Abstract
High mobility group (HMG) proteins assume important roles in regulating chromatin dynamics, transcriptional activities of genes and other cellular processes. Post-translational modifications of HMG proteins can alter their interactions with DNA and proteins, and consequently, affect their biological activities. Although the mechanisms through which these modifications are involved in regulating biological processes in different cellular contexts are not fully understood, new insights into these modification “codes” have emerged from the increasing appreciation of the functions of these proteins. In this review, we focus on the chemical modifications of mammalian HMG proteins and highlight their roles in nuclear functions.
Keywords: High mobility group protein, post-translational modification, mass spectrometry
Introduction
The genetic material within the nucleus of eukaryotic cells exists in the form of chromatin, which functions as an integrated platform for the dynamic regulation of various nuclear processes, such as transcription, replication and repair. The basic building block of chromatin is nucleosome, which consists of DNA and core histones (H2A, H2B, H3 and H4). Nucleosomes are modified and organized into higher-order structures by histone H1 and other abundant chromosomal proteins, among which high mobility group (HMG) proteins represent the largest group of non-histone components [1–3]. HMG proteins play important roles in remodeling the assembly of chromatin and regulating gene transcription in higher eukaryotic cells by distorting, bending or modifying the structure of DNA, which forms complexes with histones and transcription factors [1, 3, 4].
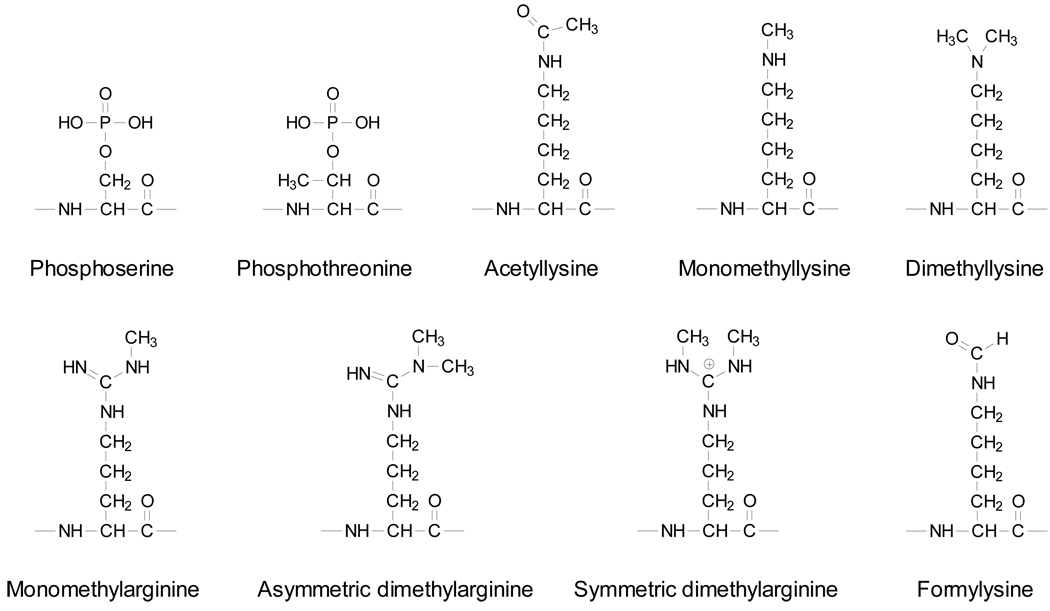
Three distinct families of HMG proteins were defined and named after the structure of their DNA binding domains as well as their substrate binding specificity, including HMG-AT-hook families (HMGA), HMG-box families (HMGB) and HMG-nucleosome binding families (HMGN) [5]. Similar to histones, HMG proteins are subject to a wide range of post-translational modifications (PTMs) as shown in Figure 1, including lysine acetylation/methylation/formylation/SUMOylation, arginine methylation, and serine/threonine phosphorylation, etc. A large body of experimental evidence indicates that HMG proteins, which are highly regulated by their PTMs, are essential players in the execution and regulation of nuclear functions. Characterization of these chemical modifications of HMG proteins has and will continue to provide significant insights into the mechanisms of action of these proteins, which may eventually lead to improved diagnosis, therapy and prognosis of human diseases. In this review, we discuss common modifications of mammalian HMG proteins, with emphasis being placed on recent findings about the roles of these modifications in nuclear functions.
Figure 1.
Mammalian HMG protein are subjected to a range of post-translational modifications, including lysine acetylation/methylation/formylation, arginine methylation, and serine/threonine phosphorylation.
PTMs of HMGA proteins
The HMGA proteins, comprising of HMGA1a, HMGA1b and HMGA2, are characterized by the possession of three DNA-binding domains known as “AT-hooks” [6] and an acidic carboxyl-terminal tail. They are involved in a wide variety of nuclear processes by regulating, positively or negatively, transcriptional activities of numerous genes in vivo [7, 8]. Through interactions with DNA and other proteins, they influence normal biological processes such as cell growth, proliferation, differentiation and death [7]. HMGA proteins are expressed abundantly in embryonic cells and exert important roles in development [9–11].
Even though HMGA proteins are hardly detectable in adult human tissues, they are overexpressed in many types of cancers [7]. Recently, blocking HMGA function has been suggested for the therapeutic interventions of cancer [12]. Aside from their oncogenic properties, HMG proteins are involved in other common diseases such as obesity [13], diabetes [14] and atherosclerosis [15].
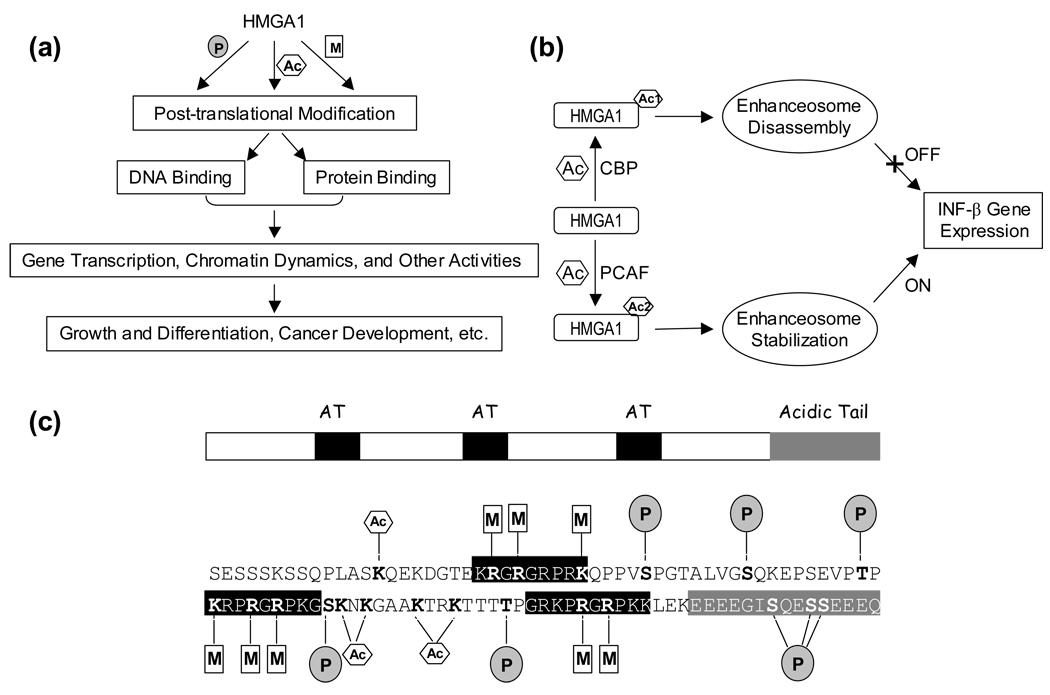
The biological activity of HMGA proteins are highly regulated by their PTMs, such as phosphorylation, acetylation, methylation, and ADP-ribosylation [16]. In response to cellular signaling events, these dynamic modifications influence HMGA’s binding toward DNA and proteins, thereby regulating gene transcription, chromatin dynamics and other nuclear functions (Figure 2a). Among all subfamilies of HMG proteins, PTMs of HMGA1 proteins have been extensively investigated for over 20 years. The domain structures and the known modifications of human HMGA1a are depicted in Figure 2c.
Figure 2.
(a) A schematic overview of the nuclear functions of post-translationally modified HMGA1 proteins. (b) An example showing how HMGA1 acetylation affects the expression of IFN-β gene [40, 41]. (c) Domain structure, primary sequence and the known major post-translational modifications of human HMGA1a with the modified residues being highlighted in bold. The three ‘AT-hook’ domains are shaded in black, and the acidic C-terminal tail is shaded in grey in the domain structure and primary sequence of the protein. Phosphorylation, methylation and acetylation are denoted by ‘P’ in shaded circle, ‘M’ in rectangle and ‘A’ in hexagon, respectively.
Phosphorylation
The phosphorylation of HMGA1 proteins was detected in Ehrlich ascite cells [17] two years after the original identification of these proteins in Hela cells [18]. Since then, HMGA1 proteins have been found to be among the most heavily phosphorylated proteins in the nucleus. It has been realized for years that HMGA proteins are actively involved in the dynamic changes of chromatin structure during various stages of a cell cycle [19].
