Abstract
Solvatochromic fluorophores possess emission properties that are sensitive to the nature of the local microenvironment. These dyes have been exploited in applications ranging from the study of protein structural dynamics to the detection of protein-binding interactions. While the solvatochromic indole fluorophore of tryptophan has been utilized extensively for in vitro studies to advance our understanding of basic protein biochemistry, the emergence of new extrinsic synthetic dyes with improved properties in conjunction with recent developments in site-selective methods to incorporate these chemical tools into proteins now open the way for studies in more complex systems. Herein we discuss recent technological advancements and their application in the design of powerful reporters, which serve critical roles in modern cell biology and assay development.
Introduction
Fluorescence-based techniques are vital to the fields of molecular cell biology and protein biochemistry. Many of these techniques rely on fundamental photophysical principles including Förster resonance energy transfer (FRET) [1], fluorescence polarization (FP) [2], and fluorescence correlation spectroscopy (FCS) [3]. These methods also draw from a diverse pool of fluorescent dyes exhibiting unique and varied photophysical properties [4]. Innovations in instrumentation have allowed fluorescence-based techniques to be applied in a wide array of formats ranging from high throughput screening assays [5] for drug discovery to the latest in state-of-the-art super-resolution microscopy [6] for in cellulo studies. Of particular interest are fluorescent dyes that possess emission properties that are responsive to physical changes in the local environment, including pH, viscosity, biological analytes, and solvent polarity. By conjugating probes of this type to biologically-relevant molecules, it is possible to obtain valuable information regarding the functions, activities and interactions of such species in the context of living systems with high spatial and temporal resolution.
This review focuses on recent advances in the development, incorporation, and application of a specific class of environment-sensitive fluorophores that display solvatochromism (Box 1). Solvatochromic fluorophores exhibit emission properties (e.g. fluorescence lifetimes, emission wavelengths, and quantum yields) that are highly sensitive to their immediate environment. Such dynamic behavior makes these species particularly well suited for investigating biomolecular interactions since it provides information on the state of a protein at a highly localized region of the protein molecule (single amino acid level). For example, if a solvatochromic fluorophore is appended to the surface of a protein at a site that is involved in a transient binding interaction or that undergoes a conformational change, then the probe will report such events provided that they are coupled to modifications in the local solvent sphere. Herein, particular attention is devoted to the incorporation of these tools into peptides and proteins for the development sensors of biomolecular dynamics. In surveying a selection of these applications, a brief analysis of the associated advantages and limitations of various probes is also presented.
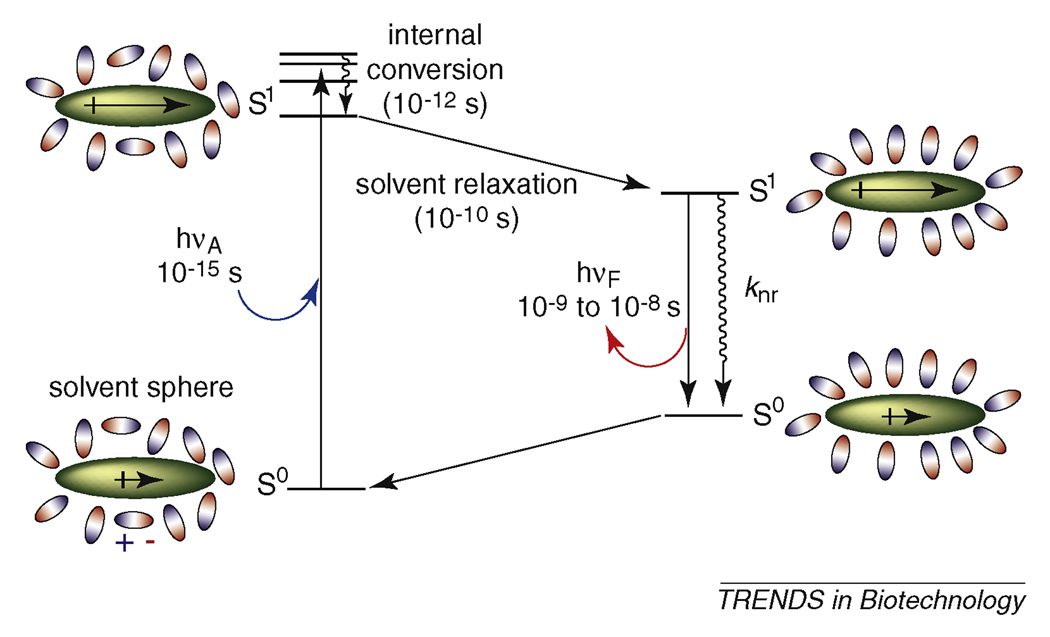
Box 1. Solvatochromism.
The effect of solvent polarity on the emission properties of a solvatochromic fluorophore are generalized in the Jablonski diagram, which depicts the energies of the different electronic states of the system (Figure I). Beginning in the bottom left corner of the diagram, the fluorophore (large oval, grey) resides in the electronic ground state (S0) surrounded by a sphere of solvent molecules (small ovals, white). The electric dipole moment of the fluorophore is indicated by the black arrow with the “+” sign at positive terminus of the dipole. Upon absorbing a photon of the appropriate energy (hνA), the system is rapidly promoted to an excited singlet state (S1). During this event, the system adopts a new electronic configuration with a dipole moment that differs significantly from that of the ground state. In this case, the dipole has increased in magnitude. This process of electronic excitation occurs on a time scale that is much faster than that of the motions of atomic nuclei (Frank-Condon principle [94]). In the picosecond time-scale, the molecules of the solvent sphere reorient dipoles to accommodate the now larger dipole of the fluorophore resulting in a more highly ordered arrangement (upper left corner). This step, termed solvent relaxation, ultimately lowers the energy of the excited singlet state while simultaneously destabilizing the ground state thereby narrowing the energetic gap between the two states. When the system finally returns to the ground state through a fluorescence event, the emitted photon is of a much longer wavelength (i.e. lower energy, hνF) than that which was originally absorbed during excitation. The degree of solvent relaxation increases with increasing solvent polarity.
In some instances, the fluorophore will return spontaneously to the ground electronic state through a thermal (non-radiative) decay process (knr) that competes with fluorescence. A common feature among many solvatochromic fluorophores is their tendency to exhibit a marked increase in non-radiative decay as the energy gap separating the S0 and S1 states is reduced. This effect is particularly apparent in polar protic solvents such as water resulting in a decrease of the fluorescence quantum yield. The mechanisms for such processes are varied and can include a range of events such as internal charge transfer (ICT), tautomerization, isomerization, andintersystem crossing (ISC) to an excited triplet state (T1). When non-radiative decay competes strongly with fluorescence in polar solvents, the fluorophore can exhibit sensitive “switch-like” emission properties upon perturbations to the ordering of the solvent sphere.
Solvatochromic fluorophores commonly used in biological studies
Protein studies based on solvatochromism have evolved with the use of both intrinsic and extrinsic fluorescence species. For example, the fluorescence of the natural amino acid tryptophan has long been known to be environment-dependent [7, 8] and has been widely used in both folding and ligand-binding studies. However, the short wavelengths required for indole excitation and the relative abundance of tryptophan in nature strongly limits its potential for applications in complex systems. This has prompted the design and application of extrinsic synthetic fluorophores with improved photophysical properties. These efforts have led to the emergence of a host of solvatochromic probes with diverse properties [9].
