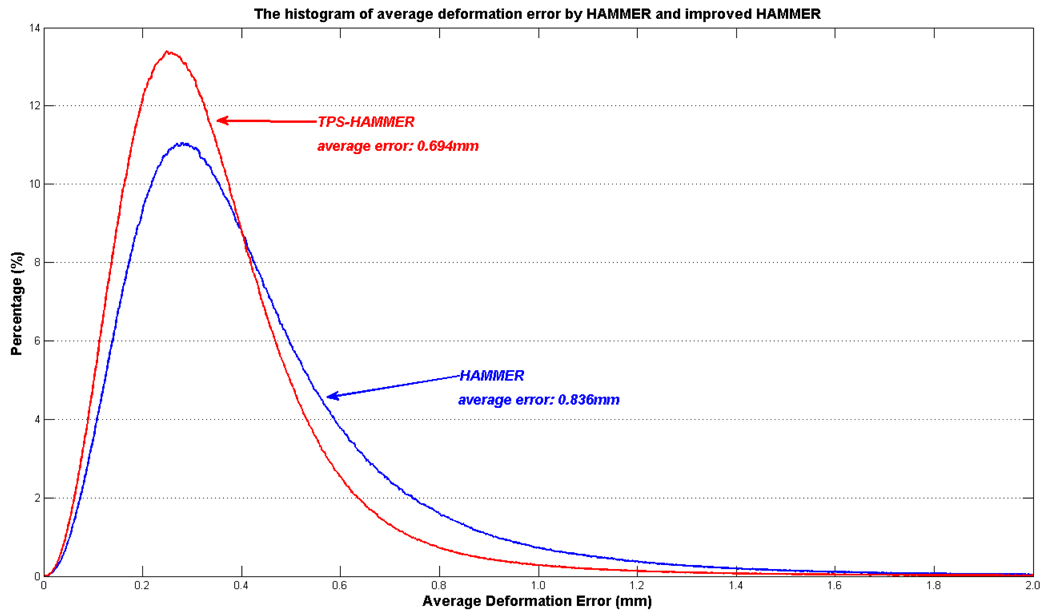
Fig. 6.
Average deformation errors estimated by HAMMER, fast HAMMER, and TPS-HAMMER at different computation times. TPS-HAMMER can achieve less registration error (0.694mm) with 25 minutes than HAMMER with 70 minutes (0.726mm). Also, performance of fast HAMMER shown in black curve is worse than TPS-HAMMER.
After determining correspondences for the driving voxels, TPS interpolation [16] is utilized to model the dense deformation field. The TPS parameters, which consist of affine and deformable counterparts, are computed by considering the driving voxels as control points. The deformation of each non-driving voxel can then be interpolated based on the estimated parameters by computing its Euclidean distances to all control points. TPS regularization is performed in overlapped blocks, and control points are uniformly sub-sampled within each block to ensure computational tractability. TPS allows generation of a dense and smooth deformation field immediately after correspondence detection. By integrating these strategies into HAMMER, promising results are achieved on both real and simulated brain images. Specially, we gain not only better registration accuracy, but also half the computation time required by HAMMER.
The paper is organized as follows. In Section 2, we formulate the registration problem and provide a solution in the form of soft correspondence matching and TPS-based deformation interpolation. Experimental results are provided in Section 3 to validate the advantages of our proposed method. Section 4 concludes this paper.
2. Methods
We first give a brief introduction to HAMMER in Section 2.1, and then describe the details of TPS-HAMMER in subsequent sections. In our method, image registration is achieved by minimizing an energy cost function, with the goal of establishing good correspondences and obtaining a smooth deformation field (Section 2.2). Specifically, to estimate the deformation field, we utilize an optimization approach which alternates between updating the sparse point correspondences and generating the dense deformation field (Section 2.3). TPS is utilized as a mechanism (Section 2.4) to estimate a dense deformation field which ensures a least-squares fit on the deformations of the driving voxels, and at the same time minimizes the bending energy. A summary of the registration algorithm will be provided in the end of this section (Section 2.5).
2.1. HAMMER – A Brief Introduction
HAMMER is a feature-based deformable registration algorithm. The overall goal of deformable registration is to find a transformation F = {f(x)|f(x) = x + h(x), x = (x1,x2,x3) ∈ ΩT}, where displacement h(x) defines the mapping of the point coordinates ΩT of template T to ΩS of subject S, where ΩT,ΩS ⊂ ℝ3. To achieve this, the attribute vectors aT(x)at location x of template T and as(y) at location y of subject S, consisting of intensity, edge type, and GMIs of each tissue type (WM, GM, VN, and CSF), are calculated and used as morphological signatures to determine point correspondences. The definitions of GMIs up to the third order can be found in Lo et al. [17] and Shen et al. [2].
