Abstract
The Ras effector Rin1 is induced concomitant with synaptogenesis in forebrain neurons, where it inhibits fear conditioning and amygdala LTP. In epithelial cells, lower levels of Rin1 orchestrate receptor endocytosis. A 945bp Rin1 promoter fragment was active in hippocampal neurons and directed accurate tissue-specific and temporal expression in transgenic mice. Regulated expression in neurons and epithelial cells was mediated in part by Snail transcriptional repressors: mutation of a conserved Snail site increased expression and endogenous Snai1 was detected at the Rin1 promoter. We also describe an element closely related to, but distinct from, the consensus site for REST, a master repressor of neuronal genes. Conversion to a consensus REST sequence reduced expression in both cell types. These results provide insight into regulated expression of a neuronal Ras effector, define a promoter useful in telencephalic neuron studies, and describe a novel REST site variant directing expression to mature neurons.
Introduction
The Rin1 gene encodes a Ras effector protein that signals through downstream Rab5 GTPases and Abl tyrosine kinases to positively regulate endocytosis and cytoskeletal remodeling (Barbieri et al., 2003; Han et al., 1997; Hu et al., 2005; Tall et al., 2001). Unlike most Ras effectors, however, Rin1 shows a highly restricted pattern of expression. Rin1 is strongly expressed in neurons of telencephalic regions of the forebrain (cortex, hippocampus, amygdala, striatum and olfactory bulb) but difficult to detect in diencephalic (thalamus and hypothalamus), midbrain and hindbrain regions (Deininger et al., 2008; Dhaka et al., 2003). Rin1 expression in the brain commences postnatally and plateaus at three weeks (Dhaka et al., 2003), suggesting functions associated with mature rather than developing neurons. Rin1 is also expressed, though at significantly lower levels, in epithelial cells (Hu et al., 2005). Consistent with this distribution, Rin1-/- mice develop normally but show altered neuronal plasticity; mutant mice exhibit elevated amygdala long term potentiation and enhanced acquisition of aversive memories (Dhaka et al., 2003). In addition, mammary epithelial cells isolated from Rin1-/- mice show changes in attachment and motility compared to matched normal cells (Hu et al., 2005).
Several transcription factors are known to regulate neuronal gene expression. REST (RE1 silencing transcription factor) is a zinc finger protein that binds to conserved sequences in the promoters of many neuronal genes and represses their transcription in non-neuronal cells and in neuronal precursor cells (Ballas et al., 2005). The role of REST in gene expression appears to be more complex, however. Deletion of a REST binding site in one gene caused reduced expression in postnatal neurons (Kallunki et al., 1998) and dominant negative REST enhanced the expression of several target genes (Otto et al., 2007). This diversity in transcriptional regulation likely reflects both deviations from the consensus REST binding site (Johnson et al., 2007; Otto et al., 2007) and the participation of multiple REST co-factors (Ballas et al., 2005).
Epithelial cell gene expression, like neuronal gene expression, is controlled through multiple factors. Among the most extensively studied regulators of epithelial genes are Snai1 (a.k.a. Snail) and related zinc finger transcription factors. Snai1 binds to E-box sequences in select promoters and directly represses genes required for epithelial cell phenotypes, such as the gene encoding adherens junction protein Cdh1 (a.k.a. E-cadherin) (Cano et al., 2000). Elevated expression of Snai1 causes broad transcriptional reprogramming with silencing of numerous epithelial determinant genes and drives an epithelial-to-mesenchymal transition (EMT) (reviewed in (Barrallo-Gimeno and Nieto, 2005)).
We demonstrate here that a 945 bp promoter fragment from the mouse Rin1 gene is sufficient to confer spatial and temporal regulation to a transgene in cultured cells and whole animals. We also highlight a role for Snai1 binding in the repression of Rin1 expression in multiple cell types. Finally, we describe a variant REST binding site that is highly conserved in mammalian Rin1 genes, but acts to enhance rather than repress expression. Our findings provide new insight into the diversity of control elements required to maintain a complex pattern of expression for a Ras effector gene. In addition, the minimal promoter defined here should be an extremely useful tool for transgene analysis requiring expression restricted primarily to postnatal forebrain neurons.
Results
Rin1 Expression is Restricted in Place and Time
To define the sequence elements controlling the restricted pattern of Rin1 expression, we first sought to establish appropriate model systems. The murine mammary gland derived-cell line NMuMG shows epithelial properties including polarity and the capacity to form three dimensional luminal structures (Hall et al., 1982). Rin1 protein was observed in NMuMG extracts, although immunoprecipitation was required prior to immunoblotting to detect the relatively low level of expression in these cells (Fig. 1A). This result was consistent with the moderate level of Rin1 found in primary mammary epithelial cells from mouse (Hu et al., 2005) and human (Milstein et al., 2007). We were unable to detect Rin1 in cultured mouse embryo fibroblast (MEF) cells (Fig. 1A).
Figure 1.
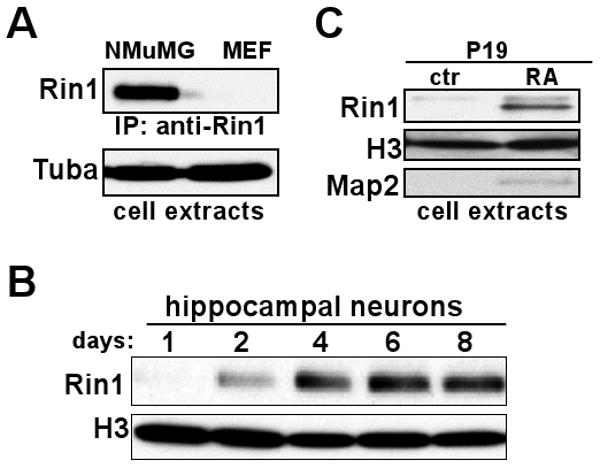
Restricted Expression of Rin1. A. Immunoprecipitation and immunoblot of cell extracts from NMuMG and primary MEF cells using anti-Rin1 (polyclonal and monoclonal, respectively). Total protein used for immunoprecipitation was normalized using Bradford assay and confirmed with an anti-α-tubulin (Tuba) blot. B. Immunoblot of extracts from mouse hippocampal neurons grown in culture for the indicated time, using anti-Rin1 and normalized using anti-histone 3 (H3). Histone expression was used because tubulin levels change during this period of extensive neurite outgrowth. C. Immunoblot of extracts from P19 cells grown under control conditions (ctr) or after neuronal differentiation with retinoic acid (RA). The anti-Rin1 signal was normalized using anti-H3, and anti-Map2 was used to validate differentiation.