Earlier work carried out by Lund [20] and Reeves [21, 22] demonstrated that the cell-cycle dependent kinase cdc2 phosphorylated the HMGA1a protein at Thr-52 and Thr-77 in vitro and in metaphase-arrested cells, resulting in a decrease in binding of HMGA1a to DNA. Recently, Fusco et al. [23] reported that the homeodomain-interacting protein kinase-2 (HIPK2) could phosphorylate HMGA1 proteins and exert a potent inhibitory effect on cell growth at the G2/M phase of the cell cycle [24]. Our recent study revealed that the HIPK2 and cdc2 phosphorylated HMGA1a at the same amino acid residues (i.e., Ser-35, Thr-52, and Thr-77) in vitro [25], though the two kinases exhibited different site preferences for the phosphorylation; the preference for HIPK2 phosphorylation followed the order of Thr-77 > Thr-52 > Ser-35, whereas the sequence for cdc2 phosphorylation was Thr-52 > Thr-77 > Ser-35. Moreover, the HIPK2-mediated phosphorylation reduced the binding affinity of HMGA1a to human germ line ε promoter, and the drop in binding affinity induced by HIPK2 phosphorylation was not as pronounced as that introduced by cdc2 phosphorylation [25], which is in line with the notion that the second AT-hook in HMGA1a is more important for DNA binding than the third AT-hook [26].
Interestingly, HMGA1 overexpression inhibits p53 activity by relocalizing HIPK2 in the cytoplasm, while HIPK2 overexpression reestablishes its nuclear localization and promotes p53-mediated apoptosis [27]. The major phosphorylation site induced by HIPK2, i.e., Thr-77, is located within the region between the second and third AT-hooks of HMGA1a, which is the region participating in the interaction of HMGA1 with p53 [28]. Thus, it is possible that the HIPK2-induced phosphorylation of HMGA1a decreased the interaction between p53 and HMGA1a and eventually promoted the p53-mediated apoptosis. On the other hand, overexpression of HMGA1a in cancer cells promotes the localization of HIPK2 to the cytoplasm, which results in decreased phosphorylation of HMGA1a, enhances its interaction with p53, and inhibits the apoptotic function of p53.
The acidic C-terminal tails of HMGA1 proteins are constitutively phosphorylated in vivo. In this regard, protein kinase CK2 catalyzes the phosphorylation of Ser-98, Ser-101 and Ser-102 in HMGA1a and the corresponding sites in HMGA1b (i.e., Ser-87, Ser-90 and Ser-91) both in vitro and in vivo [29–31]. Due to the high content of negatively charged amino acid residues, the C-terminal domain is generally believed to participate in protein-protein interactions rather than binding to DNA; however, an indirect effect of this phosphorylation on HMGA1-DNA interaction was noted [32]
Aside from the kinases described above, protein kinase C (PKC) has been demonstrated to phosphorylate HMGA1a at two major sites of Ser-43 and Ser-63, which attenuates its binding to the promoter regions of PKCγ and neurogranin/RC3 genes [33]. Similarly, Banks et al. [34] demonstrated that, upon treatment of human mammary epithelial cells with phorbol ester TPA (activator of PKC), HMGA1 proteins became highly phosphorylated, indicating that HMGA1 proteins are direct downstream targets of the intracellular PKC signaling pathway.
Recent studies also support that the phosphorylation of HMGA1 proteins may also play an important role in DNA damage response, the protein’s interaction with chromatin, and apoptosis. In this context, HMGA1 proteins were identified as novel downstream target of the ATM (ataxia-telangiectasia mutated) kinase pathway in response to DNA damage [35]. ATM phosphorylates specifically serine or threonine residues preceding a glutamine (also known as the SQ motif) [36]. Indeed, HMGA1 is phosphorylated by the ATM kinase at the SQ motif in its C-terminal region both in vitro and in vivo, and the phosphorylation site was located at Ser-87 in HMGA1b [35]. Additionally, fluorescence recovery after photobleaching studies revealed that the interaction between HMGA1a and chromatin is dynamic, and increased level of phosphorylation is correlated with elevated residence time of HMGA1a in heterochromatin and chromosomes, relative to euchromatic regions [26]. Furthermore, hyper-phosphorylation in HMGA1a was detected in early apoptotic leukemic cells (HL-60, K562, NB4, and U937), which may be associated with HMGA1a’s displacement from chromatin, whereas de-phosphorylation occurred during the formation of highly condensed chromatin in apoptotic bodies [37].
Acetylation
Other than the constitutive acetylation at the N-termini, HMGA1 proteins are post-translationally acetylated on several lysine residues in vivo [30, 34, 38, 39]. The most notable example (as shown in Figure 2b) of the biological significance of HMGA1 acetylation lies in its dynamic control of virus-induced expression of interferon-β (IFN-β) gene [40, 41]. The accurate execution of IFN-β transcriptional switch depends on the ordered acetylation of HMGA1 by two histone acetyltransferases CBP (CREB-binding protein) and PCAF (p300/CBP-associated factor)/GCN5, and the acetylation mediated by these two enzymes confers distinct biological outcomes. In particular, acetylation at Lys-64 by CBP destabilizes the enhanceosome, a higher-order nucleoprotein complex formed in response to virus infection, leading to the transcriptional turn-off of the human IFN-β gene, whereas the PCAF-induced acetylation at Lys-70 potentiates the transcription of IFN-β by stabilizing enhanceosome and preventing CBP-catalyzed acetylation of HMGA1a [40]. A recent study by Zhang et al. [39] revealed that purified PCAF and p300 could also acetylate Lys-14, Lys-66 and Lys-73 in HMGA1a, apart from the acetylation at Lys-64 and Lys-70. Moreover, all five lysine residues are also acetylated in PC-3 human prostate cancer cells, though the acetylation profile is different from either p300- or PCAF-induced acetylation in vitro [39].
In addition to its function in regulating the transcription of the IFN-β gene, the in-vivo PTM study of HMGA1a in breast cancer cells with differing metastatic potential suggested that acetylation might be correlated with the different metastatic potential of cultured human breast tumor cells; increased acetylation was observed in HMGA1a isolated from metastatic cancer cells relative to the protein purified from the non-metastatic counterparts [42].
Methylation
HMGA1 proteins are also susceptible to methylation at several sites located exclusively within their AT-hooks, indicating its potential roles in regulating DNA binding activities of HMGA1. Giancotti et al. [37] first observed arginine mono-methylation in HMGA1a during apoptosis in leukemic cells. In a subsequent study, they found that the increase in HMGA1a methylation is a distinct feature of leukemic cells induced to undergo apoptosis [43]. They next extended this observation by studying other tumor cells, and found that the degree of methylation increased with apoptosis for all tumor cells studied [44]. Moreover, mono-methylation at Arg-25 is independent of the experimental procedures employed for the induction of apoptosis, which include the use of chemical (etoposide, camptothecin, lovastatin), viral (HSV-1) and physical (UVC) agents [44]. Together, these results showed that the methylation of HMGA1a at Arg-25 could be considered as a general phenomenon of all tumor cells induced to undergo apoptosis.
Other than mono-methylation at Arg-25, Zou et al. [45] found that the same site can be both mono- and dimethylated in PC-3 human prostate cancer cells. In addition, both isoforms of arginine dimethylation, i.e., symmetric and asymmetric dimethylations (Figure 1), were detected by MS/MS based on characteristic neutral losses from the side chains of modified arginines [45].
Unlike phosphorylation and acetylation, where the kinases and acetyltransferases responsible for the corresponding modifications have been extensively examined, exploring the enzymes inducing the arginine methylation of HMGA1 proteins has emerged as an important area of investigation. In this context, arginine methylation is catalyzed by protein arginine methyltransferases (PRMTs), which are categorized into type-I and type-II enzymes; these two types of enzymes catalyze the formation of asymmetric ω-NG,NG-dimethylarginine and symmetric ω-NG,N’G-dimethylarginine, respectively [46].
Recent studies revealed that PRMT6, which is a type-I enzyme, could induce the efficient methylation of HMGA1a both in vitro and in vivo, and the sites of methylation were mapped to be Arg-57 and Arg-59, which are embedded in the second AT-hook [47, 48]. In addition, Arg-83 and Arg-85 could be methylated by PRMT6 to a lower extent than Arg-57 and Arg-59 in the third AT-hook, and a very low level of methylation occurred in the first AT-hook [48].
Since Arg-25 within the first AT-hook seems to be the major methylation site in tumor cells [45, 49], PRMTs other than PRMT6 could methylate preferentially HMGA1a at Arg-25. Indeed it was found that PRMT1 and PRMT3 exhibited preference for the methylation of the first AT-hook at Arg-25 and Arg-23 [50]. PRMT3 depicts less specific methyltransferase activity than PRMT1 and the former exists at a lower level in nucleus than in cytoplasm. Therefore, PRMT1 is more likely to be responsible for the methylation of Arg-25 in vivo [50]. However, PRMT1, being a type-I PRMT, produces only asymmetric ω-NG,NG-dimethylarginine in proteins, whereas both the symmetric and asymmetric forms of dimethylations were observed at Arg-25 [45]. Thus, other PRMT(s) catalyzing the symmetric dimethylation of HMGA1a has yet been identified.