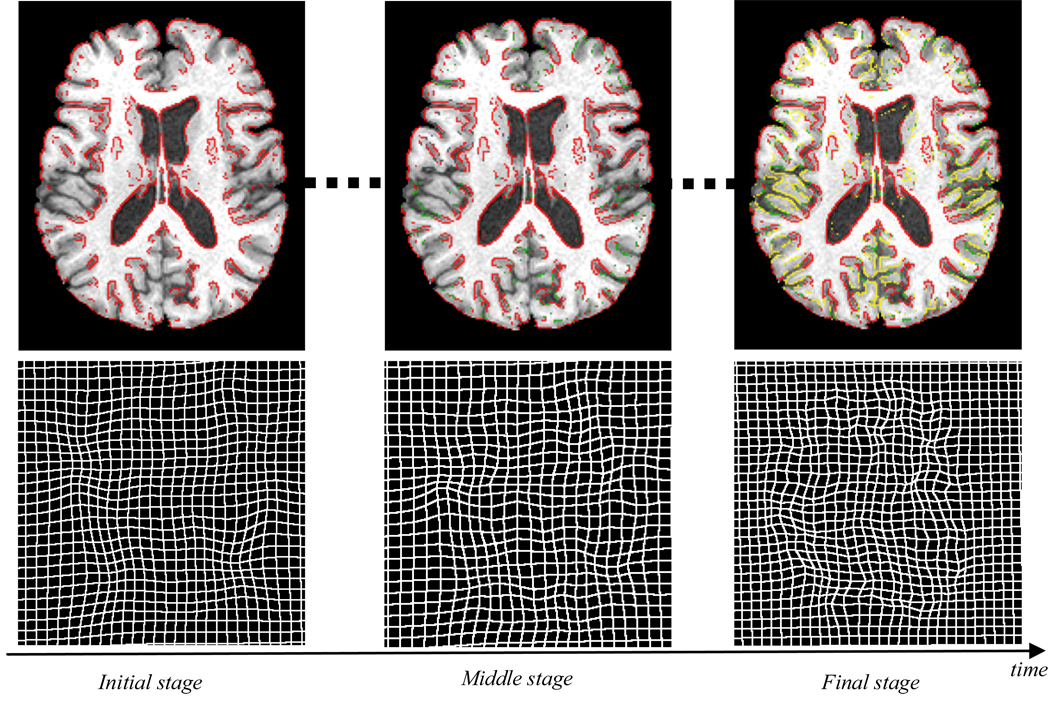
Instead of finding the correspondence for each voxel, HAMMER hierarchically selects a set of most distinguishable voxels, called driving voxels, to establish the correspondence and steer the deformation of other voxels. As shown in Fig. 1, the driving voxels (displayed in red) are located at sulcal root, gyral crown, and the ventricular boundary, and are hence able to detect the correspondence more reliably than other voxels in the brain. The deformation field guided by these distinguishable driving voxels is rough, as displayed in the bottom left of Fig. 1, while, it leads the image warping against ambiguities. With the progress of registration, more and more voxels (shown in green and yellow) are added as the driving voxels, until all voxels are included as driving voxels in the end. Note that the selection of driving voxels is controlled by applying a progressively relaxed threshold on the zero-order GMI. The threshold can be determined based on prior knowledge of anatomical information [2]. In the end of registration, all of the boundary voxels are considered as the driving voxels, thus estimating the deformation field from the coarse to fine scale (as displayed in the bottom right of Fig. 1).
Fig. 1.
The evolution of driving voxels and deformation field in different stages of registration. In the initial stage, only a small number of distinguishing voxels (displayed in red) in the brain image, which are usually located at sulcal roots, gyral crowns, and ventricular boundaries, are selected to steer the registration of other less distinctive voxels. With the development of registration, more and more voxels are gradually added as driving voxels, as shown in green and yellow, until all of the edge voxels are used as driving voxels in the end. The bottom row shows the corresponding deformation fields estimated at three different stages. It can observed that, as the number of driving voxel increases, the deformation field turns from coarse to fine scale.
The energy function that is minimized in HAMMER involves measuring the differences of attribute vectors between the template and the warped subject, as well as the Laplacian regularization term of the deformation field. The inverse transformation is also considered for registration consistency. HAMMER spreads the influence of the deformations of the driving voxels by using a Gaussian kernel. However, such a mechanism is neither able to provide a well-defined deformation field, nor to propagate the correspondence results to the entire image domain immediately. We present the details of TPS-HAMMER in the subsequent sections. In a nutshell, TPS-HAMMER solves the problems of HAMMER by integrating both robust correspondence detection and efficient dense deformation estimation.
2.2. Energy Function for Deformable Registration Problem
In order to achieve robust attribute vector matching, where displacement defines the mapping of the point coordinates of template, we adopt a region-wise, instead of voxel-wise, measure as used in HAMMER to describe the similarity between aT(x) from template T and as(y)from subject S given a deformation field f:
| (1) |
where n1(.) is a neighborhood for subvolume similarity computation, and u is a point in the neighborhood n1(.). Δs = f(u) − f(x) in the subject domain measures the corresponding radial distance (u −x) in the template domain. ‖.‖2 is the L2-norm distance of the two attribute vectors and sim(x,y) has a range of 0 to 1.
Due to the curse of dimensionality, it is difficult and computationally intractable to determine the correspondence of each voxel. As a remedy, we select subsets of voxels iteratively, i.e., the driving voxels XT = {xt |t = 1,…, M} ⊂ ΩT for template T, and YS = {ys|S = 1,…, N} ⊂ ΩS for subject S, which can be used to determine initial correspondences reliably. We show the driving voxels on template image in Fig. 1.
Therefore, the problem of registering two images is now simplified to a relatively less complex energy minimization problem. We will first solve for the forward correspondence matching by locating for the template driving voxels XT their corresponding matching voxels in the subject volume. We then solve for the backward correspondence matching in a similar manner for the subject driving voxels YS Soft correspondence is permitted in the sense that multiple candidate points are allowed. After obtaining the correspondences, we will estimate the dense deformation field f by TPS. These two steps can be mathematically incorporated into the energy function, as described next.