We next examined cultured mouse hippocampal neurons prepared from newborn (P0) animals. Rin1 was initially undetectable but was apparent by the second day (Fig. 1B). Rin1 protein levels appeared to plateau at six days in culture. Extensive synapse formation had taken place during this period, suggesting that Rin1 gene induction may be coupled with synaptogenesis. This temporal regulation of Rin1 protein parallels the post-natal induction of Rin1 mRNA observed by in situ analysis of mouse brain tissue (Dhaka et al., 2003), though with more rapid kinetics.
We considered the mouse teratocarcinoma-derived cell line P19 as a model for neuronal expression. P19 cells differentiate along a neuronal lineage, showing axonal and dendritic extensions and the expression of numerous neuron-specific genes, following retinoic acid (RA) treatment (reviewed in (Bain et al., 1994)). We found that Rin1 protein levels were induced in RA treated P19 cells, concomitant with expression of the neuronal marker Map2 (Fig. 1C) and morphological differentiation (data not shown). This result is consistent with the induction of Rin1 gene expression being part of a neuronal differentiation program.
A Minimal Rin1 Promoter Drives Expression in Cultured Neurons
To define the requirements for regulated expression of Rin1, we isolated a promoter sequence from the murine Rin1 gene. The 945 bp sequence begins immediately following the transcription stop of an upstream gene (Brms1) and includes the 51 bp 5′ untranslated region of Rin1. This sequence was placed upstream of the eGFP coding sequence to create a Rin1 expression reporter, R1pr-GFP, which was then cloned into a lentivirus construct (Fig. 2A). Because we used a self-inactivating virus vector with a defective LTR enhancer, and because no other promoter is included within the provirus, GFP expression was controlled by the Rin1 promoter sequence. Infection of wild type hippocampal cell cultures with the R1pr-GFP lentivirus resulted in an easily detected GFP signal (Fig. 2B). GFP expression was seen almost exclusively in neuronal cells, which were identified with anti-Map2, demonstrating that the cloned Rin1 promoter confers cell-type expression specificity (Fig. 2C). In parallel, we analyzed a vector containing 1,290 bp of the Camk2a (a.k.a. Cam Kinase IIαor CaMK2α) promoter to drive GFP expression (Dittgen et al., 2004). The Camk2a gene is normally expressed at high levels in both telencephalic and diencephalic regions of the forebrain and to a lesser extent in other parts of the brain (Benson et al., 1992). A Camk2a promoter fragment can limit expression of transgenes primarily to forebrain neurons and has been used to direct expression in transgenic mice for learning and memory studies (Limback-Stokin et al., 2004; Mayford et al., 1996), although little is known about the specific elements regulating transcription from this sequence. The transduced Camk2pr-GFP lentivirus gave a more intense signal but showed an expression pattern similar to that seen for R1pr-GFP; GFP fluorescence was observed in a high proportion of neurons and rarely seen in non-neuronal cells (Fig. 2B, C). In contrast to the Rin1 and Camk2a promoter constructs, a cytomegalovirus (CMV) enhancer/chicken β-actin promoter lentivirus vector expressed GFP in non-neuronal cells as well as some neurons, consistent with published observations (Dittgen et al., 2004). These results demonstrate the neuronal specificity of the cloned Rin1 promoter fragment.
Figure 2.
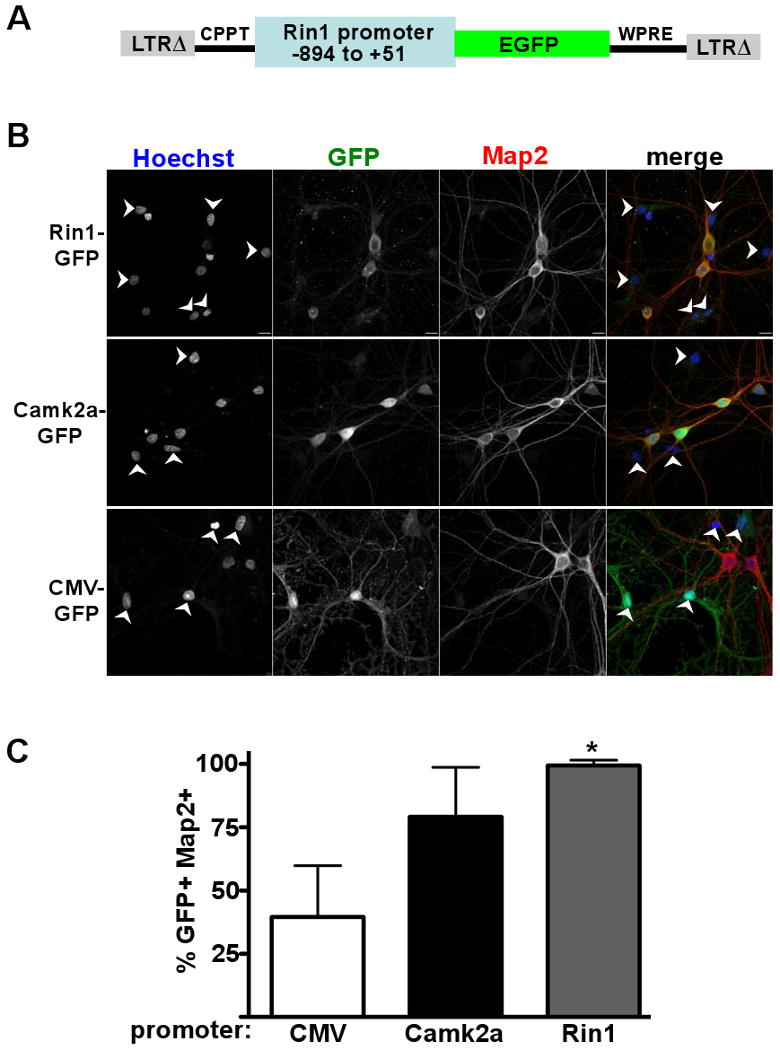
A Rin1 promoter fragment confers neuronal expression to a GFP reporter. A. Diagram of the lentivirus construct used in the expression experiments. LTRΔ = long terminal repeat with self-inactivating deletion; CPPT = central polypurine tract; GFP = green fluorescent protein; WPRE = woodchuck hepatitis virus post-transcriptional regulatory element. B. Rat hippocampal neurons infected with the promoter-GFP virus indicated at left, and visualized as indicated above with Hoechst (nuclear stain), anti-GFP, anti-Map2 and as a merged image. Arrows indicate cells not stained with anti-Map2. Scale bars (10 microns) are in bottom right of top row images. C. Statistical analysis of infected cultures shown in B. For each cell population, 12 independent fields (> 450 total cells) were evaluated. GFP and Map2 double immunofluorescent cells (GFP+ and Map2+) are given as a percentage of total GFP+ cells, with standard deviation. * p < 0.001 (comparing Rin1 and CMV promoter signals)
A Cloned Rin1 Promoter Recapitulates Restricted Expression in Transgenic Mice
To examine tissue-specific expression from the minimal Rin1 promoter in vivo, we created transgenic mice by direct infection of pronuclear stage embryos with the R1pr-GFP virus. This methodology has been used to efficiently produce transgenic animals ((Lois et al., 2002) and reviewed in (Sauvain et al., 2008)), but has not been widely used with restricted expression vectors. Four independent R1pr-GFP founder mice were established.