Besides its methylation on arginine residues, HMGA1 proteins can be methylated on lysine residues, though the biological functions of the latter methylation remain unclear. Dimethylation of lysine residues in HMGA1 protein was observed in cultured breast cancer cells with different metastatic potential [38, 42]. In an effort to examine the nature of PTMs of HMGA1 proteins isolated from cancerous/normal human breast tissues, Zou et al. [49] found that Lys-30 and Lys-54 in HMGA1a could be monomethylated in both metastatic and primary human breast tumor tissues [49].
HMGA2 protein is encoded by a different gene (HMGA2); it distinguishes from HMGA1a in the first 25 amino-acid residues and contains a short peptide located between the third AT-hook and the acidic tail, which is absent in HMGA1a [51]. Notwithstanding growing lines of evidence indicating that HMGA2 plays a crucial role in a variety of biological processes, such as mammalian growth, embryonic development and turmorigenesis [52–57], a few studies have focused on its PTMs and the roles of these PTMs in modulating the functions of HMGA2. It’s clear that HMGA2 protein can be phosphorylated, and the potential kinases involved in the phosphorylation include cdc2 [58], CK2 [59] and Nek2 [60]. Very recently, Sgarra et al. [61] demonstrated that HMGA2, similar to HMGA1, could be highly phosphorylated on the acidic C-terminal tail at Thr-96, Thr-99, Ser-100, Ser-101 and Ser-104 both in vitro and in vivo [61]. In addition, phosphorylation of Ser-43, located outside of the acidic tail, was also detected in endogenous HMGA2 [61].
Aside from phosphorylation, a study by Cao et al. [62] showed that HMGA2 protein can undergo SUMOylation, which can lead to the degradation of promyelocytic leukemia (PML) protein via the ubiquitin-proteasomal pathway; interestingly, arsenic trioxide treatment can stimulate the SUMOylation of HMGA2 and the subsequent degradation of PML.
PTMs of HMGB proteins
HMGB proteins, including HMGB1 (formerly known as HMG1), HMGB2 (previously HMG2) and HMGB3, contain two HMG-box domains and a highly acidic C-terminal tail. The HMG boxes bend DNA and bind preferentially to structurely distorted DNA [63].
In the nucleus, HMGB proteins regulate numerous activities such as transcription, replication and repair [3, 64]. The Hmgb1 knockout mice die shortly after birth, supporting the functional importance of the HMGB1 protein [65]. One mechanism for HMGB proteins to control gene transcription is through its direct interaction with nucleosomes to loosen the wrapped DNA and to promote its sliding, which allows for nucleosome remodeling [66, 67]. On the other hand, HMGB proteins can also interact with specific transcription factors, such as HOX and POU protein, NF-κB, and SREBPs (sterol-regulatory element-binding proteins), to regulate gene expression [3, 64, 68].
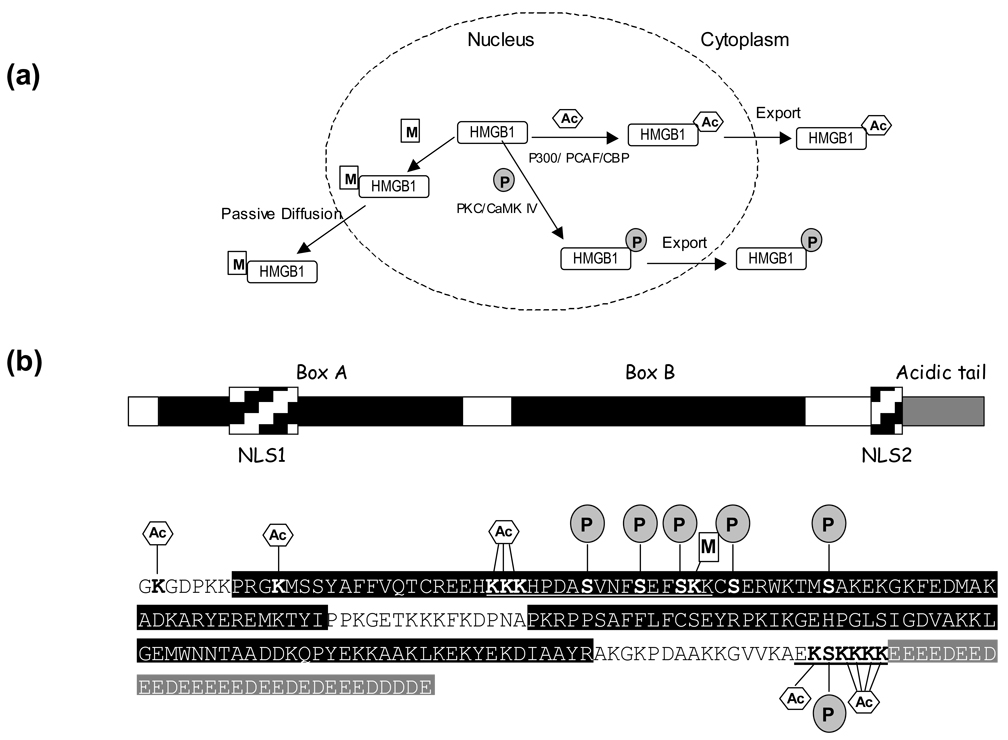
Distinguished from other HMG subfamilies, HMGB1 can be passively leaked out from cells during necrosis to trigger inflammation [69], or secreted from nucleus by certain cells such as monocytes and macrophages to serve as a proinflammatory cytokine [70]. The data accumulated in the past few years have clearly indicated that the extracelluar HMGB1 plays critical roles in inflammation and tumor progression [70–73]. Viewing the fact that HMGB1 is almost ubiquitous and only ten times less abundant than core histones [70], these observations raise the question of how a nuclear protein can reach extracellular space. Mounting evidence suggests that phosphorylation, acetylation and methylation are essential for the release of HMGB1 from nucleus to cytoplasm and its ultimate secretion (Figure 3a)
Figure 3.
(a) Subcellular distribution of HMGB1 is regulated by its PTMs [79, 81, 86]. (b) Domain structure, primary sequence, and the known post-translational modifications of human HMGB1 with the modified residues being highlighted in bold. The two NLS regions are labeled in the domain structure and underlined in the protein’s primary sequence; the two HMG box domains are shaded in black, and the acidic C-terminal tail is shaded in grey. Phosphorylation, methylation and acetylation are denoted by ‘P’ in shaded circle, ‘M’ in rectangle and ‘A’ in hexagon, respectively.
Acetylation
Reversible acetylation of HMGB1 and HMGB2 at Lys-2 and Lys-11 was first detected in 1979 by Sterner et al. [74] by incubating calf thymus homogenates with 3H-labeled acetate. The acetylated HMGB1 protein, isolated from cells grown in the presence of sodium n-butyrate, exhibited significantly enhanced ability to recognize UV light- or cisplatin-damaged DNA and four-way junction [75]. Lately, it was reported that mutation of both lysine residues to alanines abrogates its binding to four-way junction and 88-bp DNA minicircles [76]. In addition, the binding of in vivo acetylated HMGB1 to supercoiled DNA was investigated, and it was found that acetylation of Lys-2 enhanced the protein’s DNA binding affinity, supercoiling activity and its capacity to protect supercoiled DNA from relaxation by topoisomerase I [77]. Taken together, these results reveal the critical role of acetylation in structure-specific DNA binding of HMGB1 and its potential function in DNA repair.
HMGB1 acetylation was also shown to influence DNA replication. In this context, native HMGB1 extracted from tumor cells inhibits the replication of closed circular plasmid DNA in cell-free extracts of Hela-S3 cells and this effect is reduced upon its acetylation [78].
The first line of evidence supporting the involvement of HMGB1 acetylation in its cellular distribution came from the studies by Bianchi and coworkers [79]. They demonstrated that HMGB1 shuttled actively between the nucleus and the cytoplasm through a tightly controlled mechanism, and HMGB1 was acetylated extensively in monocytes to direct its release from nucleus to the cytoplasm for exocytosis. Lys-27, Lys-28, Lys-29, Lys-179, Lys-181, Lys-182, Lys-183 and Lys-184 were found to be the major acetylated residues, and all of them were within the nuclear localization signal (NLS) regions (Figure 3b). Mutation of six lysines to glutamines, which mimic acetylated lysines, caused the relocalization of a fraction of HMGB1 from the nucleus to the cytoplasm [79]. Similar results were obtained with the treatment of histone deacetylase inhibitors, indicating that the subcellular localization of HMGB1 depends on its acetylation [79].
Phosphorylation
Phosphorylation of mammalian HMGB1 and HMGB2 by the calcium-phospholipid-dependent protein kinase has been suggested 25 years ago [80]; however, its biological function has not been revealed until recently. Shin et al. [81] found that phosphorylation of HMGB1, like its acetylation, was also involved in the protein’s cytoplasmic relocation. After treatment with TNF-α or OA (okadaic acid, a phosphatase inhibitor), HMGB1 is phosphorylated in RAW264.7 cells and human monocytes, resulting in the transport of HMGB1 from the nucleus to the cytoplasm for its subsequent secretion. Moreover, phosphorylation of both NLS regions of HMGB1 is required and the major phosphorylation sites were suggested to be Ser-34, Ser-38, Ser-41, Ser-45, Ser-52 and Ser-180, mainly around NLS1 and NLS2 [81].