Forward Correspondence Matching
Allowing the driving voxels to have multiple correspondences is conducive to the robustness of the registration, especially in the case that one-to-one correspondence is hard to establish in the beginning. Therefore, for each template driving voxel xt, every subject voxel v ∈ n2(f(xt)) will be considered as a possible candidate with probability pt,v, where n2(.) denotes the search neighborhood around the estimated displaced location f(xt). The size of n2 is initially large and is gradually decreased until the registration algorithm converges. The suitability of matching of a particular candidate v can be measured by the similarity defined in Eq. 1. The distance between v and the displaced location xt, i.e., ‖f(xt) − v)‖2, is required to be as close as possible. Accordingly, denoting correspondence probability matrix P = [pt,v], the energy term for forward correspondence matching is formulated as:
| (2) |
where pt,v satisfies: 0 ≤pt,v ≤1, and Σv ∈ n2(f(xt)) pt,v = 1 for each template driving voxel xt.
Backward Correspondence Matching
We require the deformation field to be symmetric in both forward and backward transforms. Consequently, we define a symmetric energy term for measuring the backward correspondence from subject to template. Similarly, denoting qs,u as the probability of each voxel u in a search neighborhood n2(h−1(ys)) as being the correspondence of ys, the energy term of backward correspondence matching can be defined as follows:
| (3) |
where Q = [qs,u] and qs,u satisfies: 0 ≤ qs,u ≤ 1, and Σu∈n2(f−1(ys))qs,u = 1.
Control of Fuzzy Assignment
Soft assignment allows each point to have multiple correspondences in order to avoid the over reliance on one particular voxel and hence the possibility of misguidance. However, in order to increase the registration specificity and accuracy, it is necessary to resort to one-to-one correspondence in the final registration stage where the images are approximately aligned. This can be formulated by requiring the fuzziness of correspondence assignment to gradually decrease with the progress of registration. The fuzziness of correspondence can be defined as:
| (4) |
| (5) |
By controlling these two terms, we can achieve robust correspondence detection in the initial stages of registration and further resort to the one-to-one correspondence in the final registration stages.
Regularization of Deformation Field
The deformation field is required to be smooth and continuous in order to preserve the topology and avoid un-biological mapping. The smoothness of the deformation field f is measured by its bending energy, as denoted by an operator L below:
| (6) |
The above equation is the sum of the squared second order derivatives in the directions of axes x1, x2, and x3 [14].
Overall Energy Function
By combining all the energy terms, we can obtain a whole energy function:
| (7) |
where r determines the significance of the fuzzy assignment (in Eq. 4 and 5) and β is a weighting factor controlling the regularization of the deformation field.
The parameter r is called as the temperature in the nomenclature of annealing schedule. In the initial stage, the degree r is large, encouraging the multiple correspondences, which means that even faraway voxels will be considered as candidates for matching. By considering more matching points, this makes the registration algorithm robust to the ambiguity. With the progress of registration, r will be gradually decreased according to the annealing rate (please refer to [14, 18]). When r drops close to zero in the end of registration, the fuzzy correspondence becomes a one-to-one correspondence, which helps to produce accurate registration result. By requiring the correspondence to be from soft to hard matching, we can obtain not only accurate but also robust registration results.
2.3. Optimization of the Energy Function
Optimizing the energy function in Eq. 7 can be achieved by alternating between (1) the determination of the correspondence matrices P and Q by a gradient descent method, and (2) the estimation of the dense deformation field f by TPS after obtaining P and Q. These two steps are detailed below.
Update the Correspondences
The correspondence probability matrix P and Q can be computed by letting ∂E(P,Q,f)/∂pt,v = 0, and ∂E(P,Q,f)/∂qs,u = 0, which leads to the following solution:
| (8) |
| (9) |
where c is a constant. It is worth noting that normalization is necessary to satisfy the constraints on pt,v and qs,u.
Update the Deformation Field
After dropping terms independent of f, optimization of the energy function becomes a problem of estimating the dense deformation field f in a least-square sense by minimizing the following function:
| (10) |
where
| (11) |
v̂t ∈ ΩS and ûs ∈ ΩT are the means of the correspondences of the driving voxels in XT and YS, respectively. It is worth noting that both forward and backward correspondences are incorporated in constructing the dense deformation field f to ensure that it is symmetric. TPS can be utilized to optimize E(f) by considering Φ = {xt} ∪ {ûs} as the control points and Ψ = {v̂t} ∪ {ys} as their corresponding destinations, as detailed in the next section.
2.4. Thin-Plate Splines
Without loss of generality, each element φi in Φ is represented by (1,φi,1,φi,2,φi,3) the 3D coordinate system, and vice versa, ψi = (1,ψi,1,ψi,2,ψi,3). For each φi,φj ∈ Φ, their geometric distance can be calculated by:
| (12) |
and d(φi,φi) = 0. We define the matrices
and , where O is a 4×4 zero matrix, D(φi) =[di1,…,di(M+N)] by letting dij = d(φi,φj) in matrix K. It is worth noting that λ is a positive scalar which regulates the deformation field so that unphysical reflection mappings can be suppressed.
Then the TPS coefficients, which consists of the affine transform portion A4×4 and the deformable portion W(M+N)×4, can be estimated by:
| (13) |
Using the estimated TPS parameters W and A, the displacement f(x) for each point x can be interpolated by:
| (14) |
where D(x) = [d(φ1,x),d(φ2,x),…,d(φM+N,x)]1×(M+N), summarizing the contribution of each TPS control point φi with respect to the point x.
It has been proven in [16] that the dense deformation field f estimated by Eq. (12)~(14) is a solution that is able to minimize the bending energy term ‖Lf‖2 defined in Eq. 6, and it also satisfies f(φi) = ψi when λ = 0.
2.5. Summary of TPS-HAMMER and Implementation Issues
Summary of TPS-HAMMER
Our registration algorithm is briefly summarized below. Note that step 3~5 are newly added components in TPS-HAMMER.
