Abstract
EWS/FLI1 is a fusion gene product generated by a chromosomal translocation t(11; 22)(q24; q12) found in Ewing sarcoma. EWS/FLI1 encodes an aberrant transcription factor with oncogenic properties in vitro. Paradoxically, expression of EWS/FLI1 in non-transformed primary cells results in apoptosis, but the exact mechanism remains unclear. In primary mouse embryonic fibroblasts (MEFs) derived from conditional EWS/FLI1 knock-in embryos, expression of EWS/FLI1 resulted in apoptosis with concomitant increase in the endogenous Caspase 3 (Casp3) mRNA. EWS/FLI1 directly bound and activated the CASP3 promoter, while siRNA-mediated knockdown of EWS/FLI1 led to a marked decrease in CASP3 transcripts in Ewing sarcoma cell lines. Ectopic expression of EWS/FLI1 resulted in an increased expression of CASP3 protein in heterologous cell lines. Importantly, expression of EWS/FLI1 in the mouse triggered an early onset of apoptosis in kidneys and acute lethality. These findings suggest that EWS/FLI1 induces apoptosis, at least partially, through the activation of CASP3 and demonstrate the cell-context dependent roles of EWS/FLI1 in apoptosis and tumorigenesis.
Keywords: Ewing sarcoma, EWS/FLI1, Caspase 3, apoptosis
Introduction
Ewing sarcoma is a cancer found in bone and soft tissues (1). About 85% of Ewing sarcoma cases are characterized by the presence of EWS/FLI1 generated by a balanced chromosomal translocation t(11;22)(q24; q12), resulting in the fusion of EWS to FLI1 (2, 3). The remaining cases of Ewing sarcoma contain chromosomal translocations of EWS with other members of Erythroblast Transformation Specific (ETS)-family genes (4). EWS/FLI1 translocation is also found in related tumors, primitive neuroectodermal tumor and Askins tumor (5). Chromosomal rearrangements involving EWS with other transcription factors are also observed in Desmoplastic Small Round Cell Tumor, DSRCT, (fused with Wilms tumor gene, WT1) (6), Myxoid liposarcoma (fused with CHOP) (7), extrachondrosarcoma (fused with CHN) (8), and clear cell sarcoma (fused with ATF1) (9).
The oncogenic activity of EWS/FLI1 has been shown by its ability to transform NIH3T3 cells (10–12) and primary bone-marrow-derived mesenchymal stem cells of human (13) and mouse origin (14, 15), all of which formed tumors in immunodeficient mice. The DNA binding domain of FLI1 and EWS/FLI1 recognizes the same GGAA/T core motif (16, 17). Interestingly, recent reports have shown that GGAA-containing microsatellites may function as the EWS/FLI1 responsive element (18, 19). A large number of EWS/FLI1 target genes have been identified which contribute to cell proliferation and tumorigenesis (20, 21).
Though the oncogenic mechanisms of EWS/FLI1 have been studied extensively, expression of EWS/FLI1 has also been demonstrated to result in apoptosis and growth arrest in primary cells. EWS/FLI1 expression in primary MEFs resulted in rapid cell death and growth arrest, which were abolished by the inactivation of p16, p19ARF, or p53 pathway (22). In human primary fibroblasts, expression of EWS/FLI1 causes growth arrest via p53-dependent pathway but not apoptosis (23), suggesting that the effects of EWS/FLI1 may be cell-context dependent. While it is clear that EWS/FLI1 causes apoptosis in murine primary fibroblasts, the mechanism has not been fully elucidated. We now report for the identification of Caspase3 (Casp3) as a new target of EWS/FLI1.
Materials and Methods
Cell lines and antibodies
TC71, A4573, CHP100, U2OS and JN-DSRCT-1 cells were grown in standard culture condition. The following antibodies were used: rabbit polyclonal α-EWS (24), α-Caspase3 (Upstate, NY),α-Actin (Sigma), α-FLI1 (C19, Santa Cruz), α-FLAG (M2, Sigma) or α-GFP (Clontech).
Generation of Ews/FLI1Stop conditional knock-in mouse
The targeting vectors were generated by inserting a loxP-flanked transcriptional Stop signal (25) in either sense or antisense direction into Ews intron 6 containing genomic DNA (Supplement Fig. 1). A human FLI1 cDNA (exons 5–9) with a FLAG epitope at the C-terminus was PCR amplified and fused to Ews exon 7. The targeting vectors were electroporated into mouse ES cells and used to generate Ews/FLI1Stop (EFStop) knock-in mouse (see Supplemental Methods for detail). A transgenic mouse constitutively expressing the CreER allele (B6.Cg-Tg(CAG-cre/Esr1)5Amc/J) was purchased from Jackson Laboratory. EFStop heterozygous mice were crossed with CreER to generate EFStop;CreER− and EFStop;CreER+ mice. For administration of tamoxifen, mice (4–8 weeks) were injected intraperitoneally (ip) with tamoxifen at 3mg/40g body weight (b.w.) for three consecutive days. Animals were handled following the NIH Animal Research Advisory Committee guidelines.
Generation of Ews/FLI1Stop;CreER− and Ews/FLI1ΔStop;CreER+ MEFs
Ews/FLI1Stop or Ews/WT1Stop (24) heterozygotes were crossed with CreER transgenic mouse, and E12.5 embryos were used to generate mouse embryonic fibroblasts (MEFs). For cell growth analysis, MEFs were plated at 5 × 104 cells and after addition of 1 μM of 4-HT, cell number was determined daily. Caspase activity was measured using a Homogenous Caspases Assay (Roche Molecular Biochemicals, Indianapolis, IN).