Since HMGB1 is a key mediator of inflammatory diseases such as sepsis [82, 83], the kinase involved in the phosphorylation of HMGB1 during sepsis would be a good candidate to regulate its secretion. Very recently, Shin and coworkers [84] revealed that the PKC kinase, instead of CKII or cdc2, is the effector kinase for HMGB1 phosphorylation in LPS-stimulated monocytes. Interestingly, it has been revealed that another kinase, i.e., the calcium/calmodulin-dependent kinase IV (CaMK IV), was also involved in LPS-induced HMGB1 release through serine phosphorylation of HMGB1 [85]. In both cases, the HMGB1 secretion is closely correlated with intracellular Ca2+ concentration. However, it is not clear whether the LPS-stimulated HMGB1 secretion is primarily mediated by PKC, CaMK IV, or both. Determination of specific phosphorylation sites induced by each kinase may provide some important insights into this question.
Methylation
HMGB1 was found to be methylated, which can also result in its cytoplasmic localization in neutrophils [86]. In contrast to the acetylation and phosphorylation of HMGB1, which occur in the NLS regions, methylation of HMGB1 occurs at Lys-42, which resides in its HMG box domain. In this respect, mono-methylation at Lys-42 of HMGB1 alters its conformation and weakens substantially its DNA binding activity, leading to the passive diffusion of HMGB1 out of the nucleus in neutrophils [86]. Indeed, most methylated HMGB1 resides in the cytoplasm of neutrophils, whereas unmethylated HMGB1 exists in the nucleus.
PTMs of HMGN proteins
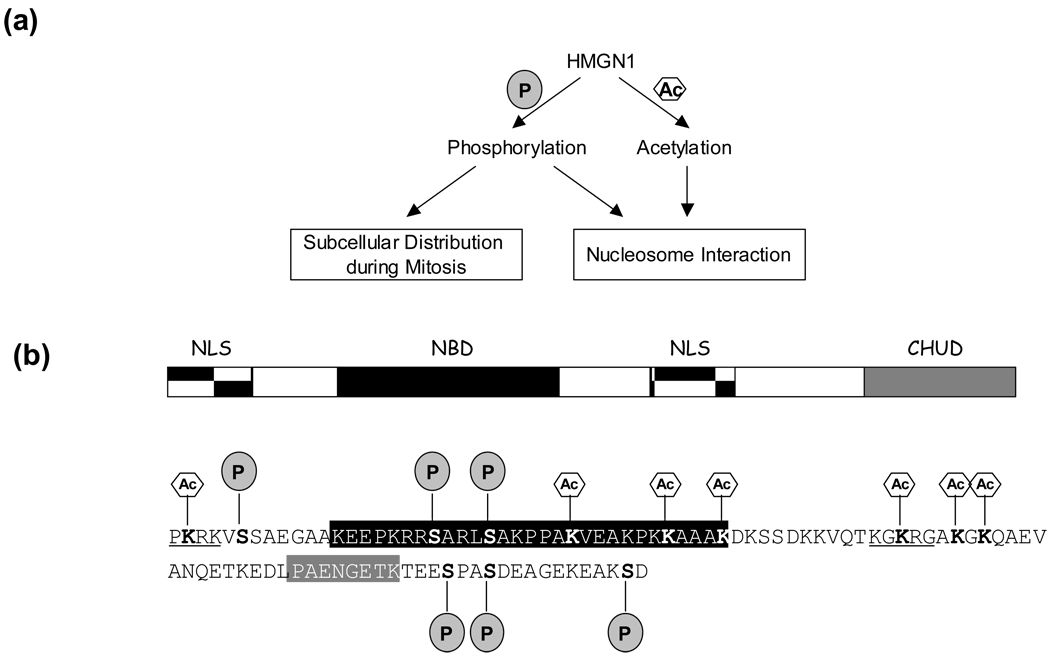
Among all HMG proteins, HMGN is the only subgroup known to bind specifically to the 147-base pair nucleosome core particle [87, 88]. The major members, HMGN1 and HMGN2, are present in the nuclei of all mammalian and most vertebrate cells [1, 2]. There are three major functional domains in HMGN proteins: a nucleosome-binding domain (NBD), a bipartite nuclear localization signal (NLS) and a chromatin-unfolding domain (CHUD) [89–91]. The NBD domain is a highly conserved and positively charged region rich in lysine, arginine and proline residues [92]. The binding of HMGN to nucleosome decreases the compactness of chromatin fiber and enhances transcription and replication from chromatin templates [2, 93]. The intracellular organization of HMG proteins is dynamic and related to both cell-cycle and transcriptional events [93–95]. Very recently, it was found that HMGN proteins repressed potently ATP-dependent chromatin remodeling to restrict nucleosome mobility in the nucleus [96]. PTMs of HMGN1 protein, including phosphorylation and acetylation, are thought to affect the interaction of HMGN1 with its chromatin targets and other proteins, thereby regulating chromatin structure, subcellular localization of HMGN1, and cellular responses to changing environmental stimuli.
Phosphorylation
Similar to HMGA proteins, phosphorylation of HMGN proteins was reported in some early studies [97–103]. Both HMGN1 and HMGN2 can be phosphorylated in vitro and in vivo, resulting in decreased interaction of HMGN proteins with the nucleosome. Several protein kinases, including PKC [80, 104], cyclic adenosine 3’,5’-monophosphate (cAMP)-dependent protein kinase (PKA) [98, 105], cGMP-dependent protein kinase [106], CK2 [107] and mitogen-and stress-activated kinases [108, 109] can phosphorylate HNGN1 protein with different site specificity. In this context, Ser-6 and Ser-24 in mammalian HMGN1 are the major and minor sites of phosphorylation catalyzed by the cyclic nucleotide-dependent protein kinases [105, 106], whereas Ser-88 and Ser-98 are the major and minor modification sites induced by protein kinase CK2 [107]. Additionally, HMGN1 isolated from calf thymus can be phosphorylated by PKC in vitro, and Ser-20 and Ser-24 were proposed to be the respective major and minor phosphorylation sites [104]. Given the fact that HMGN proteins are highly dynamic [94, 110, 111], these different kinases, and thus the distinct phosphorylated forms of HMGN may be involved in its diverse functions in the nucleus.
Phosphorylation of HMGN1 at Ser-6 and histone H3 at Ser-10 is associated with the rapid induction of immediate-early genes in response to mitogenic and stress stimuli in fibroblasts, and MSK1 and MSK2 are the major kinases responsible for the phosphorylation [108, 109]. Phosphorylation of HMGN1 leads to a transient weakening of its association with chromatin and allows protein kinases to access histone H3 [112]. Thus, phosphorylation of HMGN1 may precede that of histone H3, indicating an important role of HMGN1 in modulating the “histone code”.
The intracellular organization of HMGN proteins is related to both transcriptional activity [94] and cell cycle progression [89]. In a series of experiments, Bustin and coworkers [113] demonstrated elegantly that HMGN proteins were highly and specifically phosphorylated at its NBD domain during mitosis, and this modification abolished their nucleosome binding capability. Using immunofluorescence and nuclear import assays, they found that mitotic phosphorylation of the NBD domain (mainly at Ser-20 and Ser-24) in HMGN1, rather than inactivation of NLS domain, prevented its reentry into the newly formed nucleus in late telophase [114]. In addition, this phosphorylation promoted the binding of HMGN1 to specific isotypes of 14.3.3 proteins, which are known to affect the subcellular localization of their binding partners, and retards its entry to nucleus during telophase [114]. Collectively, these results revealed the significant roles of mitotic phosphorylation of HMGN1 in both abolishment of chromatin binding and inhibition of nuclear import.
It was also suggested that HMGN phosphorylation may play a role in its subcellular redistribution [102]. Mass spectrometric results revealed that Ser-20 and Ser-24 in HMGN1, which was isolated from K562 human cells treated with a phosphatase inhibitor (i.e., 12-O-tetradecanoylphorbol-13-acetate or okadaic acid), were the major and minor phosphorylation sites, respectively, and a third phosphorylation site in HMGN1 was located to be either Ser-6 or Ser-7. Interestingly, the proportion of phosphorylated HMGN proteins found in cytosolic pools was significantly higher than those in the nuclear fraction, suggesting that phosphorylation of HMGN proteins regulates their subcellular distribution [102]. A more recent study by Zou et al. [115] demonstrated that Ser-6, Ser-85, Ser-88, and Ser-98 were phosphorylated in MCF-7 human breast cancer cells.
Acetylation
Other than phosphorylation, acetylation of HMGN proteins was also involved in regulating their interaction with nucleosome, and possibly HMGN’s intracellular distribution. The HMGN1 and HMGN2 isolated from duck erythrocytes were acetylated mainly at Lys-2, and additional sites were also detected in cells treated with a deacetylase inhibitor [116]. Bustin et al. [117, 118] reported that the enzymes responsible for the reversible acetylation of HMGN proteins were histone acetyltransferases p300 and PCAF. P300 acetylates both HMGN proteins at multiple lysine residues, which are conserved among all HMGN family members [118]. There are seven major acetylation sites in HMGN1, which include four residues located near or at the bipartite NLS region (i.e., Lys-2, Lys-54, Lys-58 and Lys-60) and three residues in the NBD domain (i.e., Lys-30, Lys-37 and Lys-41). PCAF, on the other hand, can acetylate HMGN2, mainly at Lys-2, but not HMGN1 [118]. The most obvious consequence of both p300- and PCAF-mediated acetylation of HMGN proteins is the reduction of HMGN’s affinity to nucleosome [117, 118]. Lysine acetylation in the NLS domain also raises the possibility that acetylation may affect its transport and reentry into the nucleus [117].