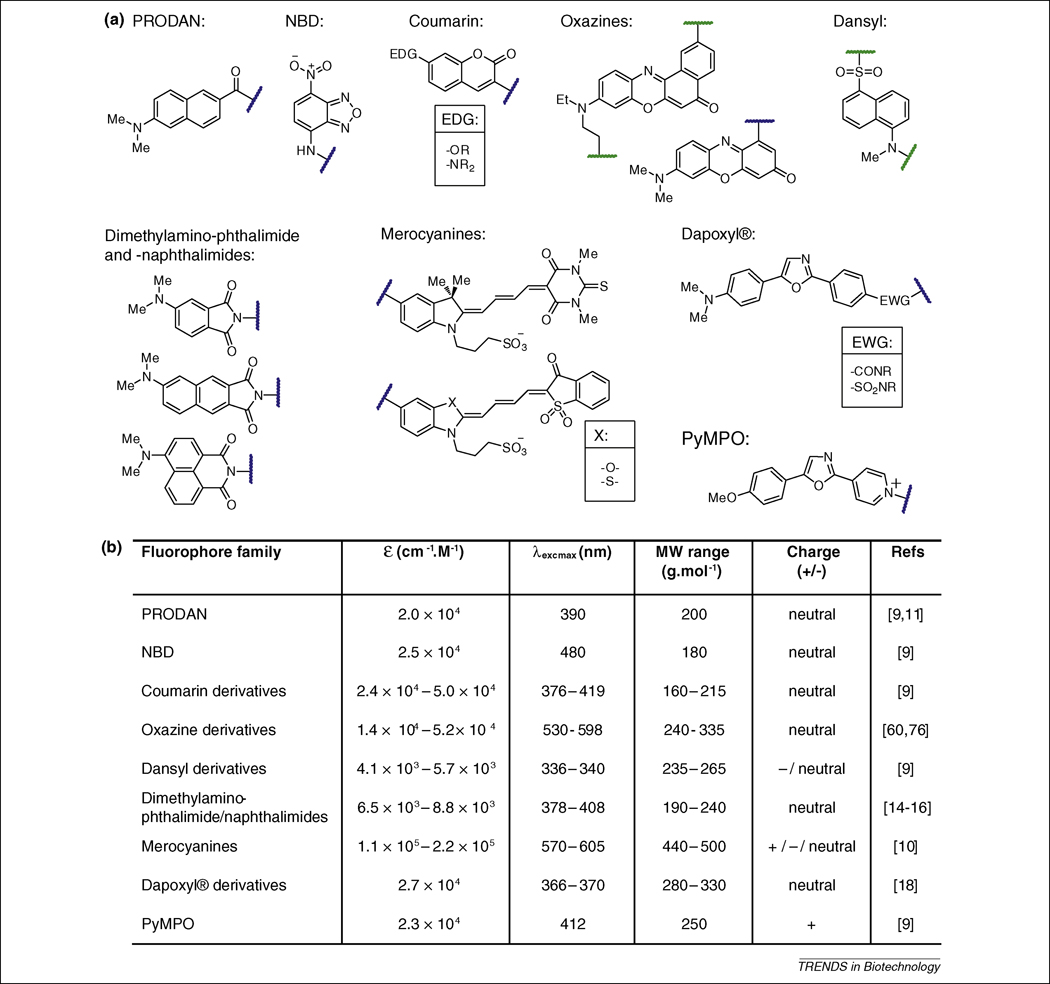
The structures and properties of some of the most common and promising solvatochromic fluorophores are summarized in Figure 1. Key physical parameters include extinction coefficients, excitation and emission wavelengths, quantum yields, size, hydrophobicity, and stability. Indeed, for specific applications, it is often challenging to identify a species that possesses all of the desired attributes. Fortunately, a number of solvatochromic fluorophores possessing overlapping properties have been developed, thereby offering researchers options in the selection of fluorophores for specific applications.
Figure 1. Established solvatochromic fluorophores commonly used in peptide and protein studies.
(a) Representative structures of the different families of solvatochromic fluorophores (PRODAN: 6-propionyl-2-(dimethylaminonaphthalene); NBD: 7-nitrobenz-2-oxa-1,3-diazol-4yl amine or nitrobenzoxadiazole; PyMPO: 1-(2-maleimidylethyl)-4-(5-(4-methoxyphenyl)oxazol-2-yl) pyridinium methanesulfonate). The position of attachment to biomolecules, either directly or via a reactive group, are indicated by wavy lines (blue when the position is unique and green when multiple alternative anchoring positions have been developed). Points of substituent variability within the structures are indicated by EDG (electron donating group), EWG (electron withdrawing group) and X (heteroatom). (b) Summary table of the solvatochromic fluorophores properties.
For example, the merocyanine dyes [10] display exceptionally large extinction coefficients with long excitation wavelengths that are ideal for in cellulo studies as this avoids the damaging effects of high-energy UV light as well as the potential for signal interference generated by auto-fluorescence. However, these dyes are also large in size and exhibit rather subtle changes in fluorescence quantum yields and emission wavelengths in response to solvent polarity. In contrast, the PRODAN fluorophore [9, 11], is much smaller thereby reducing the likelihood that the probe will negatively impact the native function or activity of the attached biomolecule and it exhibits significant changes in emission wavelength with shifts as great as 100 nm. Unlike the merocyanine derivatives, PRODAN is excited at wavelengths below 400 nm and has an appreciably smaller extinction coefficient. A report of a conjugation-extended analogue of PRODAN showed enhancement of these properties by shifting the excitation wavelength to around 460 nm [12].
The variation that exists among the emission properties of solvatochromic fluorophores can be difficult to compare directly since most of the studies have been conducted under varying conditions and using different metrics (i.e. Lippert-Mataga plots [7] versus the ET(30) scale [13]). Typically, this is done by measuring the emission properties of the fluorophores under a range of solvents conditions. However, these studies are complicated by the fact that the solubility of many of these species is restricted to a narrow range of solvents. This limitation can be overcome by conjugating the fluorophore to another molecule to enhance solubility [14]; however the need still exists for standardized approaches for comparing the solvatochromic properties of all fluorophores. With that stated, there are a number of generalities that can be made regarding each species. For example, the dimethylaminophthalimide and related dyes exhibit extremely weak fluorescence in aqueous buffers [14–16] providing the advantage of low background signal until the occurrence of an event that perturbs the local environment. This creates the effect of “switch-like” fluorescence changes with the potential to produce 1000-fold increases in emission intensity [17]. The dapoxyl® dyes are noted for the ability to exhibit shifts in emission wavelength greater than 200 nm in response to changes in solvent polarity alone [18].
Finally, in recent years, many new classes of solvatochromic fluorophores have emerged that serve to expand the range of useful properties. For example, the 2-dicyanomethylene-3-cyano-2,5-dihydrofuran (DCDHF) family of fluorophores stands out for utility in single-molecule experiments [19]; polymethyne dyes have been evolved to display near-IR emission for use in a biological environment [20]; both solvatochromic BODIPY derivatives [21] and fluorophores with reverse sensitivity to polarity have also been reported [22, 23]. While most of these fluorophores have not yet been exploited in advanced biomolecular studies, they represent interesting alternatives to those described above and illustrate the need for new tools adapted for use in complex environments that can take advantage of technological advances.