Calculate the GMIs for both template and subject images;
Select the driving voxels XT and YS template and subject images;
Determine the sparse soft correspondences for XT and YS, respectively, by Eq. (8)~Eq. (9);
Obtain the dense deformation f by Eq. 14;
If not converged, relax the criteria of driving voxel selection (i.e., relax the threshold on the zero-order GMIs) and go to step 2. (See Fig. 1 for examples of selected driving voxels.)
Implementation Issues
We allow only a small number of driving voxels (around 200,000, i.e., 2~3% of the total number of image voxels in a 256 × 256 × 124 image) to participate in the initial correspondence matching. With the progress of registration, more and more voxels will be allowed to join in refining the correspondences by relaxing the threshold on the zero-order GMIs. The searching neighborhood will, however, be gradually decreased to narrow down the search region, e.g., the search radius decreasing from 10 to 2. Therefore, not only the computation time can be reduced, but also the robustness of correspondence matching can also be improved by using this hierarchical mechanism.
In estimating the dense deformation field, operations such as matrix inversion and calculation of geometric distances (Eq. 12) in each iteration consume a lot of computation resources. For the latter, we use a look up table to save computation time. Since the complexity of matrix inversion is increased with the number of control points in the deformation field - usually up to 0.2 million driving voxels in a 256 × 256 × 124 image - we have to apply TPS fitting locally using the overlapping blocks. However, the number of control points in each block is still too large, especially at cortical and ventricular areas (see Fig. 1 for example). Therefore, we perform uniform sub-sampling on the selected driving voxels in each block to further reduce matrix Θ to a computationally feasible dimension. In all experiments, we set the block size to be 32 × 32 × 32 and limit the number of control points to be no more than 500 by sub-sampling. Furthermore, after determining the distribution of control points of each block in the initial stage, we keep the result of each matrix inverse Θ−1 for use in the following iterations, despite the fact that the driving voxels will increase as the registration progresses. The reasons are: 1) it helps reduce TPS regularization into a small number of matrix addition and multiplication operations, and thus eliminates the need of performing matrix inversion for each iteration; 2) the distribution of control points in each iteration is very similar after sub-sampling; 3) we find no significant difference in registration performance with and without updating Θ−1 in each iteration.
Advantages of TPS-HAMMER
In summary, we propose a unified framework to improve the registration accuracy of HAMMER. The two key components used in HAMMER-TPS are (1) soft correspondence detection and (2) TPS-based deformation interpolation. Specifically, the use of soft, instead of hard, correspondence detection in the initial registration procedure will alleviate the problem of mismatches caused by the ambiguity of anatomical structures. On the other hand, TPS provides a fast and effective means of obtaining a dense deformation field, while at the same time it ensures the minimal bending energy. By integrating these two components, TPS-HAMMER can achieve more accurate registration results, at only half the computation cost of HAMMER.
3. Experiments
For performance evaluation, we tested TPS-HAMMER on both real and simulated data, and compared it with HAMMER. Unless otherwise mentioned, we used the same set of parameters in all experiments and performed the experiments on the same workstation (Quad CPU@2.4GHz and 4G RAM). The results indicate that TPS-HAMMER achieved greater accuracy at a lesser amount of time, marking its potential for more accurate and rapid neuroimage analysis.
3.1. Experiment on Real Data Set
Experiment on Elderly Brains
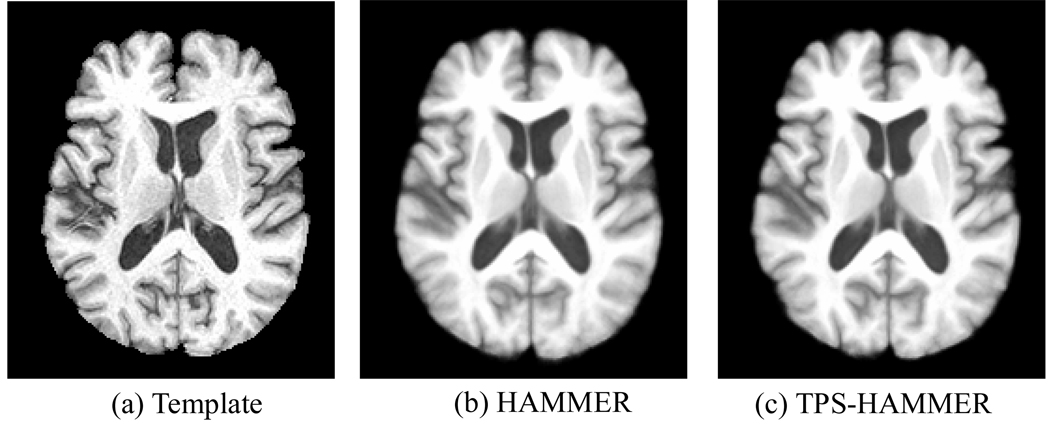
Brain images of 18 elderly subjects, which were acquired with image dimension of 256 × 256 × 124 and resolution of 0.9375 × 0.9375 × 1.5mm3, were used in this experiment. By selecting one subject as a template, we registered all other 17 subjects onto the selected template. We used an identical set of parameters for HAMMER and TPS-HAMMER. But to showcase the fact that TPS-HAMMER can achieve better performance at a lower computation cost, we reduced its number of iterations. Fig. 2 shows the mean image constructed from the images after registration. Through visual inspection, the results given by the two methods are similar. However, we note the fact that the average computation times taken by HAMMER and TPS-HAMMER to register a pair of images are 70.3 min and 25.8 min, respectively. This is equivalent to a speed performance gain of 63.3% by TPS-HAMMER, which is especially attractive for clinical studies involving large datasets.