As discussed above, phosphorylation and acetylation both affect the intracellular distribution and nucleosome interactions of HMGN proteins. Given the fact that enzymes catalyzing the modifications of histone proteins may also be responsible for the modifications of HMGN proteins, it is reasonable to predict that the local structure of the chromatin fiber is very likely modulated by a dynamic interplay between histones and HMGN proteins. Along this line, the effects of HMGN proteins on the PTMs of histones H2A and H3 have been recently examined [112, 119]. The presence of HMGN1, but not HMGN2, inhibited the phosphorylation of histone H3 at Ser-10 and Ser-28 [112, 119]. Likewise, HMGN1 could modulate the phosphorylation of histone H2A at Ser-1 [120]. In addition, the existence of HMGN1 changed the acetylation of Lys-9 and Lys-14 as well as the methylation of Lys-4 and Lys-9 in histone H3 [112, 119]. Very recently, the HMGN1-mediated acetylation of histone H3 at Lys-14 was found to modulate the interaction between ATM and chromatin both before and after the ionizing radiation-induced DNA double-strand break formation, thereby resulting in the autophosphorylation of ATM and the activation of its target genes [121]. On the grounds that the PTMs of HMGN proteins affect their interactions with different targets [93], these findings suggested that “histone code” could be finely regulated by HMGN proteins and their PTMs. Further studies on the dynamic interactions between HMGN proteins and histone proteins and how they are perturbed by the PTMs of these two groups of proteins will provide insights into the roles of HMGN proteins in chromatin activities.
Conclusions
HMG proteins participate in a wide range of cellular activities influenced by their post-translational modifications. Characterization of these modifications of HMG proteins has and will continue to yield significant insights into the biological functions of these proteins. Recent years have witnessed continuing discovery in the diversity and complexity of HMG modifications that are functionally important in distinct cellular pathways. The enzymes involved with the deposition of some of these modifications and their functions have been revealed; however, it is not clear which enzyme(s) are involved in many other modifications and how they participate in regulating the nuclear functions of HMG proteins. Understanding the upstream regulatory pathways of individual enzymes and the downstream events following the enzyme-induced chemical modifications of HMG proteins remains an important task. Additionally, HMG proteins interact with linker and core histones in nucleus to regulate chromatin activities, it is important to have a genome-wide assessment about how the chemical modifications of HMG proteins are correlated with those of the histone proteins.
Many lines of evidence support that the PTMs of HMG proteins play very important roles in various nuclear functions including transcription, DNA replication, DNA damage response, and chromatin assembly. Recent studies also revealed that all three subfamilies of HMG proteins are involved in various DNA repair pathways; however, it remains to be assessed how the covalent modifications of HMG proteins modulate their functions in DNA repair. For instance, an elevated expression of HMGA1 proteins was found to result in compromised nucleotide excision repair [122–124], and overexpression of HMGA2 suppressed non-homologous end joining repair [125]. HMGB1 protein was revealed to be an important component of protein complexes involved in DNA mismatch repair [126] and in recognizing duplex DNA containing a 6-thioguanine [127] or cisplatin-induced intrastrand crosslink [128]. The latter binding toward HMGB1 shields cisplatin-induced DNA adduct from recognition by excision repair factors [128]. By contrast, mammalian cells lacking HMGB1 are hypersensitive to DNA damage induced by UVC irradiation or psoralen plus UVA irradiation (PUVA), and the repair and removal of UVC lesions from genomic DNA is markedly reduced in cells lacking HMGB1 [129]. Lange et al. [129] further explored the role of HMGB1 in chromatin remodeling upon DNA damage and they observed that cells lacking HMGB1 displayed no histone acetylation upon DNA damage. A recent study showed that Hmgn1−/− mouse embryonic fibroblasts (MEFs) are hypersensitive to UV, and the removal rate of photoproducts from the chromatin of Hmgn1−/− MEFs is decreased when compared with the chromatin of the wild-type MEF. It was concluded that HMGN1 facilitates access of DNA repair factors to UV-damaged DNA sites and enhances the rate of DNA repair in chromatin by reducing the compactness of the higher-order chromatin structure [130]. It will be important to investigate the effects of the PTMs of HMG proteins on their roles in DNA repair.
Taken together, HMG proteins have crucial and complex roles in various activities in the nucleus. Understanding how these activities are regulated by their PTMs will contribute to improved diagnosis and therapeutic interventions of human diseases.
Figure 4.
(a) Subcellular distribution of HMGN1 during cell cycle and its interactions with nucleosome are regulated by its acetylation and phosphorylation [113, 114, 117, 118]. (b) The domain structure, primary sequence, and the known major post-translational modifications of human HMGN1 with the modified residues being highlighted in bold. The two NLS regions are underlined in the protein’s primary sequence, and the NBD and CHUD domains are shaded in black and grey, respectively. Phosphorylation and acetylation are denoted by ‘P’ in shaded circle and ‘A’ in hexagon, respectively.
Acknowledgment
The authors want to thank the National Institutes of Health for supporting this research (R01 CA116522) and Dr. Yan Zou for helpful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 2.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Abe N, Watanabe T, Masaki T, Mori T, Sugiyama M, Uchimura H, Fujioka Y, Chiappetta G, Fusco A, Atomi Y. Pancreatic duct cell carcinomas express high levels of high mobility group I(Y) proteins. Cancer Res. 2000;60:3117–3122. [PubMed] [Google Scholar]
- 5.Bustin M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 2001;26:152–153. doi: 10.1016/s0968-0004(00)01777-1. [DOI] [PubMed] [Google Scholar]
- 6.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 7.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 8.Reeves R, Beckerbauer L. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta. 2001;1519:13–29. doi: 10.1016/s0167-4781(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Schiltz JF, Ashar HR, Chada KK. HMGA1 is required for normal sperm development. Mol. Reprod. Dev. 2003;66:81–89. doi: 10.1002/mrd.10323. [DOI] [PubMed] [Google Scholar]
- 10.Fedele M, Fidanza V, Battista S, Pentimalli F, Klein-Szanto AJ, Visone R, De Martino I, Curcio A, Morisco C, Del Vecchio L, Baldassarre G, Arra C, Viglietto G, Indolfi C, Croce CM, Fusco A. Haploinsufficiency of the Hmga1 gene causes cardiac hypertrophy and myelo-lymphoproliferative disorders in mice. Cancer Res. 2006;66:2536–2543. doi: 10.1158/0008-5472.CAN-05-1889. [DOI] [PubMed] [Google Scholar]
- 11.Caron L, Bost F, Prot M, Hofman P, Binetruy B. A new role for the oncogenic high-mobility group A2 transcription factor in myogenesis of embryonic stem cells. Oncogene. 2005;24:6281–6291. doi: 10.1038/sj.onc.1208781. [DOI] [PubMed] [Google Scholar]
- 12.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 13.Anand A, Chada K. In vivo modulation of HMGIC reduces obesity. Nat. Genet. 2000;24:377–380. doi: 10.1038/74207. [DOI] [PubMed] [Google Scholar]
- 14.Foti D, Chiefari E, Fedele M, Iuliano R, Brunetti L, Paonessa F, Manfioletti G, Barbetti F, Brunetti A, Croce CM, Fusco A, Brunetti A. Lack of the architectural factor HMGA1 causes insulin resistance and diabetes in humans and mice. Nat. Med. 2005;11:765–773. doi: 10.1038/nm1254. [DOI] [PubMed] [Google Scholar]
- 15.Schlueter C, Hauke S, Loeschke S, Wenk HH, Bullerdiek J. HMGA1 proteins in human atherosclerotic plaques. Pathol. Res. Pract. 2005;201:101–107. doi: 10.1016/j.prp.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Giancotti V, Bandiera A, Sindici C, Perissin L, Crane-Robinson C. Calcium-dependent ADP-ribosylation of high-mobility-group I (HMGI) proteins. Biochem. J. 1996;317(Pt 3):865–870. doi: 10.1042/bj3170865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lund T, Holtlund J, Laland SG. On the phosphorylation of low molecular mass HMG (high mobility group) proteins in Ehrlich ascites cells. FEBS Lett. 1985;180:275–279. doi: 10.1016/0014-5793(85)81085-1. [DOI] [PubMed] [Google Scholar]