Methods of incorporation into peptides and proteins
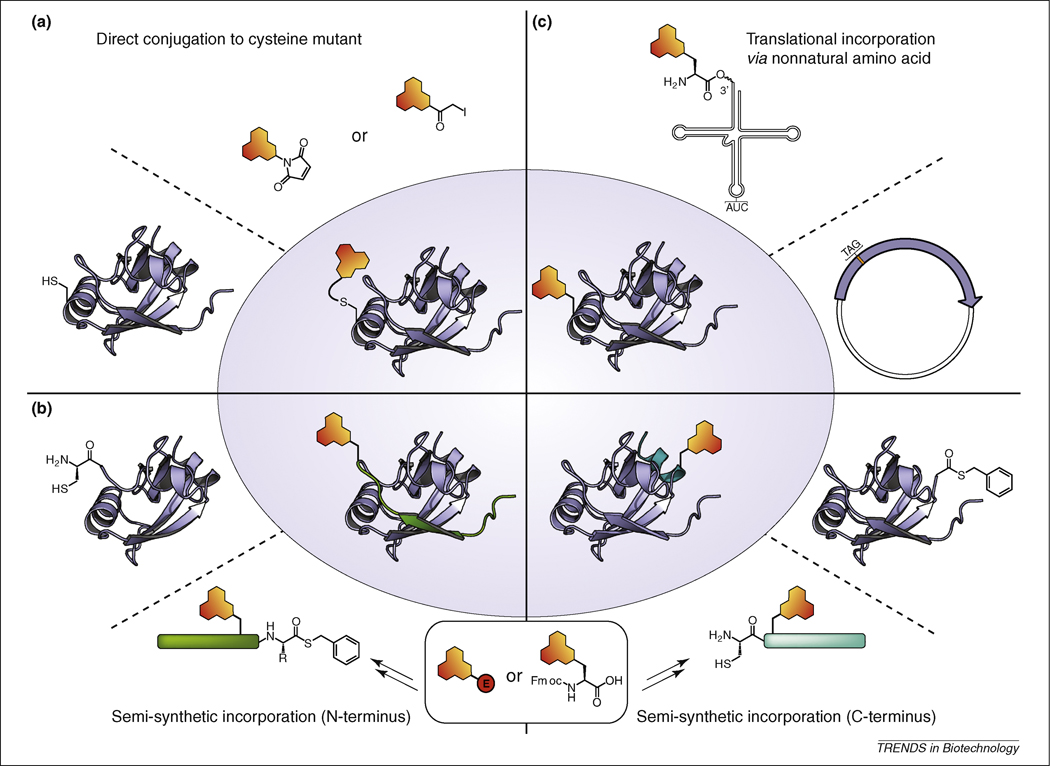
The methods for introducing solvatochromic fluorophores into peptides and proteins are based on similar approaches for incorporating other unique functionalities, such as reactive cross-linking groups and affinity tags (e.g. biotin). However, in contrast to many other species, the insertion of solvatochromic fluorophores is topologically restricted to sites in the protein that preserve function and activity while permitting the dye to make necessary contacts that will result in measurable fluorescence changes. This consideration necessitates the use of methods that offer precise control over dye placement within the peptide or protein structure of interest with minimal perturbation. In this respect, it should be noted that labeling approaches that rely on the use of a targeting module, either in the form of a protein (SNAP-tag and Halo-Tag systems [24]) or a peptide sequence (labeled via Sfp phosphopantetheinyl transferase [25], biotin ligase [26], formylglycine-generating enzyme [27], Ni-NTA- [28] or biarsenical-derived [29, 30] fluorophores) are generally limited for most applications of these tools because the target system would start to deviate too much from the native sequence. Insertion of solvatochromic fluorophores into peptides can generally be accomplished through standard solid phase synthesis (SPPS) approaches to mutate a single native residue. For proteins, the most common methods can broadly be divided into three categories: (i) direct covalent modification; (ii) incorporation of fluorescent amino acids via semi-synthesis (expressed protein ligation); and (iii) incorporation of fluorescent amino acids via suppression of the amber (TAG) stop codon. Here, only a brief description of each approach is provided as these have been thoroughly reviewed elsewhere [9, 31, 32].
Direct covalent modification of proteins
A large number of methods for site-selective chemical modification of proteins with extrinsic fluorophores have been developed [9, 31]. Cysteine and lysine reactive agents offer a convenient and direct method for labeling proteins and have been used extensively to conjugate a myriad of auxiliary groups into biomolecules. Most common among these are the thiol-selective electrophiles, such as maleimides and α-halocarbonyl compounds, along with the amine-selective acylating agents such as the O-succinimidyl esters (Figure 2a). In general, cysteine residues are ideal since this amino acid occurs relatively infrequently in proteins [32] and possesses excellent nucleophilic properties under most physiological conditions. Proteins with unique cysteines can be readily prepared using standard molecular biology techniques. In contrast, lysine residues are far more abundant [32] making selective site-specific labeling of recombinant proteins less practical.
Figure 2. Methods for site-selective and site-specific incorporation of solvatochromic fluorophores into protein.
(a) Direct labeling of a solvent exposed cysteine residue using a thiol-reactive agent [9, 31]. (b) Incorporation of an unnatural amino acid possessing a solvatochromic fluorophore as the side chain group via suppression of the amber stop codon. A tRNA molecule designed to recognize and read-through the amber codon is charged with the unnatural amino acid [32, 42]. (c) Expressed protein ligation involves a semi-synthetic approach typically requiring either the N- or C-terminal end of the protein to be prepared by solid phase peptide synthesis (SPPS). The chromophore can be inserted either as an amino acid building block (e.g. Fmoc-protected amino acid) during SPPS, or afterwards by labeling a side-chain residue with an appropriate electrophilic derivative of the chromophore (represented as a red circle). The peptides are then ligated to the portion of the protein construct that was expressed from a recombinant gene product [33, 95, 96].
Incorporation of unnatural amino acids via synthesis and semi-synthesis (expressed protein ligation)
Expressed protein ligation (EPL) [33, 34] is a powerful semisynthetic approach for incorporating non-native elements, such as solvatochromic fluorophores, site-specifically into proteins. The method involves expressing a truncated form of the target protein to which a synthetically prepared peptide is ligated at the N- or C-terminus. The synthetic peptide that is prepared constitutes the omitted portion of the native protein, but includes a fluorescent amino acid in place of another residue. The most commonly employed ligation method requires one of the two fragments to contain an N-terminal cysteine residue while the complementary fragment bears a C-terminal thioester. The N-terminal cysteine residue facilitates ligation through an initial, reversible transthioesterification step followed by an irreversible intramolecular S→N acyl transfer resulting in the formation of an amide bond (Figure 2b).
The insertion of solvatochromic fluorophores into peptide-based probes, or peptide fragments to be used in EPL to generate protein-based probes, is conducted either by the use of a prepared fluorescent unnatural amino acid building block for SPPS or by on-resin derivatization of a peptide sequence containing an orthogonally-protected amino acid with a reactive side chain. Many solvatochromic fluorophores have been developed into unnatural amino acid building blocks for subsequent incorporation into both peptides and proteins through synthetic and semisynthetic approaches [14–16, 35–39]. The complementary expressed portion of the protein is obtained either by using a protease to reveal an N-terminal cysteine or using a “defective intein”-based approach to produce the C-terminal thioester [34, 40]. While EPL requires more synthetic manipulations and some specialized expertise, the technique offers the advantage of yielding material of greater homogeneity than that typically achieved through direct chemical modification. In general, applications of EPL are confined to modification of amino acids within approximately 40 residues of the N- or C-termini of proteins due to the greater technical challenges involved in generating long peptide sequences or the development of three-segment ligation strategies [41].
Incorporation of unnatural amino acids via suppression of the amber stop codon
Methods to expand the genetic code to include unnatural amino acids have been the source of intense study by several research groups [32]. One of the more successful approaches utilizes the amber stop codon (TAG) to encode a new amino acid [42]. Early embodiments of the method involved the semisynthesis of an artificial amino acyl tRNA (AA-tRNA) molecule designed to recognize the amber codon through base pairing of the anticodon loop. Although different methods have been applied to prepare the charged suppressor AA-tRNACUA molecule, one of the more straightforward approaches utilizes a semisynthetic approach wherein a dinucleotide (pdCpA) charged with the desired unnatural amino acid is enzymatically ligated (T4 ligase) to a tRNACUA that is transcribed from a DNA template such that it lacks these two 3’-nucleotides. Once obtained, the artificially acylated tRNA is introduced into a cell extract derived from E. coli (or in other cases from yeast or rabbit reticulocyte lysates) that is rich in the molecular machinery necessary for protein synthesis. The gene for the protein of interest, which has been mutated to incorporate the amber codon site-specifically, is then translated with the unnatural amino acid integrated into the protein at the desired position (Figure 2c). Drawbacks of the in vitro translation approach include the difficulty of synthesizing the aminoacylated dinucleotide (AA-pdCpA) and the generally low overall protein yields. Furthermore, suppression efficiency can be low (20–30%) [32] and may vary widely depending on the nature of the amino acid, the gene to be expressed, the site within the gene that the amber codon is located, and the protein expression system being used.