Transgenic mice from two independent founders showed clear GFP expression in cortex, hippocampus, amygdala and striatum regions (Fig. 3A, B). This distribution closely correlated with the expression of endogenous Rin1 mRNA (Deininger et al., 2008; Dhaka et al., 2003) and Rin1 protein (Fig. 3A, B) (Deininger et al., 2008; Dhaka et al., 2003). No transgene signal above background was seen in the thalamus (Fig. 3A, B) or hindbrain (data not shown), again indicating that transgene expression from the Rin1 promoter was correctly restricted to telencephalic forebrain regions. Immunoblot analysis of brain tissue extracts from the progeny of founder T1 confirmed that GFP expression paralleled the expression of endogenous Rin1 protein; both gene products were easily observed in forebrain but undetectable in hindbrain (Fig. 3C, top left). Tissues representative of hematopoietic cells (thymus), epithelial cells (lung) and muscle cells (calf muscle) showed no detectable Rin1 and relatively weak or undetectable GFP. Progeny from an independent founder (T2) showed GFP expression that was high in forebrain, moderate in lung and thymus, and weak in hindbrain and muscle (Fig. 3C, bottom left). We next performed immunoprecipitations from larger amounts of tissue extract to amplify the immunoblot signal. Under these conditions, it was possible to detect endogenous Rin1 in epithelial cell-rich lung tissue (Fig. 3C, right). Again, GFP expression appeared to parallel Rin1 expression with the exception that some GFP was seen in skeletal muscle, where endogenous Rin1 was undetectable.
Figure 3.
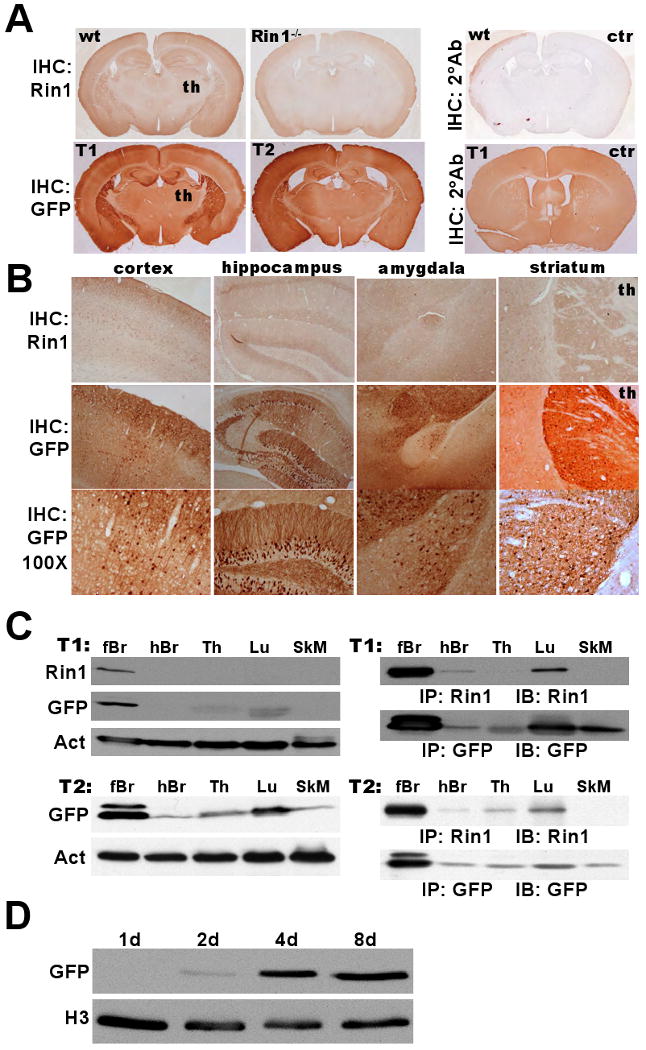
Rin1 promoter-driven GFP expression in transgenic mice. A. Top: Wild type (wt) or Rin1-/- mouse brain coronal sections subjected to immunohistochemistry (IHC) staining with anti-Rin1 or secondary antibody (2°) alone as control (ctr). Signal in thalamus (th) is near background level. Bottom: Two independently derived Rp-GFP transgenic animals (T1 & T2) stained with anti-GFP or secondary antibody alone (ctr). B. Top: Rin1 staining in designated telencephalic regions of wild type brain (th = thalamus). Middle: GFP staining from T1 tissue (40× mag). Bottom: same as above (100× mag). C. Immunoblot detection of Rin1 and GFP expression in T1 and T2 transgenic mouse tissues (forebrain (fBr), hindbrain (hBr), thymus (Th), lung (Lu) and skeletal muscle (SkM)). Left: Direct immunoblot of tissue extracts, with actin (Act) normalization. Right: Immunoblot (IB) of immunoprecipitated (IP) material from tissue extracts. D. Dissociated hippocampal neurons from a transgenic (T1) mouse cultured for the indicated time and immunoblotted with anti-GFP, normalized with anti-histone 3 (H3).
To examine whether the Rin1 promoter fragment conferred temporal regulation to the transgene, we isolated and cultured hippocampal neurons from newborn R1pr-GFP transgenic mice. GFP was undetectable until the fourth day in culture, with expression levels increasing at later time points (Fig. 3D). This closely paralleled the pattern seen for endogenous Rin1 protein in neuron cultures (Fig. 1B), suggesting that the promoter fragment includes all sequences required to properly couple expression with maturation of forebrain neurons.
Taken together, these results demonstrate that the 945 bp Rin1 promoter sequence contains the elements necessary to correctly guide temporally inducible, high level expression in telencephalic forebrain neurons as well as lower level expression in several other tissue types.
Multiple Regions Contribute to Regulated Expression in the Rin1 Promoter
The Rin1 promoter sequence is well conserved among seven mammalian suborders examined (Supplemental Fig. 1), with especially high sequence identity surrounding the mouse transcription and translation start sites. All of the mammalian Rin1 promoters appear to be TATA-less and most likely employ an initiator element. Several islands of sequence identity appear within the alignment. A particularly strong region of similarity was found between nucleotides -378 and -680 (Supplemental Fig. 1) and includes a striking 20-nucleotide perfect identity.