- 18.Lund T, Holtlund J, Fredriksen M, Laland SG. On the presence of two new high mobility group-like proteins in HeLa S3 cells. FEBS Lett. 1983;152:163–167. doi: 10.1016/0014-5793(83)80370-6. [DOI] [PubMed] [Google Scholar]
- 19.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions. Int. J. Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 20.Lund T, Laland SG. The metaphase specific phosphorylation of HMG I. Biochem. Biophys. Res. Commun. 1990;171:342–347. doi: 10.1016/0006-291x(90)91399-d. [DOI] [PubMed] [Google Scholar]
- 21.Nissen MS, Langan TA, Reeves R. Phosphorylation by cdc2 kinase modulates DNA binding activity of high mobility group I nonhistone chromatin protein. J. Biol. Chem. 1991;266:19945–19952. [PubMed] [Google Scholar]
- 22.Reeves R, Langan TA, Nissen MS. Phosphorylation of the DNA-binding domain of nonhistone high-mobility group I protein by cdc2 kinase: reduction of binding affinity. Proc. Natl. Acad. Sci. USA. 1991;88:1671–1675. doi: 10.1073/pnas.88.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YH, Choi CY, Kim Y. Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by the ubiquitin-like protein SUMO-1. Proc. Natl. Acad. Sci. USA. 1999;96:12350–12355. doi: 10.1073/pnas.96.22.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierantoni GM, Fedele M, Pentimalli F, Benvenuto G, Pero R, Viglietto G, Santoro M, Chiariotti L, Fusco A. High mobility group I (Y) proteins bind HIPK2, a serine-threonine kinase protein which inhibits cell growth. Oncogene. 2001;20:6132–6141. doi: 10.1038/sj.onc.1204635. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q, Wang Y. Homeodomain-Interacting Protein Kinase-2 (HIPK2) Phosphorylates HMGA1a at Ser-35, Thr-52, and Thr-77 and Modulates Its DNA Binding Affinity. J. Proteome Res. 2007;6:4711–4719. doi: 10.1021/pr700571d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrer M, Luhrs H, Bustin M, Scheer U, Hock R. Dynamic interaction of HMGA1a proteins with chromatin. J. Cell Sci. 2004;117:3459–3471. doi: 10.1242/jcs.01160. [DOI] [PubMed] [Google Scholar]
- 27.Pierantoni GM, Rinaldo C, Mottolese M, Di Benedetto A, Esposito F, Soddu S, Fusco A. High-mobility group A1 inhibits p53 by cytoplasmic relocalization of its proapoptotic activator HIPK2. J. Clin. Invest. 2007;117:693–702. doi: 10.1172/JCI29852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Pierantoni GM, Rinaldo C, Esposito F, Mottolese M, Soddu S, Fusco A. High Mobility Group A1 (HMGA1) proteins interact with p53 and inhibit its apoptotic activity. Cell Death Differ. 2006;13:1554–1563. doi: 10.1038/sj.cdd.4401839. [DOI] [PubMed] [Google Scholar]
- 29.Palvimo J, Linnala-Kankkunen A. Identification of sites on chromosomal protein HMG-I phosphorylated by casein kinase II. FEBS Lett. 1989;257:101–104. doi: 10.1016/0014-5793(89)81796-x. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X, Wang Y. Acetylation and phosphorylation of high-mobility group A1 proteins in PC-3 human tumor cells. Biochemistry. 2006;45:7194–7201. doi: 10.1021/bi060504v. [DOI] [PubMed] [Google Scholar]
- 31.Ferranti P, Malorni A, Marino G, Pucci P, Goodwin GH, Manfioletti G, Giancotti V. Mass spectrometric analysis of the HMGY protein from Lewis lung carcinoma. Identification of phosphorylation sites. J. Biol. Chem. 1992;267:22486–22489. [PubMed] [Google Scholar]
- 32.Wang DZ, Ray P, Boothby M. Interleukin 4-inducible phosphorylation of HMG-I(Y) is inhibited by rapamycin. J. Biol. Chem. 1995;270:22924–22932. doi: 10.1074/jbc.270.39.22924. [DOI] [PubMed] [Google Scholar]
- 33.Xiao DM, Pak JH, Wang X, Sato T, Huang FL, Chen HC, Huang KP. Phosphorylation of HMG-I by protein kinase C attenuates its binding affinity to the promoter regions of protein kinase C gamma and neurogranin/RC3 genes. J. Neurochem. 2000;74:392–399. doi: 10.1046/j.1471-4159.2000.0740392.x. [DOI] [PubMed] [Google Scholar]
- 34.Banks GC, Li Y, Reeves R. Differential in vivo modifications of the HMGI(Y) nonhistone chromatin proteins modulate nucleosome and DNA interactions. Biochemistry. 2000;39:8333–8346. doi: 10.1021/bi000378+. [DOI] [PubMed] [Google Scholar]
- 35.Pentimalli F, Palmieri D, Pacelli R, Garbi C, Cesari R, Martin E, Pierantoni GM, Chieffi P, Croce CM, Costanzo V, Fedele M, Fusco A. HMGA1 protein is a novel target of the ATM kinase. Eur. J. Cancer. 2008;44:2668–2679. doi: 10.1016/j.ejca.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 36.O'Neill T, Dwyer AJ, Ziv Y, Chan DW, Lees-Miller SP, Abraham RH, Lai JH, Hill D, Shiloh Y, Cantley LC, Rathbun GA. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J. Biol. Chem. 2000;275:22719–22727. doi: 10.1074/jbc.M001002200. [DOI] [PubMed] [Google Scholar]
- 37.Diana F, Sgarra R, Manfioletti G, Rustighi A, Poletto D, Sciortino MT, Mastino A, Giancotti V. A link between apoptosis and degree of phosphorylation of high mobility group A1a protein in leukemic cells. J. Biol. Chem. 2001;276:11354–11361. doi: 10.1074/jbc.M009521200. [DOI] [PubMed] [Google Scholar]
- 38.Edberg DD, Adkins JN, Springer DL, Reeves R. Dynamic and differential in vivo modifications of the isoform HMGA1a and HMGA1b chromatin proteins. J. Biol. Chem. 2005;280:8961–8973. doi: 10.1074/jbc.M407348200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Q, Zhang K, Zou Y, Perna A, Wang Y. A quantitative study on the in-vitro and in-vivo acetylation of high mobility group A1 proteins. J. Am. Soc. Mass Spectrom. 2007;18:1569–1578. doi: 10.1016/j.jasms.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 2001;293:1133–1136. doi: 10.1126/science.293.5532.1133. [DOI] [PubMed] [Google Scholar]
- 41.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol. Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 42.Edberg DD, Bruce JE, Siems WF, Reeves R. In vivo posttranslational modifications of the high mobility group A1a proteins in breast cancer cells of differing metastatic potential. Biochemistry. 2004;43:11500–11515. doi: 10.1021/bi049833i. [DOI] [PubMed] [Google Scholar]
- 43.Sgarra R, Diana F, Rustighi A, Manfioletti G, Giancotti V. Increase of HMGA1a protein methylation is a distinctive characteristic of leukaemic cells induced to undergo apoptosis. Cell Death Differ. 2003;10:386–389. doi: 10.1038/sj.cdd.4401184. [DOI] [PubMed] [Google Scholar]
- 44.Sgarra R, Diana F, Bellarosa C, Dekleva V, Rustighi A, Toller M, Manfioletti G, Giancotti V. During apoptosis of tumor cells HMGA1a protein undergoes methylation: identification of the modification site by mass spectrometry. Biochemistry. 2003;42:3575–3585. doi: 10.1021/bi027338l. [DOI] [PubMed] [Google Scholar]
- 45.Zou Y, Wang Y. Tandem mass spectrometry for the examination of the posttranslational modifications of high-mobility group A1 proteins: symmetric and asymmetric dimethylation of Arg25 in HMGA1a protein. Biochemistry. 2005;44:6293–6301. doi: 10.1021/bi0475525. [DOI] [PubMed] [Google Scholar]
- 46.McBride AE, Silver PA. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 47.Miranda TB, Webb KJ, Edberg DD, Reeves R, Clarke S. Protein arginine methyltransferase 6 specifically methylates the nonhistone chromatin protein HMGA1a. Biochem. Biophys. Res. Commun. 2005;336:831–835. doi: 10.1016/j.bbrc.2005.08.179. [DOI] [PubMed] [Google Scholar]
- 48.Sgarra R, Lee J, Tessari MA, Altamura S, Spolaore B, Giancotti V, Bedford MT, Manfioletti G. The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J. Biol. Chem. 2006;281:3764–3772. doi: 10.1074/jbc.M510231200. [DOI] [PubMed] [Google Scholar]
- 49.Zou Y, Wang Y. Mass spectrometric analysis of high-mobility group proteins and their post-translational modifications in normal and cancerous human breast tissues. J. Proteome Res. 2007;6:2304–2314. doi: 10.1021/pr070072q. [DOI] [PubMed] [Google Scholar]
- 50.Zou Y, Webb K, Perna AD, Zhang Q, Clarke S, Wang Y. A mass spectrometric study on the in vitro methylation of HMGA1a and HMGA1b proteins by PRMTs: methylation specificity, the effect of binding to AT-rich duplex DNA, and the effect of C-terminal phosphorylation. Biochemistry. 2007;46:7896–7906. doi: 10.1021/bi6024897. [DOI] [PubMed] [Google Scholar]
- 51.Manfioletti G, Giancotti V, Bandiera A, Buratti E, Sautiere P, Cary P, Crane-Robinson C, Coles B, Goodwin GH. cDNA cloning of the HMGI-C phosphoprotein, a nuclear protein associated with neoplastic and undifferentiated phenotypes. Nucleic Acids Res. 1991;19:6793–6797. doi: 10.1093/nar/19.24.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monzen K, Ito Y, Naito AT, Kasai H, Hiroi Y, Hayashi D, Shiojima I, Yamazaki T, Miyazono K, Asashima M, Nagai R, Komuro I. A crucial role of a high mobility group protein HMGA2 in cardiogenesis. Nat. Cell Biol. 2008;10:567–574. doi: 10.1038/ncb1719. [DOI] [PubMed] [Google Scholar]