Currently, a great deal of attention is being focused on expanding the scope of this approach by evolving novel AA-tRNA synthetase/suppressor tRNA pairs in E. coli [43], yeast [44] and mammalian cells [45] that may recognize unnatural amino acids. This in vivo approach shows great promise for yielding practical quantities of protein and has been utilized for the incorporation of a wide range of tyrosine derivatives. However, the unnatural amino acid of interest must first meet several criteria. It must be passively or actively transported into the host cell and lack any toxic activity. Furthermore, it should be orthogonal to the native AA-tRNA synthetases of the host such that it is not recognized as a substrate and used to misacylate one of the endogenous tRNAs. Despite these challenges, there are significant opportunities for future development of this methodology, which would provide exceptional control over the placement of solvatochromic amino acids into native proteins.
Applications
The development of new solvatochromic fluorophores and methods to insert them into proteins of interest has greatly expanded the scope of potential applications for these unique chemical tools. Since the initial in vitro studies of proteins containing a single tryptophan, advances in this field have enabled researchers to conduct experiments in living cells to investigate the dynamics of protein activities in their native environment. Herein, we present an overview of the main applications of these tools to illustrate their potential to advance our understanding of essential biomolecular events.
Protein folding
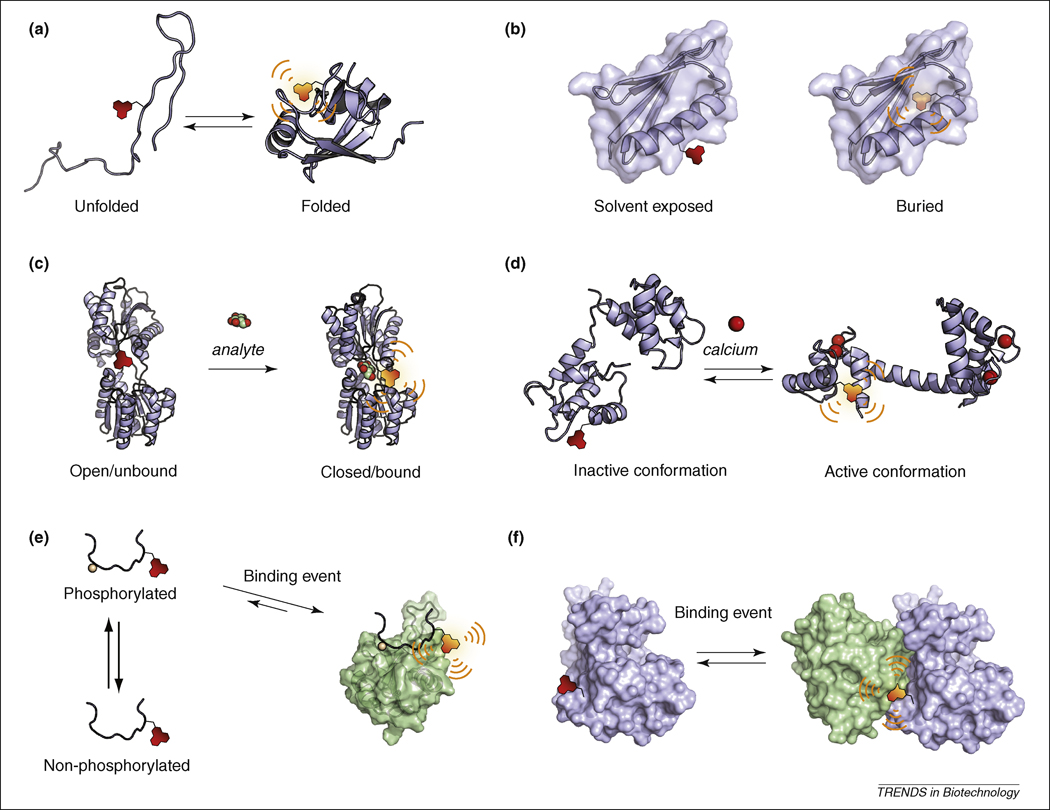
Accurate folding is essential to protein function and valuable information regarding folding processes can be obtained by monitoring changes in the fluorescence properties of solvatochromic fluorophores incorporated into the primary sequence of a protein of interest [7]. In this case, the unfolded state would result in maximal exposure of the probe to the polar solvent environment while intermediate states or the final tertiary structure will result in increased fluorescence emission by lowering the solvent accessibility and the local polarity (Figure 3a). Examples of such applications of solvatochromism include folding studies that employ chemical or thermal methods to promote denaturation/refolding of proteins for steady-state [46] or time-resolved measurements [47, 48].
Figure 3. Applications of solvatochromism for biomolecular protein studies.
(a) Folding studies. (b) Mapping of a protein local environment and/or solvatation. (c) Sensors for small molecule analytes. The sensors are generally designed by exploiting protein domains that intrinsically bind a select analyte resulting in fluorescence change due to a conformational change in the protein or the displacement of the fluorophore (illustrated here with the d-glucose/d-galactose-binding protein from E. Coli, 2FW0 for the apo state and 2FVY for the glucose-bound state). (d) Reporting protein structural changes. In response to a signaling event, the protein of interest will undergo a conformational change that corresponds to a different functional state, which may be monitored by an appropriately positioned solvatochromic fluorophore (illustrated with calmodulin, 1DMO apo state and 1UP5 calcium-bound state). (e) Fragment- or peptide-based probes for the monitoring of protein interactions. A minimal fragment of one of the binding partners can be labeled with a solvatochromic fluorophore to report interactions. In the case of transient interactions, the signaling event that will induce the interaction can affect either one of the partners thus providing information on function or activity. This approach is illustrated here with the Crk SH2 domain (1JU5) that binds to phosphopeptides sequences and that can be used to report either the activity of a kinase or the phosphorylated state of a substrate. (f) Reporting protein-protein interactions. A similar approach to panel (e) can be applied to full-length proteins instead of fragments. In this case advantage can be taken of the larger interaction interface between two proteins compared to peptide-based probes that do not adopt secondary and tertiary structures.
Structural information
The sensitive emission properties of solvatochromic dyes can be used to obtain localized structural information on proteins by reporting the nature of the environment immediately surrounding the incorporation site (Figure 3b). This approach is complementary to structural methods such as crystallography and NMR since it can yield detailed information regarding highly localized structural dynamics under physiologically relevant conditions or in environments that are beyond the scope of X-ray and NMR-based methods (e.g. large protein complexes, extremely dilute conditions, and with elements that exhibit significant dynamics). The method can also provide information on hydration [49, 50], degree of solvent exposure or electrostatic environment of specific residues [37], and insight into interactions with membrane lipid bilayers [51]. An illustration of the potential of the approach is provided by the fluorescence mapping of a transmembrane complex involved in protein translocation across the mitochondrial inner membrane [52]. Analysis of the fluorescent properties of various 4-nitrobenzoxadiazole (NBD)-labeled mutants of a specific transmembrane segment of the complex afforded high-resolution information (amino acid level) concerning the protein–conducting channel.
Sensors for small molecules
Sensors for small molecules or ions can be developed by incorporating environment-sensitive fluorophores into proteins or protein domains that naturally bind target analytes such as bacterial periplasmic binding proteins [53]. In this application, strategic placement of the chromophore into the macromolecule affords fluorescence changes that are directly coupled either with a displacement of the fluorophore by the analyte or to conformational changes that occur in response to analyte binding (Figure 3c). This approach has been utilized to develop sensors for various classes of analytes including carbohydrates (glucose [54–57] and maltose [58–60]), ions (nickel [61], zinc [62, 63], sulfate [64], and inorganic phosphate [65]), amino acids [66], signaling small molecules (autoinducer-2 involved in quorum sensing [67] or acyl-CoA [68]), steroids [69] and peptide-oligonucleotides [70].
Reporting conformational states
As a common regulatory mechanism, many proteins –in particular enzymes and ion channels-alternate between different stable conformations, which constitute different functional states. These modulations are typically the consequence of various signaling mechanisms, which may include interactions with binding partners (e.g. protein, small molecule ligand or ion) or post-translational modification. Solvatochromic fluorophores can be utilized to report these dynamic changes in protein structure providing direct information on protein activity (Figure 3d). The activation of proteins has thus been monitored for various calcium-binding proteins [71, 72]. Conversely, monitoring the inactivated state of certain enzymes by this approach can be of interest for inhibitor screening. For example, acrylodan modification of a cysteine in a critical regulatory loop region of the cSrc kinase enabled development of a direct binding assay for identifying small molecule inhibitors that specifically stabilize the inactive conformation of the kinase [73].