We next created a luciferase reporter construct to quantify the activity of the 945 bp mouse Rin1 promoter in representative epithelial (NMuMG) and neuronal (differentiated P19) cell types. Luciferase reporter signals were moderate in NMuMG and high in differentiated P19 cells (Fig. 4A). These results are in agreement with the tissue-specific GFP transgene expression observed using the same Rin1 promoter fragment (Fig. 3).
Figure 4.
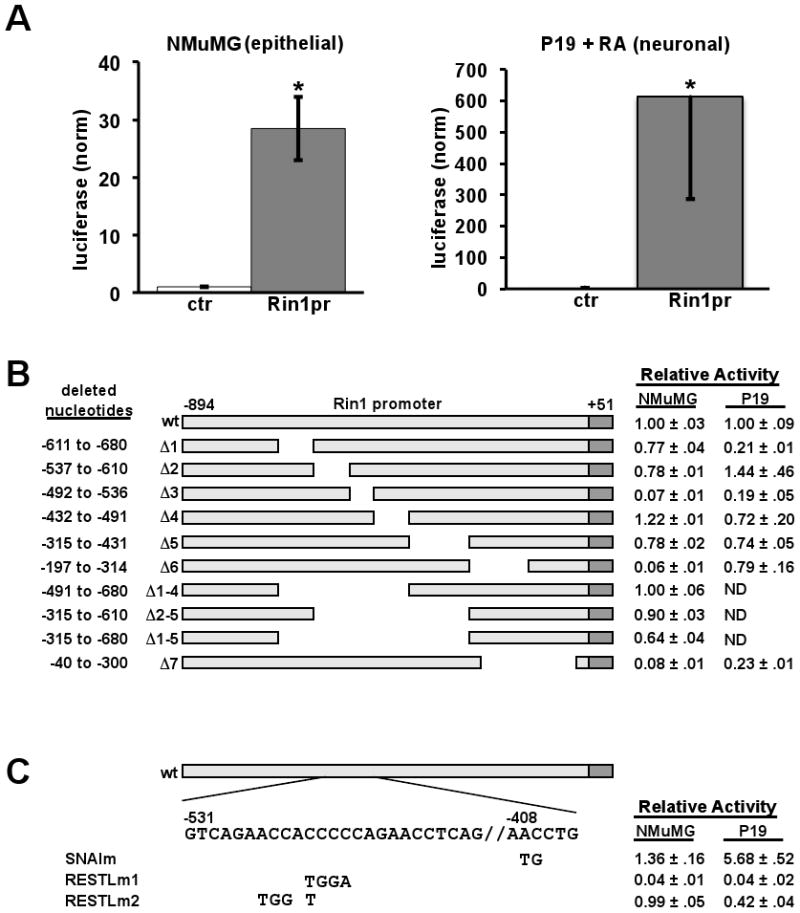
Identification of regulatory elements in the Rin1 promoter. A. Normalized luciferase expression from a promoterless (ctr) or Rin1 promoter vector in NMuMG cells (left) or differentiated P19 cells (P19 + RA, right). B. Rin1 promoter deletion mutants pictured with deletion range shown at left and luciferase expression in NMuMG and P19+RA cells, normalized to the wild type promoter, at right. C. Point mutations within a putative Snail element and a REST-like element. Normalized luciferase expression in NMuMG and P19+RA cells at right
A series of deletion mutations was used to identify promoter regions with the greatest influence on expression (Fig. 4B). Deletion of the -492 to -536 (Δ3), -197 to -314 (Δ6) and -40 to -300 (Δ7) regions decreased expression by more than ten fold in NMuMG, an epithelial cell line that showed moderate levels of endogenous Rin1 protein (Fig. 1A). When tested in differentiated P19 cells that model neurons, reporter activities from the Δ1, Δ3 and Δ7 constructs showed the most pronounced diminution. These results suggested that the Rin1 promoter contains elements that influence expression primarily in epithelial cells (Δ6) and elements that regulate expression primarily in neuronal cells (Δ1). There also appear to be regions that control expression levels across cell types (Δ3 and Δ7). Interestingly, few of the promoter deletion constructs showed significant increases in reporter gene expression, suggesting that positive cis-acting sequences predominate in this promoter.
SNAI1, a Repressor of Epithelial Cell Genes, Inhibits Rin1 Expression
The 945 bp mouse Rin1 promoter has 12 putative binding sites for the Snail transcription repressor (Snai1-3 and related Zn finger transcription factors). This is the largest number of identified sites for any transcription factor in the mouse Rin1 promoter sequence, an observation that holds true for the equivalent sequence in the rat (19 Snail sites) and human (21 Snail sites) Rin1 gene promoters. Three of the best-conserved Snail sites reside within the Δ1, Δ2 and Δ4 regions. Only the Δ4 deletion construct showed an increase in expression, however, and this was modest. The absence of large expression increases corresponding to deletion of predicted repressor sites could be due to confounding effects from the large deletions with consequent juxtaposition of boundary sequences. We therefore created a double point mutation (Snaim) to specifically disrupt the Snail site located in the Δ4 region. We again observed a modest increase in reporter gene expression in NMuMG cells (Fig. 4C).
Reasoning that loss of an individual Snail binding site may have relatively little effect on epithelial cell expression due to Snail binding at several other sites on the promoter, we next looked for evidence of Snai1 transcription factor binding on the Rin1 promoter using chromatin immuno-precipitation (ChIP). Using the human cell line MDA-MB-231 because of its epithelial cell origin and relatively high Snail levels, these experiments revealed an association of endogenous Snai1 with the chromosomal Rin1 promoter (Fig. 5A). The results are consistent both with our promoter mutation studies (Fig. 4) and with the inverse correlation between human SNAI1 and RIN1 expression in a breast tumor cell line (Milstein et al., 2007).
Figure 5.