Fig. 2.
(a) The template image, (b) average image produced by HAMMER, and (c) average image produced by the TPS-HAMMER. Through visual inspection, average images by both methods are quite similar. TPS-HAMMER, however, requires less than half of the computational time needed by HAMMER.
Since segmentation had been performed on the images, we can calculate the voxel-wise overlap ratios between the warped subjects and the template for different tissue types (WM, GM, VN and CSF). Table 1 reports the overlap ratios of each tissue type (WM, GM, VN, and CSF) by HAMMER and TPS-HAMMER, respectively, and each algorithm is performed with two different computation times. It is obvious that TPS-HAMMER obtains better registration results within 30 min for all tissue types, compared to HAMMER which needs over 1 hour. For further quantifying registration consistency, we computed the entropy of tissue probability map across all subjects voxel by voxel. A higher entropy value indicates less consistency. For the TPS-HAMMER with average computation time of 25.8 min, the average entropy is 0.107, nearly 12% improvement compared to 0.121 obtained by HAMMER with average computation time 70.3 min.
Table 1.
Overlap ratios of WM, GM, VN, and CSF produced by HAMMER and TPS-HAMMER.
| Computation Time | WM | GM | VN | CSF | Overall | |
|---|---|---|---|---|---|---|
| HAMMER | 30.0 min (average) | 53.10% | 75.28% | 60.53% | 69.56% | 70.17% |
| HAMMER | 70.3 min (average) | 55.90% | 81.25% | 63.66% | 76.86% | 74.88% |
| TPS-HAMMER | 25.8 min (average) | 56.52% | 82.15% | 64.59% | 77.14% | 75.69% |
| TPS-HAMMER | 68.2 min (average) | 56.82% | 83.19% | 64.86% | 77.51% | 76.02% |
Fig. 3 demonstrates that TPS-HAMMER gives significantly improved registration, especially when ventricle sizes between subject and template brains is large. We registered a number of subjects with large ventricles to a template with a small ventricle. It can be observed that TPS-HAMMER yields more consistent results than HAMMER.
Fig. 3.
The registration results by HAMMER and TPS-HAMMER. The left two columns show, respectively, three subject brains and a template brain. The difference of ventricular sizes between subject and template brains is large. The right two columns show the warping results by HAMMER and TPS-HAMMER, respectively. It can be observed that the registration results by TPS-HAMMER are much better than HAMMER, particularly in ventricular corners and boundaries (i.e., the top one).
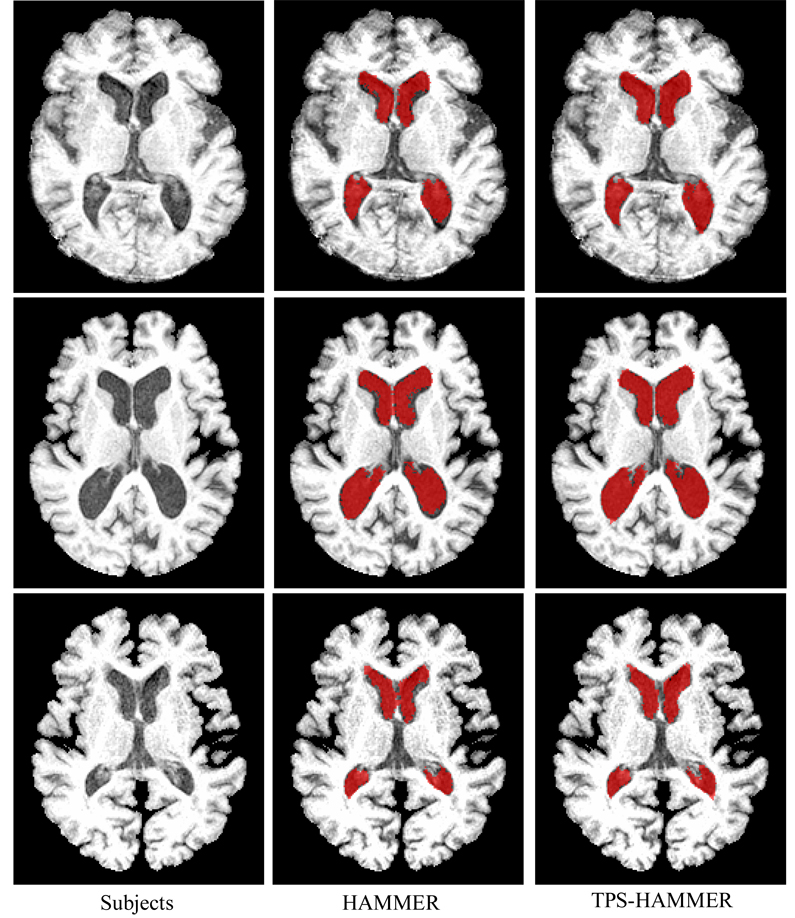
Atlas-based parcellation is highly dependent on the accuracy of the registration algorithm. In this experiment, we employed HAMMER and TPS-HAMMER to first warp a brain atlas, developed by Noor Kabani at Montreal Neurological Institute, onto a number of subjects (i.e., those shown in the left column of Fig. 4). The atlas is then warped according to the estimated deformation fields and the labeling results for the left and right ventricles are overlaid on the subject images (i.e., in the right two columns of Fig. 4). By checking the labeling results on the ventricular corners, it is apparent that TPS-HAMMER achieves better results than HAMMER.
Fig. 4.