An elegant application of this general approach has been pioneered by Isacoff with the development of voltage/patch-clamp fluorometry for the study of voltage- or ligand-gated ion channels [74–76]. By using site-directed labeling with an environment-sensitive fluorophore, subtle conformational changes in a specific region of the channels could be monitored using fluorescence in conjunction with the modulation of the currents resulting from the control of the gating state either with a voltage/patch clamp or by introduction of an effector ligand. The capacity of these tools to respond rapidly to events that occur over extremely small time-scales and yield local information at the amino acid level has provided new insight into the structural dynamics that underlie channel activity [77, 78].
Direct reporting of post-translational modifications
Post-translational modifications such as phosphorylation can be directly monitored using a proximal solvatochromic fluorophore, which reports on local changes in polarity. This approach has been utilized to monitor the activity of various kinases (myosin kinase [79] and protein kinase C [80]) by appending an environment-sensitive fluorophore to the N-terminus of the Ser/Thr kinase substrate sequence. A complementary approach has also been reported for reporting phosphatase activity [81]. Similarly, assays for protein prenyltransferase activity have been developed by using peptides incorporating the environment-sensitive dansyl group into a cysteine-containing substrate sequence [82].
Reporting binding interactions
Specific protein-protein interactions can be reported by monitoring solvatochromic fluorophores positioned within or near an interaction interface that undergoes significant changes to the local environment upon the formation of new contacts between residues (e.g. via hydrogen bonding and/or displacement of solvent molecules) (Figure 3e and f). This approach has been widely exploited by labeling small protein fragments or short peptide sequences derived from one of the binding partners while preserving the specificity determinants required to establish the interaction (Figure 3e). While the use of such abridged constructs offers a convenient starting point for the design and optimization of probes that will later include the full-length protein, these tools can also provide valuable information, such as affinity constants. Such peptide-based probes may also serve as useful sensors for a particular state of a specific binding partner [14] or may be evolved as tag-probe pairs for protein labeling applications [83]. Furthermore, since short peptides often lack the ability to form complex secondary and tertiary structures, the fluorophores generally remain largely exposed to the solvent environment until the probe interacts with a binding partner. This minimizes the potential for background fluorescence, which often occurs when solvatochromic dyes are appended on globular proteins. Probes that report binding to common protein-interaction domains have been developed for SH2 [16] and PDZ domains [84] by incorporating the solvatochromic dyes within peptides sequences derived from cognate ligands while maintaining their native specificity across different domain family members. Using the same general principle, phosphorylation of specific sequences can be monitored indirectly via the resulting interaction with a cognate phosphopeptide binding domain as illustrated with 14-3-3 [85] and SH2 domains [86]. Similarly, protein activation can be observed when the active state induces binding to other proteins. Hence, by labeling a recognition domain that only binds the GTP-bound, activated form of Cdc42, the Hahn laboratory developed a sensor for monitoring the activation of the endogenous GTPase [87].
Despite the advantages of solvatochromic fluorophores over alternative more perturbing approaches, the use of these tools to monitor the binding of full-length proteins is still limited. This is largely due to the technical challenges that are presented when studying molecular interactions within complex systems such as living cells in addition to the difficulties met in when designing efficient probes. Examples of successfully employed probes, in which one of the binding partners is a peptide, include fluorescent reporters for class II MHC complexes [17], opioid receptors [88] and the OppA protein of L. lactis [89]. The development of fluorescent constructs used to detect the polymerization of actin filaments [90] as well as the design and application of antibody based-biosensors [91] have also been reported.
Design considerations when incorporating solvatochromic fluorophores into proteins
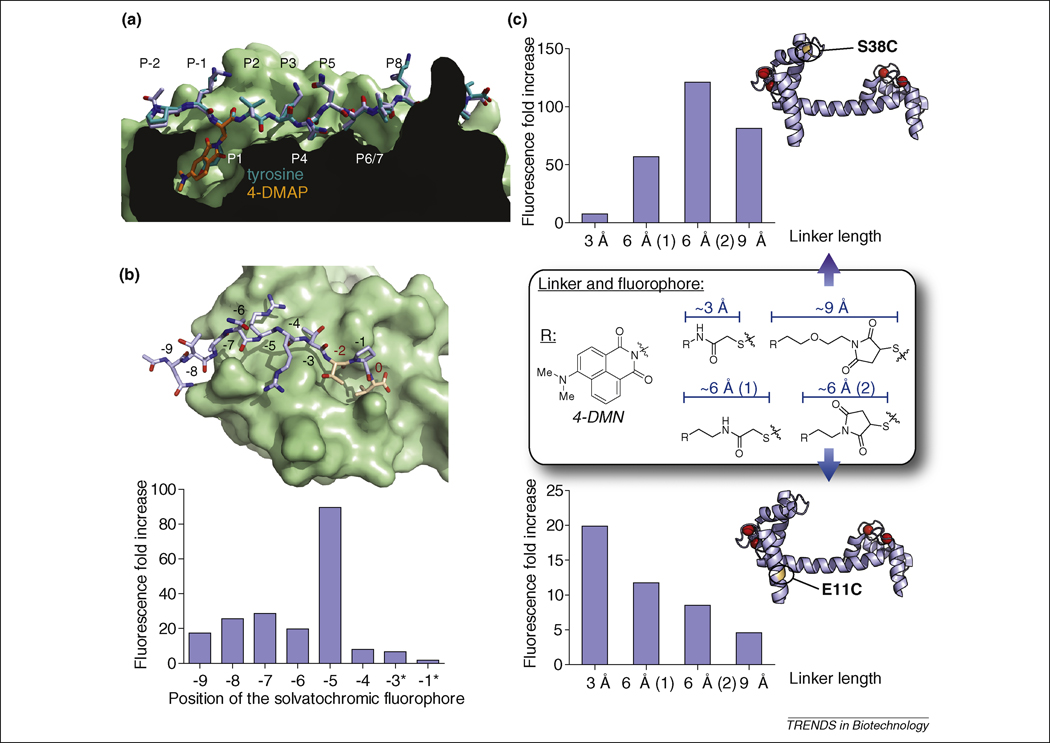
For certain applications, the precise placement of a solvatochromic fluorophore within the structure of a protein is restricted to a particular region of interest. Examples include investigations of protein-folding dynamics, for which detailed information regarding the stability of discrete protein domains is required, as well as structural studies to map the location of solvent-exposed residues. Alternatively, the optimal placement of the solvatochromic fluorophore in a binding partner might be readily identified in that a well-defined hydrophobic interaction is already known. An excellent example of this is in the development of a series of fluorogenic probes to detect peptide loading onto class II MHC proteins that are involved in the activation of the adaptive immune response (Figure 4a) [17]. Crystallographic studies of the class II MHC protein, HLA-DR1, in a complex with a short peptide fragment (HA) derived from influenza revealed a pronounced hydrophobic pocket (P1) that could bind large aliphatic or aromatic side chains such as tyrosine. This position in the HA peptide chain was replaced with the fluorescent amino acids 4-N,N-dimethylaminophthalimidoalanine (4-DAPA) [15] and 6-N,N-dimethylamino-2,3-naphthalimidoalanine (6-DMNA) [16]. Upon binding to HLA-DR1, the peptide probes exhibited 102 to 103-fold increases in fluorescence intensity without perturbing the affinity or biological activity of the native complex.
Figure 4. Design considerations.