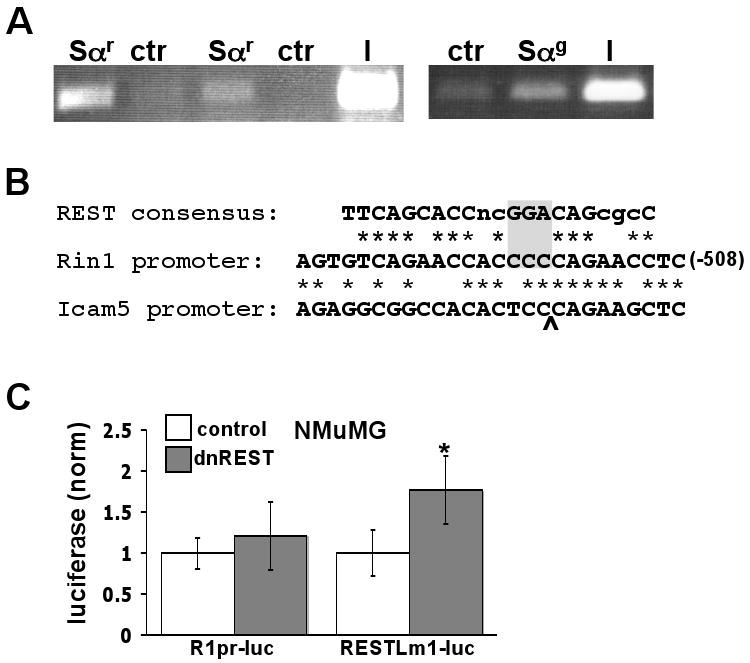
Contributions of Snail and REST-like elements in the Rin1 promoter. A. Chromatin immunoprecipitation analysis of SNAI1 binding to the RIN1 promoter in human MDA-MB-231 cells. Sαr = rabbit anti-SNAI1; Sαg = goat anti-SNAI1; ctr = anti-Flag; I = input (10% of pre-immunoprecipitation material). ImageQuant analysis of the two independent Sαr experiments gave a normalized signal of 2.3 ± 0.5. B. Alignment of the REST binding site consensus sequence, REST-like element of the Rin1 promoter and related sequence in the Icam5 (a.k.a. telencephalin) promoter. “*” denotes sequence identity. Gray box highlights nucleotides altered in RESTLm1. Carrot indicates position of sequence gap in alignment with Icam5. C. Expression of dominant negative REST (dnREST) represses luciferase expression from the RESTLm1 mutant promoter but not from the wild type Rin1 promoter in NMuMG cells. Luciferase assay values are normalized to control (no dnREST) and represent the mean and standard deviation from 9 independent assays. * p < 0.01
Surprisingly, we observed increased expression from the Snaim construct in differentiated P19 cells. Although these cells are not known to express Snai1, they do express the related transcriptional repressor Scrt1 (a.k.a. scratch (Nakakura et al., 2001). It should also be noted that while this conserved site (AACCTG) is scored as a Snail site by transcription factor algorithms, it varies from the most common type of E-box (CAxxTG), and could represent a specialized element for transcriptional repression in neurons.
A Novel REST-Like Element Contributes to Regulation of Rin1 Expression
The longest stretch of sequence identity among the ten mammalian Rin1 genes examined starts at position −528 and, with a T/C variation at two positions, extends almost perfectly over 26 nucleotides (Supplemental Fig. 1). The sequence bares a striking resemblance to the consensus recognition site for the transcription repressor REST (Fig. 5B). Moreover, Rin1 was identified in a large-scale screen for REST target genes (Otto et al., 2007), suggesting that this conserved element might mediate REST binding in the Rin1 promoter.
Deletion of a 45 bp region including the REST-like sequence (Δ3) led to a reduction in reporter gene expression in NMuMG epithelial cells, suggesting that this region includes elements that enhance expression. This was somewhat surprising because REST normally represses transcription of neuronal genes in non-neuronal cells (Chong et al., 1995). Because the Δ3 result may have been influenced by the removal of bordering sequences, we examined the function of the REST-like element using point mutations. RESTLm1 carries four nucleotide mutations that create a stronger match with the consensus REST element. These changes led to a marked decrease in reporter gene expression in NMuMG cells (Fig. 4C), consistent with more efficient silencing by endogenous REST. We further tested the potential role of REST in repression of the Rin1 promoter using a dominant negative REST construct. While the wild type Rin1 promoter was unaffected, the RESTLm1 mutant promoter showed a modest though significant induction when co-transfected with dominant negative REST (Fig. 5C). These data suggested that the Rin1 promoter REST-like element is at best a weakly responsive site for repression by REST in epithelial cells.
When examined in differentiated P19 cells, the RESTLm1 mutant promoter gave reduced expression similar to that observed in NMuMG cells. Differentiated P19 cells have low levels of endogenous REST (Okamoto et al., 1999), however, suggesting that in this cell context the Rin1 promoter REST-like element may bind a repressor with similar sequence preference to REST, or may recruit a previously uncharacterized transcription activator.
To resolve how the REST-like element contributes to regulated expression we created RESTLm2, which changes four nucleotides identical across mammalian Rin1 promoters (Supplemental Fig. 1) while reducing overall sequence relatedness to the consensus REST binding site. This mutant was essentially unchanged in reporter expression levels compared to the wild type promoter in NMuMG cells (Fig. 4C). By contrast, the RESTLm2 mutant promoter had significantly reduced expression compared to the wild type Rin1 promoter in differentiated P19 (Fig. 4C).
Discussion
Although substantial progress has been made in defining transcription machinery components and the sequence elements they recognize, it remains difficult to define promoters and predict patterns of expression from their sequence. We show here that expression of Rin1 is restricted by cell type and, in neurons, is temporally regulated. We also show that a 945 bp Rin1 promoter fragment is sufficient to confer a similar pattern of expression on a reporter gene in cultured cells and transgenic animals. It remains possible, of course, that distal elements or 3′ UTR sequences not included in this fragment may contribute to regulated expression of Rin1 in vivo. Any such contribution would appear to be minor, but might, for instance, dictate the level of expression in neuron subtypes. Still, the promoter fragment described here should prove useful for moderate transgene expression restricted primarily to mature telencephalic neurons.
Localization of high level Rin1 expression to the telencephalic regions of the forebrain is unusual but not unique. A search of the Allen Brain Atlas (mouse.brain-map.org) for similar patterns of expression identified Icam5 (a.k.a. telencephalin) and Ptk2b, both of which appear to have TATA-less promoters, as the highest matches (data not shown). Indeed, a 1.1 kb fragment of the Icam5 promoter was used successfully in conventionally constructed transgenic mice to direct expression to the telencephalon (Mitsui et al., 2007). Comparison of the 945 bp Rin1 and 1.1 kb Icam5 promoter fragments revealed a partial, interrupted, alignment within the REST-like sequence (Fig. 5B). Otherwise, these promoter fragments showed only modest similarity, highlighting the limitation of using sequence alignment for expression pattern prediction.
The presence of multiple E-box type Snail binding sites within the Rin1 promoter, and the detection of Snai1 binding to the promoter, strongly suggests a role for Snail family transcription factors in the repression of Rin1 transcription. This interpretation is also consistent with the observation that human SNAI1 is overexpressed and RIN1 is silenced in the breast tumor cell line ZR75-1, and that SNAI1 silencing restores RIN1 expression in these cells (Milstein et al., 2007). We found, however, that deletion or mutation of individual Snail binding sites at best moderately increased expression from the Rin1 promoter. This may indicate that Snail family transcriptional repressors normally bind at multiple sites along the promoter, perhaps working cooperatively, and that any individual site contributes only modestly to repression.