The parcellation results on ventricles by HAMMER and TPS-HAMMER. Three real brain images of the left column are used as examples. After warping an atlas with ventricular labels onto these three subject images, the ventricles can be labeled as shown in red on each subject image. By inspecting the ventricular corners, it can be observed that the results yielded by TPS-HAMMER are better than those of HAMMER (particularly for the top one). (This figure is best viewed with color.)
Experiment on NIREP Dataset
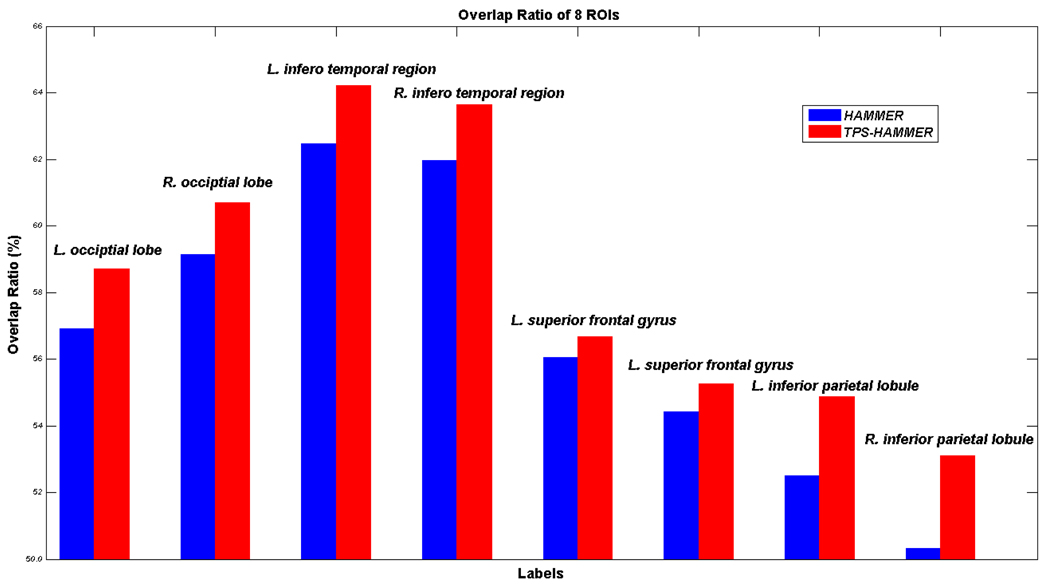
We also evaluated HAMMER and TPS-HAMMER on the 16 images of the NIREP dataset [19], which can be obtained from www.nirep.org. After registration, the overlap ratio can be calculated for each of the 32 manually delineated labels. Fig. 5 shows the comparison results of 8 ROIs by HAMMER and TPS-HAMMER, in blue and red bars, respectively. From left to right, the 8 ROIs denote the left and right occipital lobes, the left and right inferior temporal regions, the left and right superior frontal gyri, and the left and right inferior parietal lobules, respectively. In these 8 ROIs, the TPS-HAMMER achieves nearly 2% improvement over HAMMER, with computation time less than half of that used by HAMMER.
Fig. 5.
The overlap ratios of the 8 ROIs for the 16 aligned images from the NIREP dataset are shown for HAMMER (blue) and TPS-HAMMER (red), respectively. From left to right, the bars show the values for the left and right occipitial lobes, the left and right inferior temporal region, the left and right superior gyri, and the left and right inferior parietal lobules, respectively.
3.2. Experiment on Simulated Data Set
The simulated data are generated by the method presented in Xue et al. [20], where a statistical model is built from each wavelet sub-band of a sample of deformation fields. In this experiment, 10 simulated subjects, as well as the ground truth deformations, were constructed by randomly sampling the deformation statistical distribution. These 10 simulated images have a dimension of 256 × 256 × 198 and resolution of 1.0 × 1.0 × 1.0mm3.
By warping the simulated brain images onto the template using HAMMER and TPS-HAMMER, we can quantify the registration accuracy by calculating the average error between the ground truths and the estimated deformation fields voxel by voxel. Fig. 6 shows a comparison of the average deformation errors yielded by each registration algorithm, with respect to different computation times. We also compared our method (TPS-HAMMER) with the fast registration algorithm developed in Shen [15], called as fast HAMMER in this paper. The curves of deformation error vs. computation time by HAMMER, fast HAMMER, and TPS-HAMMER are displayed in blue, black, and red, respectively. Under the same computation time, TPS-HAMMER achieves better registration accuracy than the other two algorithms. In fact, TPS-HAMMER is able to achieve less registration error (0.694mm) in 25 min than HAMMER in 70 min (0.726mm). Also, under the same computation time (25 min), the average deformation error yielded by fast HAMMER is over 1.09mm. These results demonstrate that TPS-HAMMER is not only fast but also accurate.
We further compared the distributions of the deformation errors yielded by HAMMER and TPS-HAMMER, both with computation time of 25 min. It is apparent from Fig. 7 that the distribution yielded by TPS-HAMMER is more concentrated towards zero, indicating more accurate registration by TPS-HAMMER.
Fig. 7.
The distributions of average deformation errors obtained by HAMMER (blue) and TPS-HAMMER (red). The distribution of TPS-HAMMER is concentrated more towards zero, indicating better accuracy. This comparison is made at the point where both methods require similar computation time.