- 53.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 54.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, Elliott KS, Hackett R, Guiducci C, Shields B, Zeggini E, Lango H, Lyssenko V, Timpson NJ, Burtt NP, Rayner NW, Saxena R, Ardlie K, Tobias JH, Ness AR, Ring SM, Palmer CN, Morris AD, Peltonen L, Salomaa V, Davey Smith G, Groop LC, Hattersley AT, McCarthy MI, Hirschhorn JN, Frayling TM. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaidi MR, Okada Y, Chada KK. Misexpression of full-length HMGA2 induces benign mesenchymal tumors in mice. Cancer Res. 2006;66:7453–7459. doi: 10.1158/0008-5472.CAN-06-0931. [DOI] [PubMed] [Google Scholar]
- 56.Fedele M, Battista S, Kenyon L, Baldassarre G, Fidanza V, Klein-Szanto AJ, Parlow AF, Visone R, Pierantoni GM, Outwater E, Santoro M, Croce CM, Fusco A. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene. 2002;21:3190–3198. doi: 10.1038/sj.onc.1205428. [DOI] [PubMed] [Google Scholar]
- 57.Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, Ciarmiello A, Pallante P, Arra C, Melillo RM, Helin K, Croce CM, Fusco A. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–471. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 58.Schwanbeck R, Manfioletti G, Wisniewski JR. Architecture of high mobility group protein I-C.DNA complex and its perturbation upon phosphorylation by Cdc2 kinase. J. Biol. Chem. 2000;275:1793–1801. doi: 10.1074/jbc.275.3.1793. [DOI] [PubMed] [Google Scholar]
- 59.Goodwin G. The high mobility group protein, HMGI-C. Int. J. Biochem. Cell Biol. 1998;30:761–766. doi: 10.1016/s1357-2725(98)00016-8. [DOI] [PubMed] [Google Scholar]
- 60.Di Agostino S, Fedele M, Chieffi P, Fusco A, Rossi P, Geremia R, Sette C. Phosphorylation of high-mobility group protein A2 by Nek2 kinase during the first meiotic division in mouse spermatocytes. Mol. Biol. Cell. 2004;15:1224–1232. doi: 10.1091/mbc.E03-09-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sgarra R, Maurizio E, Zammitti S, Lo Sardo A, Giancotti V, Manfioletti G. Macroscopic differences in HMGA oncoproteins post-translational modifications: C-terminal phosphorylation of HMGA2 affects its DNA binding properties. J. Proteome Res. 2009;8:2978–2989. doi: 10.1021/pr900087r. [DOI] [PubMed] [Google Scholar]
- 62.Cao X, Clavijo C, Li X, Lin HH, Chen Y, Shih HM, Ann DK. SUMOylation of HMGA2: selective destabilization of promyelocytic leukemia protein via proteasome. Mol. Cancer Ther. 2008;7:923–934. doi: 10.1158/1535-7163.MCT-07-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas JO, Travers AA. HMG1 and 2, and related 'architectural' DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 64.Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 65.Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein HMG1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 66.Bonaldi T, Langst G, Strohner R, Becker PB, Bianchi ME. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 2002;21:6865–6873. doi: 10.1093/emboj/cdf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Travers AA. Priming the nucleosome: a role for HMGB proteins? EMBO Rep. 2003;4:131–136. doi: 10.1038/sj.embor.embor741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Najima Y, Yahagi N, Takeuchi Y, Matsuzaka T, Sekiya M, Nakagawa Y, Amemiya-Kudo M, Okazaki H, Okazaki S, Tamura Y, Iizuka Y, Ohashi K, Harada K, Gotoda T, Nagai R, Kadowaki T, Ishibashi S, Yamada N, Osuga J, Shimano H. High mobility group protein-B1 interacts with sterol regulatory element-binding proteins to enhance their DNA binding. J. Biol. Chem. 2005;280:27523–27532. doi: 10.1074/jbc.m414549200. [DOI] [PubMed] [Google Scholar]
- 69.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 70.Muller S, Scaffidi P, Degryse B, Bonaldi T, Ronfani L, Agresti A, Beltrame M, Bianchi ME. New EMBO members' review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. EMBO J. 2001;20:4337–4340. doi: 10.1093/emboj/20.16.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campana L, Bosurgi L, Rovere-Querini P. HMGB1: a two-headed signal regulating tumor progression and immunity. Curr. Opin. Immunol. 2008;20:518–523. doi: 10.1016/j.coi.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 72.Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, Tanji N, Lu Y, Lalla E, Fu C, Hofmann MA, Kislinger T, Ingram M, Lu A, Tanaka H, Hori O, Ogawa S, Stern DM, Schmidt AM. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405:354–360. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 73.Apetoh L, Tesniere A, Ghiringhelli F, Kroemer G, Zitvogel L. Molecular interactions between dying tumor cells and the innate immune system determine the efficacy of conventional anticancer therapies. Cancer Res. 2008;68:4026–4030. doi: 10.1158/0008-5472.CAN-08-0427. [DOI] [PubMed] [Google Scholar]
- 74.Sterner R, Vidali G, Allfrey VG. Studies of acetylation and deacetylation in high mobility group proteins. Identification of the sites of acetylation in HMG-1. J. Biol. Chem. 1979;254:11577–11583. [PubMed] [Google Scholar]
- 75.Ugrinova I, Pasheva EA, Armengaud J, Pashev IG. In vivo acetylation of HMG1 protein enhances its binding affinity to distorted DNA structures. Biochemistry. 2001;40:14655–14660. doi: 10.1021/bi0113364. [DOI] [PubMed] [Google Scholar]
- 76.Assenberg R, Webb M, Connolly E, Stott K, Watson M, Hobbs J, Thomas JO. A critical role in structure-specific DNA binding for the acetylatable lysine residues in HMGB1. Biochem. J. 2008;411:553–561. doi: 10.1042/BJ20071613. [DOI] [PubMed] [Google Scholar]
- 77.Ugrinova I, Pashev IG, Pasheva EA. Post-synthetic acetylation of HMGB1 protein modulates its interactions with supercoiled DNA. Mol. Biol. Rep. 2009;36:1399–1404. doi: 10.1007/s11033-008-9327-z. [DOI] [PubMed] [Google Scholar]
- 78.Topalova D, Ugrinova I, Pashev IG, Pasheva EA. HMGB1 protein inhibits DNA replication in vitro: a role of the acetylation and the acidic tail. Int. J. Biochem. Cell Biol. 2008;40:1536–1542. doi: 10.1016/j.biocel.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 79.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22:5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramachandran C, Yau P, Bradbury EM, Shyamala G, Yasuda H, Walsh DA. Phosphorylation of high-mobility-group proteins by the calcium-phospholipid-dependent protein kinase and the cyclic AMP-dependent protein kinase. J. Biol. Chem. 1984;259:13495–13503. [PubMed] [Google Scholar]
- 81.Youn JH, Shin JS. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J. Immunol. 2006;177:7889–7897. doi: 10.4049/jimmunol.177.11.7889. [DOI] [PubMed] [Google Scholar]
- 82.Mantell LL, Parrish WR, Ulloa L. HMGB-1 as a therapeutic target for infectious and inflammatory disorders. Shock. 2006;25:4–11. doi: 10.1097/01.shk.0000188710.04777.9e. [DOI] [PubMed] [Google Scholar]
- 83.Wang H, Zhu S, Zhou R, Li W, Sama AE. Therapeutic potential of HMGB1-targeting agents in sepsis. Expert Rev. Mol. Med. 2008;10:e32. doi: 10.1017/S1462399408000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oh YJ, Youn JH, Ji Y, Lee SE, Lim KJ, Choi JE, Shin JS. HMGB1 is phosphorylated by classical protein kinase C and is secreted by a calcium-dependent mechanism. J. Immunol. 2009;182:5800–5809. doi: 10.4049/jimmunol.0801873. [DOI] [PubMed] [Google Scholar]
- 85.Zhang X, Wheeler D, Tang Y, Guo L, Shapiro RA, Ribar TJ, Means AR, Billiar TR, Angus DC, Rosengart MR. Calcium/calmodulin-dependent protein kinase (CaMK) IV mediates nucleocytoplasmic shuttling and release of HMGB1 during lipopolysaccharide stimulation of macrophages. J. Immunol. 2008;181:5015–5023. doi: 10.4049/jimmunol.181.7.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ito I, Fukazawa J, Yoshida M. Post-translational methylation of high mobility group box 1 (HMGB1) causes its cytoplasmic localization in neutrophils. J. Biol. Chem. 2007;282:16336–16344. doi: 10.1074/jbc.M608467200. [DOI] [PubMed] [Google Scholar]