The REST-like element described in this study extends over 26 base pairs and includes nucleotides that diverge from the consensus REST site. Although this element may act in part as a weak REST binding site for transcription silencing in non-neuronal cells (allowing low level expression in mammary epithelial cells), its striking conservation among mammalian Rin1 gene promoters suggests that it carries out another function. The reduced expression seen for both the RESTLm1 and RESTLm2 mutant promoters in differentiated P19 cells suggested that this element may positively regulate expression in neuronal cells. Indeed, other transcription factors have been shown to utilize subsets of distinct binding sites to achieve different outcomes (Blauwkamp et al., 2008; So et al., 2007). It should also be noted that REST functions in neuronal stem/progenitor cells (Johnson et al., 2008; Su et al., 2006) and is expressed in some mature neurons (Koenigsberger et al., 2000; Palm et al., 1998; Shimojo and Hersh, 2004). Further, multiple REST isoforms (Palm et al., 1998) and REST-containing protein complexes (Belyaev et al., 2004) have been described. The Rin1 promoter REST-like element may work through recruitment of a specialized REST complex in neurons, perhaps without the CoREST factor needed for silencing (Andres et al., 1999), and this may positively regulate expression. Alternatively, an unidentified neuronal transcription factor with binding site specificity similar to REST may function at this site.
The Rin1 gene product is an established Ras effector that stimulates Rab5-mediated endocytosis and Abl-mediated actin remodeling. The restriction of Rin1 protein to specific cell types and by stage of postnatal development should both confine the effects of these signaling pathways and prevent unwanted interference with Ras signaling through alternate effectors (e.g. Raf and PI3K). The combination of multiple Snail binding sites and a highly conserved REST-like element are likely to be instrumental in directing proper expression, but distal sequences and chromatin structure undoubtedly contribute additional regulation to Rin1 gene expression.
Experimental Methods
Plasmid Construction and Mutagenesis
The 945 bp mouse Rin1 promoter fragment was isolated from M99RB (Dhaka et al., 2003) and ligated to the eGFP coding region, which shares an in-frame NcoI restriction site surrounding the translation start site, to create pKS-RP-GFP. The same promoter fragment was cloned into pGL3B to create the luciferase reporter plasmid GL3B-R1pr. The Rin1 promoter-driven GFP lentivirus, M4-RP-GFP, was created by replacing the internal CMV promoter of the M4 vector (Hu et al., 2005) with the 945 bp Rin1 promoter fragment and downstream eGFP coding sequence from pKS-R1pr-GFP. Point mutations and Δ1 through Δ6 were created using PCR primers shown in Table 1. Deleting between a HindIII site introduced in Δ6 and a natural SacI site created the Δ7 mutation. The Renilla luciferase construct pRL-CMV (Promega) was used to normalize luciferase readings from transfected NMuMG cells. The Camk2a-GFP construct (gift of Pavel Osten) has been previously described (Dittgen et al., 2004).
Table 1.
Nucleotide sequence of the primers used to create deletions and point mutations in the mouse Rin1 promoter. The mutated nucleotides are in bold and lower case. Newly created restriction sites (RESTLm1= Nco I, RESTLm2= BamH, SNAILm1= Sph I, Δ5= HindIII) as a result of the mutation are underlined.
| mutant | Primers (5′ → 3′) |
|---|---|
| RESTLm1 | F: 5′-GTGAGTGTCAGAACCAtggaCAGAACCTCAGTTAAGC-3′ R: 5′-GCTTAACTGAGGTTCTGtccaTGGTTCTGACACTCAC-3′ |
| RESTLm2 | F: 5′-CTGGTTAACAGTGAGTGTCAGAtggAtCCCCAGAACCTCAGTTAAGCAAAG-3′ R: 5′-CTTTGCTTAACTGAGGTTCTGGGGaTccaTCTGACACTCACTGTTAACCAG-3′ |
| SNAILm | F: 5′-CTTTCTCCATAGCAtgCTGAAATCACAAAAC-3′ R: 5′-GTTTTGTGATTTCAGcaTGCTATGGAGAAAG-3′ |
| Δ1 | F: 5′-TGGTGAAGGGACTTTTGGACATGCCCCTGTGTGTGGGAGGCAGCTAGCAG-3′ R: 5′-CTGCTAGCTGCCTCCCACACACAGGGGCATGTCCAAAAGTCCCTTCACCA-3′ |
| Δ2 | F: 5′-TGGCAAGGCAGGCTAGCTAGCAGGGGTGAGTGTCAGAACCACCCCCAGAA-3′ R: 5′-TTCTGGGGGTGGTTCTGACACTCACCCCTGCTAGCTAGCCTGCCTTGCCA-3′ |
| Δ3 | F: 5′-AGCCACACTCCCCCTCTGGTTAACATAGTTTCCATGGAGACACAGCATCC-3′ R: 5′-GGATGCTGTGTCTCCATGGAAACTATGTTAACCAGAGGGGGAGTGTGGCT-3′ |
| Δ4 | F: 5′-CAGAACCTCAGTTAAGCAAAGAGCTTTAGGTTCAGGAATGAGTCATCTGC-3′ R: 5′-GCAGATGACTCATTCCTGAACCTAAGCTCTTTGCTTAACTGAGGTTCTG-3′ |
| Δ5 | F: 5′-GCAACCTGAAATCACAAAACCCAAGCTTCTTAAGGTGACCCAGGGGTGTG-3′ R: 5′-CACACCCCTGGGTCACCTTAAGAAGCTTGGGTTTTGTGATTTCAGGTTGC-3′ |
| Δ6 | F: 5′-GAGCCGATGAGCCAGTGACCACAGATGGTCCTCCCTTAGCCGGGGCCTGA-3′ R: 5′-TCAGGCCCCGGCTAAGGGAGGACCATCTGTGGTCACTGGCTCATCGGCTC-3′ |
| Δ2→Δ5 | F: 5′-TGGCAAGGCAGGCTAGCTAGCAGGGCTTCTTAAGGTGACCCAGGGGTGTG-3′ R: 5′-CACACCCCTGGGTCACCTTAAGAAGCCCTGCTAGCTAGCCTGCCTTGCCA-3′ |
| Δ1→Δ4 | F: 5′-TGGTGAAGGGACTTTTGGACATGCCTTAGGTTCAGGAATGAGTCATCTGC-3′ R: 5′-GCAGATGACTCATTCCTGAACCTAAGGCATGTCCAAAAGTCCCTTCACCA-3′ |
| Δ1→Δ5 | F: 5′-TGGTGAAGGGACTTTTGGACATGCCCTTCTTAAGGTGACCCAGGGGTGTG-3′ R: 5′-CACACCCCTGGGTCACCTTAAGAAGGGCATGTCCAAAAGTCCCTTCACCA-3′ |
Cell Culture and Virus Production
Mouse embryo fibroblasts (MEF, gift of Hong Wu, UCLA) were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). NMuMG were obtained from Dr. Tim Lane and were grown in DMEM with 10% FBS. P19 were obtained from Dr. Robert Chui and were propagated in MEM-α (MediaTech) with 7.5% FBS and 2.5% calf serum, and differentiated by the addition of 2 μM retinoic acid (RA) according to an established protocol (Jones-Villeneuve et al., 1983). Primary hippocampal neurons were generated from newborn (P0) wild type rats or transgenic mice. Rat neurons were cultured for several weeks in serum-free medium supplemented with B27 prior to infection and immunofluorescence analysis. Mouse neurons were cultured for the indicated time prior to lysis and immunoblot analysis. HEK293T cells were maintained in DMEM plus 10% fetal calf serum. Lentivirus was produced using an established protocol (Hu et al., 2005).