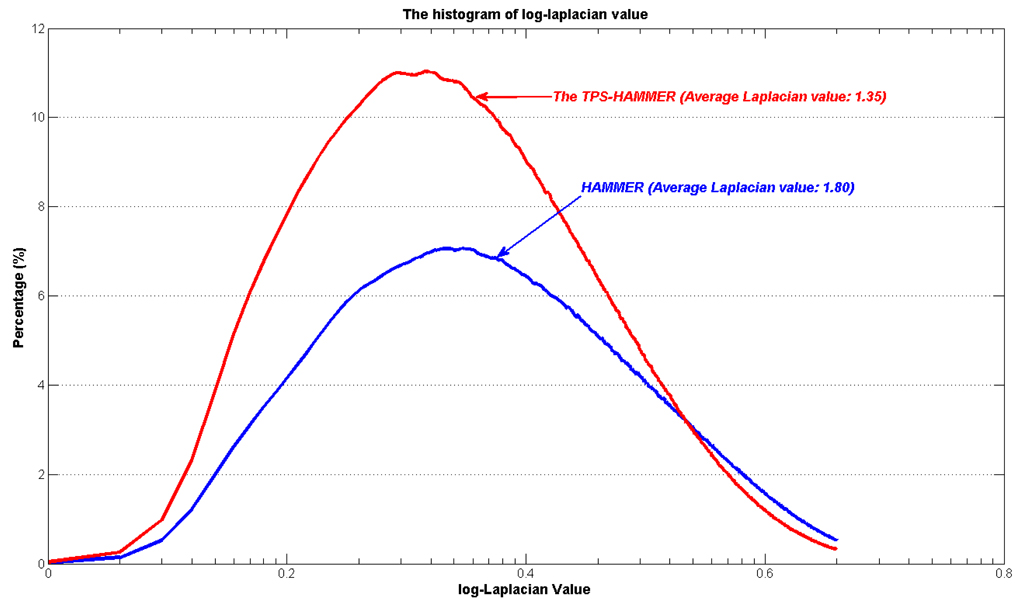
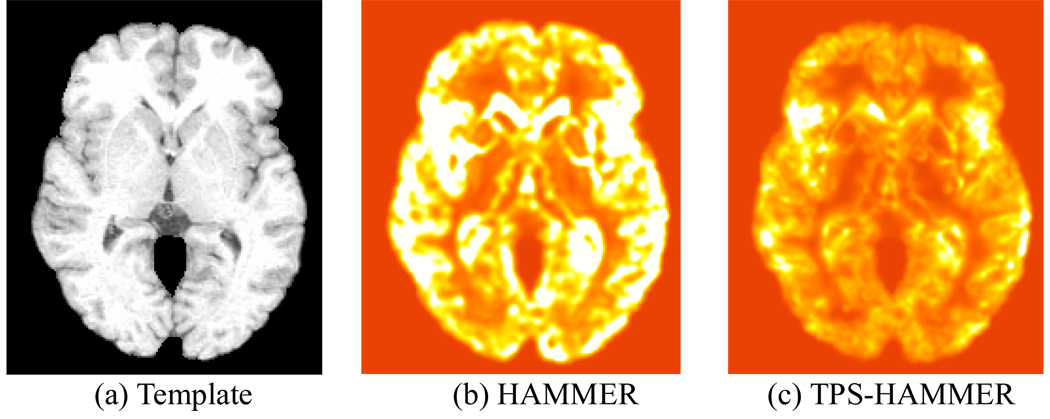
We further evaluated the smoothness of the estimated deformation field by evaluating its Laplacian value. Fig. 8 shows the histograms of average log-Laplacian values given by HAMMER and TPS-HAMMER using the same set of parameters. The average log-Laplacian value given by TPS-HAMMER is 1.35, compared to 1.80 by HAMMER, indicating that TPS-HAMMER produces smoother deformation fields. Fig. 9 shows the average Laplacian maps of all subjects, where greater brightness indicates larger Laplacian values. The Laplacian values yielded by the TPS-HAMMER are much smaller than those of HAMMER in most brain regions, again validating the merit of our method (TPS-HAMMER).
Fig. 8.
Histograms of log-Laplacian values of the deformation field yielded by HAMMER (blue) and TPS-HAMMER (red). The average Laplacian value yielded by the TPS-HAMMER (1.35) is less than that of HAMMER (1.80), indicating that the deformation field produced by TPS-HAMMER is smoother and hence better preserves the anatomical topology after registration.
Fig. 9.
(a) The template and the Laplacian maps yielded by (b) HAMMER and (c) TPS-HAMMER. Greater brightness indicates larger Laplacian values. For most brain regions, the Laplacian values yielded by the TPS-HAMMER are much smaller than HAMMER, validating the fact that TPS-HAMMER generates smoother deformation fields.
4. Conclusion
In this paper, we address the shortcomings of HAMEMR and re-formulate it using a unified framework which takes advantage of a soft correspondence mechanism and TPS regularization. The deformation field between two images can be solved by alternatively identifying the sparse point correspondences and estimating the dense deformation field. Incorporating the proposed improvements into HAMMER brings forth promising results, not only in registration accuracy, but also in computation time. TPS-HAMMER produces better registration results at less than half the computation time required by HAMMER, indicating its potential for more accurate and rapid neuroimage analysis.
The research on volume-preserving diffeomorphism has become more and more important in computational anatomy community, where both invertability of deformation field and one-to-one correspondence are highlighted [21, 22]. However, TPS is not able to afford the diffeomorphism since it does not enforce boundary conditions on the spline functions [22]. Therefore, our future work will include: 1) investigating an efficient algorithm that can produce dense diffeomorphic deformation field; and 2) extending our registration framework for improving the performance of other existing registration algorithms, such as Shen [23] and Wu et al. [24].