(a) Replacement of a conserved hydrophobic/aromatic residue by a solvatochromic fluorophore. Alignment of crystal structures of the (4-DAPA)-HA (2IPK, stick representation of the peptide in light blue with 4-DAPA highlighted in orange) and HA (1JWU, stick representation in teal) peptides bound to HLA-DR1 protein (class II MHC protein, surface representation), adapted from [17]. The replacement of the conserved ligand aromatic residue that occupies the P1 pocket of the HLA-DR protein by the 4-DAPA and 6-DMNA amino acids yielded highly efficient fluorogenic probes (over 1000-fold increase in fluorescence upon binding) without significantly affecting the specificity or affinity compared to the native interaction [17]. The crystal structures illustrate the ability of the small size 4-DAPA solvatochromic amino acid to replace the tyrosine of the native ligand. The fluorogenic probes have enabled the monitoring of in vivo regulation of cell-surface peptide-binding activity of class II MHC proteins in primary dendritic cells. (b) Screening for optimal positioning of the environment-sensitive fluorophore. Top: crystal structure of the third PDZ domain of PSD-95 with a modeled bound decapeptide ligand derived from the PDZ domain-binding motif of Stargazin in stick representation (NTANRRTTPV, adapted from 1TP3). Critical residues for PDZ domain-mediated interactions (at position 0 and -2) are represented in orange with red numbering. The 4-DMAP fluorophore was inserted systematically in each non-critical position with a diaminobutyric acid linker (*: except for -3 and -1, where a diaminopropionic acid linker was used instead). The respective fluorescence increases observed upon binding to the cognate PDZ domain are presented in the bar graph. The screening approach yielded a probe with a ~90-fold fluorescence increase by insertion of the fluorophore at position -5 [84]. (c) Screening for optimal linker length between the protein backbone and the environment-sensitive fluorophore 4-DMN. Cysteine labeling agent analogues derived from 4-DMN were incorporated into monocysteine mutants of calmodulin (illustrated with S38C and E11C mutants in the calcium-bound state, 1UP5) and compared at each position for the effect of the linker length on the ability of the solvatochromic fluorophore to report changes in its local protein environment upon binding of calcium [92].
Alternatively, the choice of dye placement must be determined empirically and if possible guided by structural information regarding the interaction sites of interest. However, in most cases, determining the optimal site of fluorophore placement is challenging even when high-quality structural data is available. When considering a binding event, the pockets and clefts that form the interaction interface can differ by size, shape and charge thereby accommodating some fluorophores while limiting others. This has lead many researchers to apply screening approaches to simultaneously optimize both placement and fluorophore type. This is particularly valuable with peptide-based ligands, since the highly modular nature of solid-phase peptide synthesis enables the rapid preparation of peptide libraries, which allows critical variables such as position, linker length and nature of the chromophore to be optimized. This approach has proven highly successful in the development of fluorogenic peptide-based sensors designed to report specific PDZ domains (Figure 4b) [84].
In applications that involve more complex interactions such as monitoring structural changes within a large protein, however, the use of peptide-based probes might be inadequate. In these cases, screens can be performed by using site-directed mutagenesis to introduce uniquely-reactive cysteine residues, which can be labeled with thiol-reactive dyes. In these experiments, the goal is to identify residues that could be mutated without significantly disrupting the native structure or function of the protein. Furthermore, it is important that the appended fluorophore is positioned appropriately such that it is capable of producing a measurable signal without negatively interfering with the interaction of interest. Recently, a screening approach that produced an effective fluorescent sensor for the detection of a signaling molecule used in bacterial quorum sensing has been reported [67]. A variety of cysteine mutants were prepared from two protein receptors known to bind different forms of the bacterial autoinducer II (AI-2). The receptors, LuxP and LsrB, exhibit structural similarity and consist of two domains linked together by a hinge region with the ligand-binding site located at the interface. The cysteine residues introduced at the periphery of this site were labeled with an assortment of solvatochromic fluorophores including derivatives of dansyl, PRODAN, NBD, dapoxyl, and PyMPO. One construct, a LuxP mutant (T137C) labeled with a dapoxyl derivative, was identified to possess the desired properties.
As with peptide probes, the linker length of a thiol-reactive dye also represents an essential variable to be explored when optimizing the fluorescent response of a biosensor. A systematic study conducted using the calcium binding protein calmodulin revealed that linker-type can exert a dramatic influence on both the measured fluorescence change and the degree of background emission generated by the construct (Figure 4c) [92].
Perspectives
Since the early protein studies that exploited the advantages of solvatochromism by relying on the intrinsic fluorescence properties of tryptophan [7], significant advancements have been made in this field. These include the design of new extrinsic synthetic fluorophores with improved fluorescence properties together with important developments in the methods for site-selectively incorporating the dyes into proteins. Recent applications have shown that unique information can be obtained on the dynamics of proteins involving changes in conformation or activity, which makes these environment-sensitive tools particularly useful with respect to other common dyes or genetically encoded fluorescent proteins. In particular, the small size of many of these chromophores minimizes any perturbation introduced by their presence, which allows studies to be conducted on quasi-native proteins. Furthermore, unlike FRET-based [1] or fluorescent protein complementation approaches [93], only a single chromophore is required, therefore providing access to systems involving multiple binding partners as well as offering a means to monitor the activity of endogenous proteins. Finally, the high sensitivity to changes in the local environment provides information regarding discrete regions of protein structure. However, the use of solvatochromic fluorophores to study biomolecular systems is still limited by the challenges encountered when trying to incorporate them into complex systems. In this context, with new extrinsic small chromophores that can display up to a 1000-fold increase in fluorescence [17] and methods that illustrate the viability of these tools in live cells [87], it can be anticipated that integration of the recent advances and improvements in the methods to incorporate fluorophores into proteins (as well as to deliver the labeled protein into cells) will foster studies that rely on these potent tools in cellular environments.
Figure I.
The origin of solvatochromic effects on fluorescence.
Acknowledgments
We apologize to the colleagues whose work we were unable to cite owing to space constraints. Research in Imperiali group is supported by the Cell Migration Consortium (GM64346) and the NSF (CHE-0414243), The support of Biotechnology Training Program (T32-GM08334) to G.S.L, and the Marie Curie postdoctoral Fellowship program to M.S. (PICK-CPP) are also gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jares-Erijman EA, Jovin TM. FRET imaging. Nat. Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 2.Goulko AA, et al. Fluorescence polarization: recent bioanalytical applications, pitfalls, and future trends. In: Wolfbeis OS, editor. Springer Series on Fluorescence. 1st edn. Springer; 2008. pp. 303–322. [Google Scholar]
- 3.Krichevsky O, Bonnet G. Fluorescence correlation spectroscopy: the technique and its applications. Rep. Prog. Phys. 2002;65:251–297. [Google Scholar]
- 4.Lavis LD, Raines RT. Bright ideas for chemical biology. ACS Chem. Biol. 2008;3:142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janzen WP. High throughput screening: methods and protocols. Humana Press; 2002. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat. Rev. Mol. Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 7.Lakowicz JR. Principles of fluorescence spectroscopy. 3rd edn. Springer; 2006. Protein fluorescence; pp. 529–575. [Google Scholar]