Immunoprecipitations, Immunoblots, Immunohistochemistry and Immunofluorescence
Immunoprecipitations from transgenic mouse tissue extracts were performed with polyclonal anti-mouse Rin1 (Hu et al., 2005) or anti-GFP (polyclonal, Molecular Probes #A11122), as indicated. Immunoblots were performed with monoclonal anti-mouse Rin1 (Hu et al., 2005), anti-GFP (monoclonal, Covance #MMS-118P), anti-Map2 (Sigma, M9942s), anti-histone H3 (polyclonal, Millipore, 07690) or anti-actin (Sigma-Aldrich, A5060). Immunohistochemical staining of brain tissue was performed with anti-GFP (Molecular Probes) on frozen sections or with polyclonal anti-mouse Rin1 [Dhaka, 2003 #347] on paraffin-embedded sections. The cultured neuron lysates were probed with the Covance antibody. Immunofluorescence of cultured neurons was performed with anti-GFP (polyclonal, gift of Dr. Greg Payne) and anti-Map2.
Luciferase assays
Firefly luciferase expression assays were performed using a standard protocol (Hu et al., 2008). For NMuMG cells, co-transfected Renilla luciferase was used to normalize results. For stably transfected (neoR) P19 cells, luciferase values were normalized to total protein. Mean values and standard deviations are derived from at least three independent experiments.
Transgenic mice
To create transgenic mice, concentrated lentiviruses (approx. 1 × 108 virus/ml) were microinjected into the perivitelline space of 0.5-day pronuclear stage embryos. A FemtoJet microinjector (Brinkmann) was used at settings Pi=120 and Pc=20. Injected embryos (n = 113 for M4-R1pr-GFP; n = 75 for CMV-GFP control) were implanted into pseudopregnant foster mice. From the M4-R1pr-GFP injected embryos, 34 mice were born and of these seven carried the RpGFP construct. Four independent founder mice transmitted the transgene.
Chromatin immunoprecipitation analysis
MDA-MB-231 cells were serum-starved overnight and treated with 10 μM of both SB216763 (GSK3β inhibitor, BioMol) and MG-132 (proteosome inhibitor, Sigma) for 6 hours. Cells were fixed with 1% formaldehyde at room temperature for 15 minutes before sonicated in lysis buffer (1% SDS, 10mM EDTA, 50mM Tris pH 8.1) to yield 200-1000 bp fragments. Equal amounts of sheared chromatin were immunoprecipitated with goat anti-SNAI1 (Santa Cruz Biotechnology), anti-RNA polymerase II (Millipore), or anti-Flag (Sigma) overnight at 4°C. One ChIP experiment was also completed successfully using a rabbit anti-SNAI1 (Santa Cruz Biotechnology). The samples were washed with low salt wash buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl pH 8.1, 150mM NaCl), with high salt wash buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl pH 8.1, 500mM NaCl), with LiCl wash buffer (0.25M LiCl, 1% NP-40, 1% deoxycholic acid, 1mM EDTA, 10mM Tris pH 8.1), and 2× with TE buffer before eluting (1% SDS, 100mM NaHCO3) and incubating at 65°C overnight to reverse formaldehyde cross-linking. Samples were treated with 1 mg/ml RNAse at 37°C for 30 min, then 0.5M EDTA, 1M Tris-HCl, 100 μg proteinase K at 42°C for 2 hours. DNA was purified using the QiaQuick PCR Purification Kit (Qiagen) and PCR-amplified: 1 cycle of 95°C for 10 min; 32 cycles of 95°C for 30 s, 55°C for 30s, 72°C for 30s; 1 cycle of 72°C for 2 min. Primers used were 5′- TACTAGCGGTTTTACGGGCG-3′ (GAPDH-f), 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′ (GAPDH-r), 5′-CTTGGAGCCTGGGTCATG-3′ (RIN1pr-f) and 5′-GACTCATTCCTGAACCTG-3′ (RIN1pr-r).
Sequence analysis
Identification of transcription factor binding sites was carried out using the Consite program (asp.ii.uib.no:8090/cgi-bin/CONSITE/consite). Multi-sequence alignments were performed using ClustalW (align.genome.jp).
Supplementary Material
Rin1 promoter sequences from representatives of the mammalian suborders Primates (Homo sapiens, Hs and Macaca mulatta, Ma), Glires (Rattus novegicus, Rn and Mus Musculus, Mm), Carnivora (Canis familiaris, Cf), Cetartiodactyla (Bos Taurus, Bt and Tursiops truncatus), Hyracoidea (Procavia capensis), Proboscidea (Loxodonta africana, La) and Dasypodidea (Dasypus novemcinctus, Dn). Perfect conservation is denoted by “*”. Near perfect conservation (9/10) is denoted by “:”. In the segment for which L. africana sequence is not available, conservation is based on the remaining sequences. The start codon is shown in bold. The mouse REST-like sequence is shaded in gray, and the 12 putative Snail sites (Consite analysis) are underlined.
Acknowledgments
The authors thank the following for generosity in providing reagents and advice: Robert Chiu, Jun Song, Chris Colwell, Noriyuki Kasahara, Renata Stipecke, Steven Dubinett, Hong Wu, Gail Mandel, Desmond Smith and Andy Lin. Peter Lin assisted with neuron culture experiments.