References
- 1.Evans AC, Dai W, Collins L, Neelin P, Marrett S. Warping of a computerized 3D atlas to match brain image volumes for quantitative neuroanatomical and functional analysis. SPIE Proc., Image Proc. Algorithms and Techniques. 1991;vol. 1445:236–246. [Google Scholar]
- 2.Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE Transactions on Medical Imaging. 2002 November;vol. 21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 3.Christensen GE. Consistent linear-elastic transformations for image matching. MICCAI. 1999:224–237. [Google Scholar]
- 4.Davatzikos C, Prince JL. Brain image registration based on curve mapping. Proceedings of the IEEE Workshop on Biomedical Image Analysis. 1994:245–254. [Google Scholar]
- 5.Kelemen A, Szekely G, Gerig G. Elastic model-based segmentation of 3-D neuroradiological data sets. IEEE Transactions on Medical Imaging. 1999 October;vol. 18:828–839. doi: 10.1109/42.811260. [DOI] [PubMed] [Google Scholar]
- 6.Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage. 2004;vol. 23 Suppl 1:S151–S160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 7.Shen DG, Davatzikos C. Very high resolution morphometry using mass-preserving deformations and HAMMER elastic registration. NeuroImage. 2003 January;vol. 18:28–41. doi: 10.1006/nimg.2002.1301. [DOI] [PubMed] [Google Scholar]
- 8.Shen D, Liu D, Liu H, Clasen L, Giedd J, Davatzikos C. Automated Morphometric Study of Brain Variation in XXY Males. NeuroImage. 2004;vol. 23:648–653. doi: 10.1016/j.neuroimage.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 9.Davatzikos C, Genc A, Xu D, Resnick SM. Voxel-Based Morphometry Using the RAVENS Maps: Methods and Validation Using Simulated Longitudinal Atrophy. NeuroImage. 2001;vol. 14:1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- 10.Lao Z, Shen D, Xue Z, Karacali B, Resnick SM, Davatzikos C. Morphological classification of brains via high-dimensional shape transformations and machine learning methods. Neuroimage. 2004;vol. 21:46–57. doi: 10.1016/j.neuroimage.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 11.Beresford T, Arciniegas D, Alfers J, Clapp L, Martin B, Beresford H, Du Y, Liu D, Shen D, Davatzikos C, Laudenslager M. Hypercortisolism In Alcohol Dependence and Its Relation to Hippocampal Volume Loss. Journal of Studies on Alcohol. 2006;vol. 67:861–867. doi: 10.15288/jsa.2006.67.861. [DOI] [PubMed] [Google Scholar]
- 12.Beresford TP, Arciniegas DB, Alfers J, Clapp L, Martin B, Du Y, Liu D, Shen D, Davatzikos C. Hippocampus Volume Loss Due to Chronic Heavy Drinking. Alcoholism: Clinical and Experimental Research. 2006;vol. 30:1866–1870. doi: 10.1111/j.1530-0277.2006.00223.x. [DOI] [PubMed] [Google Scholar]
- 13.Shen D, Davatzikos C. Measuring Temporal Morphological Changes Robustly in Brain MR Images Via 4-Dimensional Template Warping. NeuroImage. 2004 April;vol. 21:1508–1517. doi: 10.1016/j.neuroimage.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Chui H, Rangarajan A. A new point matching algorithm for non-rigid registration. Computer Vision and Image Understanding. 2003;vol. 89:114–141. [Google Scholar]
- 15.Shen D. Fast image registration by hierarchical soft correspondence detection. Pattern Recognition. 2009;vol. 42:954–961. doi: 10.1016/j.patcog.2008.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bookstein FL. Principal Warps: Thin-Plate Splines and the Decomposition of Deformations. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1989;vol. 11:567–585. [Google Scholar]
- 17.Lo CH, Don HS. 3-D Moment Forms: Their Construction and Application to Object Identification and Positioning. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1989;vol. 11:1053–1064. 1989. [Google Scholar]
- 18.Suman B, Kumar P. A survey of simulated annealing as a tool for single and multiobjective optimization. J Oper Res Soc. 2005;vol. 57:1143–1160. [Google Scholar]
- 19.Christensen GE, Geng X, Kuhl JG, Bruss J, Grabowski TJ, Pirwani IA, Vannier MW, Allen JS, Damasio H. Introduction to the Non-rigid Image Registration Evaluation Project (NIREP); 3rd International Workshop on Biomedical Image Registration; 2006. pp. 128–135. [Google Scholar]
- 20.Xue Z, Shen D, Karacali B, Stern J, Rottenberg D, Davatzikos C. Simulating deformations of MR brain images for validation of atlas-based segmentation and registration algorithms. NeuroImage. 2006 Nov 15;vol. 33:855–866. doi: 10.1016/j.neuroimage.2006.08.007. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pennec X. Intrinsic Statistics on Riemannian Manifolds: Basic Tools for Geometric Measurements. Journal of Mathematical Imaging and Vision. 2006;vol. 25:127–154. [Google Scholar]
- 22.Marsland S, Twining CJ. Constructing diffeomorphic representations for the groupwise analysis of nonrigid registrations of medical images. Medical Imaging, IEEE Transactions on. 2004;vol. 23:1006–1020. doi: 10.1109/TMI.2004.831228. [DOI] [PubMed] [Google Scholar]
- 23.Shen D. Image Registration by Local Histogram Matching. Pattern Recognition. 2007;vol. 40:1161–1171. [Google Scholar]
- 24.Wu G, Qi F, Shen D. IPMI. The Netherlands: 2007. Learning Best Features and Deformation Statistics for Hierarchical Registration of MR Brain Images; pp. 160–171. [DOI] [PubMed] [Google Scholar]