- 8.Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haugland RP, et al. The handbook : a guide to fluorescent probes and labeling technologies. Molecular Probes; 2005. [Google Scholar]
- 10.Toutchkine A, et al. Solvent-sensitive dyes to report protein conformational changes in living cells. J. Am. Chem. Soc. 2003;125:4132–4145. doi: 10.1021/ja0290882. [DOI] [PubMed] [Google Scholar]
- 11.Weber G, Farris FJ. Synthesis and spectral properties of a hydrophobic fluorescent-probe - 6-propionyl-2-(dimethylamino)naphthalene. Biochemistry. 1979;18:3075–3078. doi: 10.1021/bi00581a025. [DOI] [PubMed] [Google Scholar]
- 12.Lu Z, et al. Long-wavelength analogue of PRODAN: synthesis and properties of Anthradan, a fluorophore with a 2,6-donor-acceptor anthracene structure. J. Org. Chem. 2006;71:9651–9657. doi: 10.1021/jo0616660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reichardt C. Solvatochromic dyes as solvent polarity indicators. Chem. Rev. 1994;94:2319–2358. [Google Scholar]
- 14.Loving G, Imperiali B. A versatile amino acid analogue of the solvatochromic fluorophore 4-N,N-dimethylamino-1,8-naphthalimide: A powerful tool for the study of dynamic protein interactions. J. Am. Chem. Soc. 2008;130:13630–13638. doi: 10.1021/ja804754y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vazquez ME, et al. A new environment-sensitive fluorescent amino acid for Fmoc-based solid phase peptide synthesis. Org. Biomol. Chem. 2004;2:1965–1966. doi: 10.1039/b408001g. [DOI] [PubMed] [Google Scholar]
- 16.Vazquez ME, et al. Photophysics and biological applications of the environment-sensitive fluorophore 6-N,N-Dimethylamino-2,3-naphthalimide. J. Am. Chem. Soc. 2005;127:1300–1306. doi: 10.1021/ja0449168. [DOI] [PubMed] [Google Scholar]
- 17.Venkatraman P, et al. Fluorogenic probes for monitoring peptide binding to class II MHC proteins in living cells. Nat. Chem. Biol. 2007;3:222–228. doi: 10.1038/nchembio868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diwu Z, et al. Fluorescent molecular probes .2. The synthesis, spectral properties and use of fluorescent solvatochromic Dapoxyl(TM) dyes. Photochem. Photobiol. 1997;66:424–431. [Google Scholar]
- 19.Lord SJ, et al. DCDHF fluorophores for single-molecule imaging in cells. Chemphyschem. 2009;10:55–65. doi: 10.1002/cphc.200800581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berezin MY, et al. Near infrared dyes as lifetime solvatochromic probes for micropolarity measurements of biological systems. Biophys. J. 2007;93:2892–2899. doi: 10.1529/biophysj.107.111609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunahara H, et al. Design and synthesis of a library of BODIPY-based environmental polarity sensors utilizing photoinduced electron-transfer-controlled fluorescence ON/OFF switching. J. Am. Chem. Soc. 2007;129:5597–5604. doi: 10.1021/ja068551y. [DOI] [PubMed] [Google Scholar]
- 22.Demelo JSS, et al. Photophysical behavior of coumarins as a function of substitution and solvent - experimental-evidence for the existence of a lowest lying 1(n,Π*) state. J. Phys. Chem. 1994;98:6054–6058. [Google Scholar]
- 23.Uchiyama S, et al. Environment-sensitive fluorophore emitting in protic environments. Org. Lett. 2006;8:5869–5872. doi: 10.1021/ol062490r. [DOI] [PubMed] [Google Scholar]
- 24.O'Hare HM, et al. Chemical probes shed light on protein function. Curr. Opin. Struct. Biol. 2007;17:488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Yin J, et al. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc. 2006;1:280–295. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 26.Chen I, et al. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 27.Carrico IS, et al. Introducing genetically encoded aldehydes into proteins. Nat. Chem. Biol. 2007;3:321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- 28.Lata S, et al. Specific and stable fluorescence labeling of histidine-tagged proteins for dissecting multi-protein complex formation. J. Am. Chem. Soc. 2006;128:2365–2372. doi: 10.1021/ja0563105. [DOI] [PubMed] [Google Scholar]
- 29.Griffin BA, et al. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 30.Nakanishi J, et al. Imaging of conformational changes of proteins with a new environment-sensitive fluorescent probe designed for site-specific labeling of recombinant proteins in live cells. Anal. Chem. 2001;73:2920–2928. doi: 10.1021/ac001528p. [DOI] [PubMed] [Google Scholar]
- 31.Hermanson GT. Bioconjugate techniques. Academic Press; 1996. [Google Scholar]
- 32.de Graaf AJ, et al. Nonnatural amino acids for site-specific protein conjugation. Bioconjug. Chem. 2009;20:1281–1295. doi: 10.1021/bc800294a. [DOI] [PubMed] [Google Scholar]
- 33.Muir TW, et al. Expressed protein ligation: A general method for protein engineering. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu. Rev. Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 35.Summerer D, et al. A genetically encoded fluorescent amino acid. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, et al. A genetically encoded fluorescent amino acid. J. Am. Chem. Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 37.Cohen BE, et al. Probing protein electrostatics with a synthetic fluorescent amino acid. Science. 2002;296:1700–1703. doi: 10.1126/science.1069346. [DOI] [PubMed] [Google Scholar]
- 38.Turcatti G, et al. Probing the structure and function of the tachykinin neurokinin-2 receptor through biosynthetic incorporation of fluorescent amino acids at specific sites. J. Biol. Chem. 1996;271:19991–19998. doi: 10.1074/jbc.271.33.19991. [DOI] [PubMed] [Google Scholar]
- 39.Dufau I, Mazarguil H. Design of a fluorescent amino acid derivative usable in peptide synthesis. Tetrahedron Lett. 2000;41:6063–6066. [Google Scholar]
- 40.Noren CJ, et al. Dissecting the chemistry of protein splicing and its applications. Angew. Chem. Int. Ed. Engl. 2000;39:450–466. [PubMed] [Google Scholar]
- 41.Cotton GJ, et al. Insertion of a synthetic peptide into a recombinant protein framework: A protein biosensor. J. Am. Chem. Soc. 1999;121:1100–1101. [Google Scholar]
- 42.Ellman J, et al. Biosynthetic method for introducing unnatural amino-acids site-specifically into proteins. Meth. Enzymol. 1991;202:301–336. doi: 10.1016/0076-6879(91)02017-4. [DOI] [PubMed] [Google Scholar]
- 43.Wang L, et al. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 44.Chin JW, et al. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 45.Liu W, et al. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 46.Smith CJ, et al. Detection and characterization of intermediates in the folding of large proteins by the use of genetically inserted tryptophan probes. Biochemistry. 1991;30:1028–1036. doi: 10.1021/bi00218a021. [DOI] [PubMed] [Google Scholar]
- 47.Mayor U, et al. The complete folding pathway of a protein from nanoseconds to microseconds. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- 48.Neuweiler H, et al. Downhill versus barrier-limited folding of BBL 2: mechanistic insights from kinetics of folding monitored by independent tryptophan probes. J. Mol. Biol. 2009;387:975–985. doi: 10.1016/j.jmb.2008.12.056. [DOI] [PubMed] [Google Scholar]
- 49.Pal SK, et al. Biological water at the protein surface: Dynamical solvation probed directly with femtosecond resolution. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1763–1768. doi: 10.1073/pnas.042697899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang LY, et al. Mapping hydration dynamics around a protein surface. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18461–18466. doi: 10.1073/pnas.0707647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen AH, et al. Single-molecule visualization of environment-sensitive fluorophores inserted into cell membranes by staphylococcal γ-hemolysin. Biochemistry. 2006;45:2570–2576. doi: 10.1021/bi0514156. [DOI] [PubMed] [Google Scholar]
- 52.Alder NN, et al. Quaternary structure of the mitochondrial TIM23 complex reveals dynamic association between Tim23p and other subunits. Mol. Biol. Cell. 2008;19:159–170. doi: 10.1091/mbc.E07-07-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Lorimier RM, et al. Construction of a fluorescent biosensor family. Protein Sci. 2002;11:2655–2675. doi: 10.1110/ps.021860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marvin JS, Hellinga HW. Engineering biosensors by introducing fluorescent allosteric signal transducers: Construction of a novel glucose sensor. J. Am. Chem. Soc. 1998;120:7–11. [Google Scholar]
- 55.Salins LLE, et al. A novel reagentless sensing system for measuring glucose based on the galactose/glucose-binding protein. Anal. Biochem. 2001;294:19–26. doi: 10.1006/abio.2001.5131. [DOI] [PubMed] [Google Scholar]