This work was supported by NIH grant RO1 NS046787 (JC), RO1 MH077022 (KCM) and UCLA Chancellor's Bioscience Initiative (MJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Ray WJ, Yao M, Gottlieb DI. From embryonal carcinoma cells to neurons: the P19 pathway. Bioessays. 1994;16:343–348. doi: 10.1002/bies.950160509. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Barbieri MA, Kong C, Chen PI, Horazdovsky BF, Stahl PD. The SRC homology 2 domain of Rin1 mediates its binding to the epidermal growth factor receptor and regulates receptor endocytosis. The Journal of biological chemistry. 2003;278:32027–32036. doi: 10.1074/jbc.M304324200. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Belyaev ND, Wood IC, Bruce AW, Street M, Trinh JB, Buckley NJ. Distinct RE-1 silencing transcription factor-containing complexes interact with different target genes. The Journal of biological chemistry. 2004;279:556–561. doi: 10.1074/jbc.M310353200. [DOI] [PubMed] [Google Scholar]
- Benson DL, Isackson PJ, Gall CM, Jones EG. Contrasting patterns in the localization of glutamic acid decarboxylase and Ca2+/calmodulin protein kinase gene expression in the rat central nervous system. Neuroscience. 1992;46:825–849. doi: 10.1016/0306-4522(92)90188-8. [DOI] [PubMed] [Google Scholar]
- Blauwkamp TA, Chang MV, Cadigan KM. Novel TCF-binding sites specify transcriptional repression by Wnt signalling. The EMBO journal. 2008;27:1436–1446. doi: 10.1038/emboj.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nature cell biology. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Deininger K, Eder M, Kramer ER, Zieglgansberger W, Dodt HU, Dornmair K, Colicelli J, Klein R. The Rab5 guanylate exchange factor Rin1 regulates endocytosis of the EphA4 receptor in mature excitatory neurons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105 doi: 10.1073/pnas.0801174105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaka A, Costa RM, Hu H, Irvin DK, Patel A, Kornblum HI, Silva AJ, O'Dell TJ, Colicelli J. The Ras effector Rin1 modulates the formation of aversive memories. J Neuroscience. 2003;23:748–757. doi: 10.1523/JNEUROSCI.23-03-00748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittgen T, Nimmerjahn A, Komai S, Licznerski P, Waters J, Margrie TW, Helmchen F, Denk W, Brecht M, Osten P. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall HG, Farson DA, Bissell MJ. Lumen formation by epithelial cell lines in response to collagen overlay: a morphogenetic model in culture. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:4672–4676. doi: 10.1073/pnas.79.15.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Wong D, Dhaka A, Afar D, White M, Xie W, Herschman H, Witte O, Colicelli J. Protein binding and signaling properties of RIN1 suggest a unique effector function. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:4954–4959. doi: 10.1073/pnas.94.10.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Bliss JM, Wang Y, Colicelli J. RIN1 is an ABL tyrosine kinase activator and a regulator of epithelial-cell adhesion and migration. Curr Biol. 2005;15:815–823. doi: 10.1016/j.cub.2005.03.049. [DOI] [PubMed] [Google Scholar]
- Hu H, Milstein M, Bliss JM, Thai M, Malhotra G, Colicelli J. Integration of TGFbeta and RAS Signaling Silences a RAB5 GEF and Enhances Growth Factor-DIrected Cell Migration. Mol Cell Biol. 2008;28:1573–1583. doi: 10.1128/MCB.01087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, Cooper ML, Volta M, Chan SS, Lipovich L, Pollard SM, et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Villeneuve EM, Rudnicki MA, Harris JF, McBurney MW. Retinoic acid-induced neural differentiation of embryonal carcinoma cells. Mol Cell Biol. 1983;3:2271–2279. doi: 10.1128/mcb.3.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki P, Edelman GM, Jones FS. The neural restrictive silencer element can act as both a repressor and enhancer of L1 cell adhesion molecule gene expression during postnatal development. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3233–3238. doi: 10.1073/pnas.95.6.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenigsberger C, Chicca JJ, 2nd, Amoureux MC, Edelman GM, Jones FS. Differential regulation by multiple promoters of the gene encoding the neuron-restrictive silencer factor. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:2291–2296. doi: 10.1073/pnas.050578797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limback-Stokin K, Korzus E, Nagaoka-Yasuda R, Mayford M. Nuclear calcium/calmodulin regulates memory consolidation. J Neurosci. 2004;24:10858–10867. doi: 10.1523/JNEUROSCI.1022-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein M, Mooser CK, Patel A, Hu H, Fejzo M, Slamon DJ, Goodglick L, Dry S, Colicelli J. RIN1 is a Breast Tumor Suppressor Gene. Cancer Res. 2007;67:11510–11516. doi: 10.1158/0008-5472.CAN-07-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui S, Saito M, Mori K, Yoshihara Y. A transcriptional enhancer that directs telencephalon-specific transgene expression in mouse brain. Cereb Cortex. 2007;17:522–530. doi: 10.1093/cercor/bhj177. [DOI] [PubMed] [Google Scholar]
- Nakakura EK, Watkins DN, Sriuranpong V, Borges MW, Nelkin BD, Ball DW. Mammalian Scratch participates in neuronal differentiation in P19 embryonal carcinoma cells. Brain research. 2001;95:162–166. doi: 10.1016/s0169-328x(01)00246-7. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Sherman K, Lipton SA. Absence of binding activity of neuron-restrictive silencer factor is necessary, but not sufficient for transcription of NMDA receptor subunit type 1 in neuronal cells. Brain research. 1999;74:44–54. doi: 10.1016/s0169-328x(99)00250-8. [DOI] [PubMed] [Google Scholar]
- Otto SJ, McCorkle SR, Hover J, Conaco C, Han JJ, Impey S, Yochum GS, Dunn JJ, Goodman RH, Mandel G. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J Neurosci. 2007;27:6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvain MO, Dorr AP, Stevenson B, Quazzola A, Naef F, Wiznerowicz M, Schutz F, Jongeneel V, Duboule D, Spitz F, Trono D. Genotypic features of lentivirus transgenic mice. J Virol. 2008;82:7111–7119. doi: 10.1128/JVI.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo M, Hersh LB. Regulation of the cholinergic gene locus by the repressor element-1 silencing transcription factor/neuron restrictive silencer factor REST/NRSF. Life sciences. 2004;74:2213–2225. doi: 10.1016/j.lfs.2003.08.045. [DOI] [PubMed] [Google Scholar]
- So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS genetics. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, Snyder E, Eberhart CG, Majumder S. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26:1666–1678. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rin1 promoter sequences from representatives of the mammalian suborders Primates (Homo sapiens, Hs and Macaca mulatta, Ma), Glires (Rattus novegicus, Rn and Mus Musculus, Mm), Carnivora (Canis familiaris, Cf), Cetartiodactyla (Bos Taurus, Bt and Tursiops truncatus), Hyracoidea (Procavia capensis), Proboscidea (Loxodonta africana, La) and Dasypodidea (Dasypus novemcinctus, Dn). Perfect conservation is denoted by “*”. Near perfect conservation (9/10) is denoted by “:”. In the segment for which L. africana sequence is not available, conservation is based on the remaining sequences. The start codon is shown in bold. The mouse REST-like sequence is shaded in gray, and the 12 putative Snail sites (Consite analysis) are underlined.