- 87.Bustin M, Lehn DA, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim. Biophys. Acta. 1990;1049:231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- 88.Sandeen G, Wood WI, Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980;8:3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hock R, Scheer U, Bustin M. Chromosomal proteins HMG-14 and HMG-17 are released from mitotic chromosomes and imported into the nucleus by active transport. J. Cell Biol. 1998;143:1427–1436. doi: 10.1083/jcb.143.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trieschmann L, Postnikov YV, Rickers A, Bustin M. Modular structure of chromosomal proteins HMG-14 and HMG-17: definition of a transcriptional enhancement domain distinct from the nucleosomal binding domain. Mol. Cell. Biol. 1995;15:6663–6669. doi: 10.1128/mcb.15.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ding HF, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol. Cell. Biol. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crippa MP, Alfonso PJ, Bustin M. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J. Mol. Biol. 1992;228:442–449. doi: 10.1016/0022-2836(92)90833-6. [DOI] [PubMed] [Google Scholar]
- 93.Bustin M. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci. 2001;26:431–437. doi: 10.1016/s0968-0004(01)01855-2. [DOI] [PubMed] [Google Scholar]
- 94.Hock R, Wilde F, Scheer U, Bustin M. Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. EMBO J. 1998;17:6992–7001. doi: 10.1093/emboj/17.23.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 96.Rattner BP, Yusufzai T, Kadonaga JT. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol. Cell. 2009;34:620–626. doi: 10.1016/j.molcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bhorjee JS. Differential phosphorylation of nuclear nonhistone high mobility group proteins HMG 14 and HMG 17 during the cell cycle. Proc. Natl. Acad. Sci. USA. 1981;78:6944–6948. doi: 10.1073/pnas.78.11.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spaulding SW, Fucile NW, Bofinger DP, Sheflin LG. Cyclic adenosine 3',5'-monophosphate-dependent phosphorylation of HMG 14 inhibits its interactions with nucleosomes. Mol. Endocrinol. 1991;5:42–50. doi: 10.1210/mend-5-1-42. [DOI] [PubMed] [Google Scholar]
- 99.Palvimo J, Maenpaa PH. Binding of high-mobility-group proteins HMG 14 and HMG 17 to DNA and histone H1 as influenced by phosphorylation. Biochim. Biophys. Acta. 1988;952:172–180. doi: 10.1016/0167-4838(88)90113-6. [DOI] [PubMed] [Google Scholar]
- 100.Palvimo J, Linnala-Kankkunen A, Maenpaa PH. Phosphorylation alters the affinity of high mobility group protein HMG 14 for single-stranded DNA. Biochem. Biophys. Res. Commun. 1985;133:343–346. doi: 10.1016/0006-291x(85)91881-9. [DOI] [PubMed] [Google Scholar]
- 101.Lund T, Berg K. Metaphase-specific phosphorylations weaken the association between chromosomal proteins HMG 14 and 17, and DNA. FEBS Lett. 1991;289:113–116. doi: 10.1016/0014-5793(91)80921-o. [DOI] [PubMed] [Google Scholar]
- 102.Louie DF, Gloor KK, Galasinski SC, Resing KA, Ahn NG. Phosphorylation and subcellular redistribution of high mobility group proteins 14 and 17, analyzed by mass spectrometry. Protein Sci. 2000;9:170–179. doi: 10.1110/ps.9.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saffer JD, Glazer RI. The phosphorylation of high mobility group proteins 14 and 17 and their distribution in chromatin. J. Biol. Chem. 1982;257:4655–4660. [PubMed] [Google Scholar]
- 104.Palvimo J, Mahonen A, Maenpaa PH. Phosphorylation of high-mobility-group chromatin proteins by protein kinase C from rat brain. Biochim. Biophys. Acta. 1987;931:376–383. doi: 10.1016/0167-4889(87)90229-1. [DOI] [PubMed] [Google Scholar]
- 105.Walton GM, Spiess J, Gill GN. Phosphorylation of high mobility group 14 protein by cyclic nucleotide-dependent protein kinases. J. Biol. Chem. 1982;257:4661–4668. [PubMed] [Google Scholar]
- 106.Palvimo J, Linnala-Kankkunen A, Maenpaa PH. Differential phosphorylation of high mobility group protein hmg 14 from calf thymus and avian erythrocytes by a cyclic gmp-dependent protein kinase. Biochem. Biophys. Res. Commun. 1983;110:378–382. doi: 10.1016/0006-291x(83)91159-2. [DOI] [PubMed] [Google Scholar]
- 107.Walton GM, Spiess J, Gill GN. Phosphorylation of high mobility group protein 14 by casein kinase II. J. Biol. Chem. 1985;260:4745–4750. [PubMed] [Google Scholar]
- 108.Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 1999;18:4779–4793. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol. Cell. Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol. Cell. 2004;15:573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 113.Prymakowska-Bosak M, Misteli T, Herrera JE, Shirakawa H, Birger Y, Garfield S, Bustin M. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol. 2001;21:5169–5178. doi: 10.1128/MCB.21.15.5169-5178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prymakowska-Bosak M, Hock R, Lim JH, Birger Y, Shirakawa H, Lee K, Bustin M. Mitotic phosphorylation of chromosomal protein HMGN1 inhibits nuclear import and promotes interaction with 14.3.3 proteins. Mol. Cell. Biol. 2002;22:6809–6819. doi: 10.1128/MCB.22.19.6809-6819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zou Y, Jiang X, Wang Y. Identification of novel in vivo phosphorylation sites in high mobility group N1 protein from the MCF-7 human breast cancer cells. Biochemistry. 2004;43:6322–6329. doi: 10.1021/bi0362828. [DOI] [PubMed] [Google Scholar]
- 116.Sterner R, Vidali G, Allfrey VG. Studies of acetylation and deacetylation in high mobility group proteins. Identification of the sites of acetylation in high mobility group proteins 14 and 17. J. Biol. Chem. 1981;256:8892–8895. [PubMed] [Google Scholar]
- 117.Bergel M, Herrera JE, Thatcher BJ, Prymakowska-Bosak M, Vassilev A, Nakatani Y, Martin B, Bustin M. Acetylation of novel sites in the nucleosomal binding domain of chromosomal protein HMG-14 by p300 alters its interaction with nucleosomes. J. Biol. Chem. 2000;275:11514–11520. doi: 10.1074/jbc.275.15.11514. [DOI] [PubMed] [Google Scholar]
- 118.Herrera JE, Sakaguchi K, Bergel M, Trieschmann L, Nakatani Y, Bustin M. Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol. Cell. Biol. 1999;19:3466–3473. doi: 10.1128/mcb.19.5.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ueda T, Postnikov YV, Bustin M. Distinct domains in high mobility group N variants modulate specific chromatin modifications. J. Biol. Chem. 2006;281:10182–10187. doi: 10.1074/jbc.M600821200. [DOI] [PubMed] [Google Scholar]
- 120.Postnikov YV, Belova GI, Lim JH, Bustin M. Chromosomal protein HMGN1 modulates the phosphorylation of serine 1 in histone H2A. Biochemistry. 2006;45:15092–15099. doi: 10.1021/bi0613271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, Kraemer KH, Shiloh Y, Bustin M. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat. Cell Biol. 2009;11:92–96. doi: 10.1038/ncb1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Adair JE, Kwon Y, Dement GA, Smerdon MJ, Reeves R. Inhibition of nucleotide excision repair by high mobility group protein HMGA1. J. Biol. Chem. 2005;280:32184–32192. doi: 10.1074/jbc.M505600200. [DOI] [PubMed] [Google Scholar]
- 123.Maloney SC, Adair JE, Smerdon MJ, Reeves R. Gene-specific nucleotide excision repair is impaired in human cells expressing elevated levels of high mobility group A1 nonhistone proteins. DNA Repair. 2007;6:1371–1379. doi: 10.1016/j.dnarep.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Adair JE, Maloney SC, Dement GA, Wertzler KJ, Smerdon MJ, Reeves R. High-mobility group A1 proteins inhibit expression of nucleotide excision repair factor xeroderma pigmentosum group A. Cancer Res. 2007;67:6044–6052. doi: 10.1158/0008-5472.CAN-06-1689. [DOI] [PubMed] [Google Scholar]
- 125.Li AY, Boo LM, Wang SY, Lin HH, Wang CC, Yen Y, Chen BP, Chen DJ, Ann DK. Suppression of nonhomologous end joining repair by overexpression of HMGA2. Cancer Res. 2009;69:5699–5706. doi: 10.1158/0008-5472.CAN-08-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yuan F, Gu L, Guo S, Wang C, Li GM. Evidence for involvement of HMGB1 protein in human DNA mismatch repair. J. Biol. Chem. 2004;279:20935–20940. doi: 10.1074/jbc.M401931200. [DOI] [PubMed] [Google Scholar]
- 127.Krynetski EY, Krynetskaia NF, Bianchi ME, Evans WE. A nuclear protein complex containing high mobility group proteins B1 and B2, heat shock cognate protein 70, ERp60, and glyceraldehyde-3-phosphate dehydrogenase is involved in the cytotoxic response to DNA modified by incorporation of anticancer nucleoside analogues. Cancer Res. 2003;63:100–106. [PubMed] [Google Scholar]
- 128.Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc. Natl. Acad. Sci. USA. 1994;91:10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc. Natl. Acad. Sci. USA. 2008;105:10320–10325. doi: 10.1073/pnas.0803181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Birger Y, West KL, Postnikov YV, Lim JH, Furusawa T, Wagner JP, Laufer CS, Kraemer KH, Bustin M. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 2003;22:1665–1675. doi: 10.1093/emboj/cdg142. [DOI] [PMC free article] [PubMed] [Google Scholar]