- 56.Amiss TJ, et al. Engineering and rapid selection of a low-affinity glucose/galactose-binding protein for a glucose biosensor. Protein Sci. 2007;16:2350–2359. doi: 10.1110/ps.073119507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian Y, et al. Structure-based design of robust glucose biosensors using a Thermotoga maritime periplasmic glucose-binding protein. Protein Sci. 2007;16:2240–2250. doi: 10.1110/ps.072969407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marvin JS, et al. The rational design of allosteric interactions in a monomeric protein and its applications to the construction of biosensors. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4366–4371. doi: 10.1073/pnas.94.9.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dattelbaum JD, et al. Analysis of allosteric signal transduction mechanisms in an engineered fluorescent maltose biosensor. Protein Sci. 2005;14:284–291. doi: 10.1110/ps.041146005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherman DB, et al. Synthesis of thiol-reactive, long-wavelength fluorescent phenoxazine derivatives for biosensor applications. Bioconjug. Chem. 2006;17:387–392. doi: 10.1021/bc050309d. [DOI] [PubMed] [Google Scholar]
- 61.Salins LLE, et al. A fluorescence-based sensing system for the environmental monitoring of nickel using the nickel binding protein from Escherichia coli. Anal. Bioanal. Chem. 2002;372:174–180. doi: 10.1007/s00216-001-1169-7. [DOI] [PubMed] [Google Scholar]
- 62.Walkup GK, Imperiali B. Fluorescent chemosensors for divalent zinc based on zinc finger domains. Enhanced oxidative stability, metal binding affinity, and structural and functional characterization. J. Am. Chem. Soc. 1997;119:3443–3450. [Google Scholar]
- 63.Walkup GK, Imperiali B. Design and evaluation of a peptidyl fluorescent chemosensor for divalent zinc. J. Am. Chem. Soc. 1996;118:3053–3054. [Google Scholar]
- 64.Shrestha S, et al. Rationally designed fluorescently labeled sulfate-binding protein mutants: Evaluation in the development of a sensing system for sulfate. Biotechnol. Bioeng. 2002;78:517–526. doi: 10.1002/bit.10221. [DOI] [PubMed] [Google Scholar]
- 65.Lundgren JS, et al. A dynamical investigation of acrylodan-labeled mutant phosphate binding protein. Anal. Chem. 1999;71:589–595. doi: 10.1021/ac980800g. [DOI] [PubMed] [Google Scholar]
- 66.Lee HS, et al. Genetic incorporation of a small, environmentally sensitive, fluorescent probe into proteins in Saccharomyces cerevisiae. J. Am. Chem. Soc. 2009;131:12921–12923. doi: 10.1021/ja904896s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu J, Pei D. A LuxP-based fluorescent sensor for bacterial autoinducer II. ACS Chem. Biol. 2008;3:110–119. doi: 10.1021/cb7002048. [DOI] [PubMed] [Google Scholar]
- 68.Wadum MCT, et al. Fluorescently labelled bovine acyl-CoA-binding protein acting as an acyl-CoA sensor: Interaction with CoA and acyl-CoA esters and its use in measuring free acyl-CoA esters and non-esterified fatty acids. Biochem. J. 2002;365:165–172. doi: 10.1042/BJ20011727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tamrazi A, et al. Molecular sensors of estrogen receptor conformations and dynamics. Mol. Endocrinol. 2003;17:2593–2602. doi: 10.1210/me.2003-0239. [DOI] [PubMed] [Google Scholar]
- 70.Shvadchak VV, et al. Sensing peptide-oligonucleotide interactions by a two-color fluorescence label: application to the HIV-1 nucleocapsid protein. Nucleic Acids Res. 2009;37:e25. doi: 10.1093/nar/gkn1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hahn K, et al. Patterns of elevated free calcium and calmodulin activation in living cells. Nature. 1992;359:736–738. doi: 10.1038/359736a0. [DOI] [PubMed] [Google Scholar]
- 72.Garrett SC, et al. A biosensor of S100A4 metastasis factor activation: Inhibitor screening and cellular activation dynamics. Biochemistry. 2008;47:986–996. doi: 10.1021/bi7021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simard JR, et al. A new screening assay for allosteric inhibitors of cSrc. Nat. Chem. Biol. 2009;5:394–396. doi: 10.1038/nchembio.162. [DOI] [PubMed] [Google Scholar]
- 74.Mannuzzu LM, et al. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 75.Gandhi CS, Isacoff EY. Shedding light on membrane proteins. Trends Neurosci. 2005;28:472–479. doi: 10.1016/j.tins.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 76.Cohen BE, et al. A fluorescent probe designed for studying protein conformational change. Proc. Natl. Acad. Sci. U.S.A. 2005;102:965–970. doi: 10.1073/pnas.0409469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pathak MM, et al. Closing in on the resting state of the shaker K+ channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 78.Savalli N, et al. Voltage-dependent conformational changes in human Ca2+- and voltage-activated K+ channel, revealed by voltage-clamp fluorometry. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12619–12624. doi: 10.1073/pnas.0601176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Post PL, et al. A genetically-engineered, protein-based optical biosensor of myosin-II regulatory light-chain phosphorylation. J. Biol. Chem. 1994;269:12880–12887. [PubMed] [Google Scholar]
- 80.Yeh RH, et al. Real time visualization of protein kinase activity in living cells. J. Biol. Chem. 2002;277:11527–11532. doi: 10.1074/jbc.M111300200. [DOI] [PubMed] [Google Scholar]
- 81.Noble JE, et al. Fluorescent peptide probes for high-throughput measurement of protein phosphatases. Anal. Chem. 2003;75:2042–2047. doi: 10.1021/ac025838e. [DOI] [PubMed] [Google Scholar]
- 82.Cassidy PB, et al. Lipid Modifications of Proteins. Academic Press Inc; 1995. Continuous fluorescence assay for protein prenyltransferases; pp. 30–43. [DOI] [PubMed] [Google Scholar]
- 83.Tsutsumi H, et al. Fluorogenically active leucine zipper peptides as tag-probe pairs for protein imaging in living cells. Angew. Chem. Int. Ed. Engl. 2009 doi: 10.1002/anie.200903183. DOI: 10.1002/anie.200903183. [DOI] [PubMed] [Google Scholar]
- 84.Sainlos M, et al. A general screening strategy for peptide-based fluorogenic ligands: Probes for dynamic studies of PDZ domain-mediated interactions. J. Am. Chem. Soc. 2009;131:6680–6682. doi: 10.1021/ja900371q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vazquez ME, et al. Fluorescent caged phosphoserine peptides as probes to investigate phosphorylation-dependent protein associations. J. Am. Chem. Soc. 2003;125:10150–10151. doi: 10.1021/ja0351847. [DOI] [PubMed] [Google Scholar]
- 86.Wang QZ, Lawrence DS. Phosphorylation-driven protein-protein interactions: A protein kinase sensing system. J. Am. Chem. Soc. 2005;127:7684–7685. doi: 10.1021/ja050789j. [DOI] [PubMed] [Google Scholar]
- 87.Nalbant P, et al. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 88.Vazquez ME, et al. 6-N,N-dimethylamino-2,3-naphthalimide: A new environment-sensitive fluorescent probe in δ- and μ-selective opioid peptides. J. Med. Chem. 2006;49:3653–3658. doi: 10.1021/jm060343t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lanfermeijer FC, et al. On the binding mechanism of the peptide receptor of the oligopeptide transport system of Lactococcus lactis. EMBO J. 2000;19:3649–3656. doi: 10.1093/emboj/19.14.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marriott G, et al. Spectroscopic and functional-characterization of an environmentally sensitive fluorescent actin conjugate. Biochemistry. 1988;27:6214–6220. doi: 10.1021/bi00417a004. [DOI] [PubMed] [Google Scholar]
- 91.Renard M, et al. Knowledge-based design of reagentless fluorescent biosensors from recombinant antibodies. J. Mol. Biol. 2002;318:429–442. doi: 10.1016/S0022-2836(02)00023-2. [DOI] [PubMed] [Google Scholar]
- 92.Loving G, Imperiali B. Thiol-reactive derivatives of the solvatochromic 4-N,N-dimethylamino-1,8-naphthalimide fluorophore: a highly sensitive toolset for the detection of biomolecular interactions. Bioconjug. Chem. 2009 doi: 10.1021/bc900319z. DOI: 10.1021/bc900319z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shyu YJ, Hu CD. Fluorescence complementation: an emerging tool for biological research. Trends Biotechnol. 2008;26:622–630. doi: 10.1016/j.tibtech.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 94.Condon EU. Nuclear motions associated with electron transitions in diatomic molecules. Phys. Rev. 1928;32:858–872. [Google Scholar]
- 95.Dawson PE, et al. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 96.Hackeng TM, et al. Protein synthesis by native chemical ligation: Expanded scope by using straightforward methodology. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10068–10073. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]