Chromatin Immunoprecipitation (ChIP) assay
ChIP assay was performed as previously described (26). Chromatin was immnoprecipitated with either α-FLI1 antibody or rabbit IgG antibody and amplified by PCR (see Supplemental Methods for primers).
Cloning of the human CASP3 promoter and luciferase reporter assay
Human CASP3 promoter was amplified by PCR (see Supplemental Methods for primers), and inserted in the pGL3-Basic promoterless luciferase vector (Promega, Madison, WI). CASP3 promoter constructs (P1–P4) were transfected into TC71, CHP100, A4573 and JN-DSRCT-1 cells along with Renilla luciferase reporter plasmid, and luciferase activities were measured using Dual-Luciferase Reporter Assay System (Promega).
siRNA knockdown
TC71, CHP100 and A4573 cells were transfected with 50 nmol of FLI1 siRNA, 5′-GUA CCC UUC UGA CAU CUC CdTdT-3′, (MWG biotech) (27) or scrambled siRNA using Lipofectamine 2000 (Invitrogen). Total RNAs were reverse-transcribed and the expression levels of CASP3 and GAPDH were analyzed by qRT-PCR using TaqMan probes (Applied Biosystems, Foster City, CA). Data were analyzed by comparative Ct method using GAPDH as an endogenous control.
Electrophoretic Mobility Shift Assay (EMSA)
Annealed wildtype or M5 oligonucleotides were end-labeled with P32-γATP (Perkin Elmer, Waltham, MA) and T4 polynucleotide kinase (NE Biolabs, Ipswich, MA). pcDNA3-EWS/FLI1 or pcDNA3 empty vector were transcribed and translated in vitro using T7 TNT Quick Coupled Transcription/Translation system (Promega) and incubated with P32-labeled oligonucleotides (25,000 cpm) in the binding buffer (4mM Hepes, 50mM NaCl, 0.5mM DTT, 0.5mM EDTA, 1mM MgCl2) containing 1μg of poly dIdC, 0.2 μg of BSA, 12% glycerol and either with or without α-FLI1 or α-WT1 antibodies. For the competition assay, unlabeled wildtype or M5 probes were added at 25-, 50- or 100-fold excess to the binding reaction.
TUNEL staining
Tamoxifen-injected Ews/FLI1ΔStop;CreER+ and Wt;CreER+ kidneys (4–8 weeks old) were fixed, embedded in paraffin and sectioned. For TUNEL staining of sectioned kidneys, TdT-FragEL™ Fragmentation Detection kit (Calbiochem) and ApopTag Red In Situ Apoptosis Detection kit (Millipore) were used. Representative images of TUNEL stained slides were captured with Leica DMLB microscope (Leica Microsystems, Bannockburn, IL) or with a confocal LSM 5 Live/Axio Observer.Z1 microscope using 20x objective lens (Carl Zeiss MicroImaing. Inc).
Results
Generation of Ews/FLI1 knock-in mouse
To study the in vivo functions of EWS/FLI1, we used a knock-in strategy to direct the expression of Ews/FLI1 under the native Ews transcriptional elements. To accomplish this, we amplified the human FLI1 cDNA (exons 5–9) with a FLAG-epitope and fused it with the mouse genomic DNA containing the Ews exon 7 breakpoint (Fig. 1). When we screened for the mouse ES cells harboring the correctly targeted Ews/FLI1 (mEF) knock-in allele, we failed to obtain any positive clones, indicating that the constitutive expression of Ews/FLI1 is incompatible with ES cells.
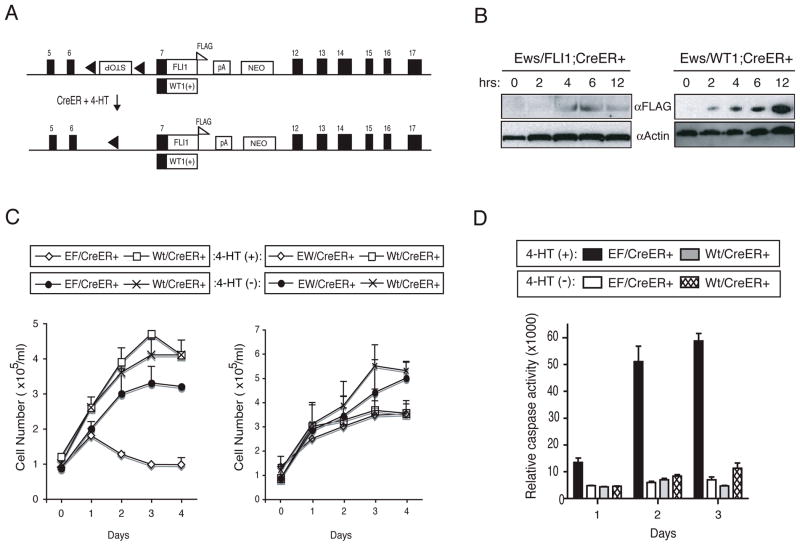
Figure 1.
Inducible expression of mEF. (A) Ews/FLI1 and Ews/WT1(+KTS) knock-in alleles with the C-terminal FLAG. The numbers indicate Ews exons. pA: poly A signal, NEO: neomycin resistant gene. (B) EF;CreER+ and EW;CreER+ MEFs were cultured with 1μM 4-HT and immunoblotted with α-FLAG and α-Actin antibodies. (C) EF;CreER+, EW;CreER+, and Wt;CreER+ MEFs were grown with (+) or without (−) 1μM 4-HT and cell number was determined daily. (D) EF;CreER+ and Wt;CreER+ MEFs were grown with or without 1μM 4-HT and Caspase activity was measured.
To circumvent this, we inserted a transcriptional termination signal (Stop) flanked by LoxP sequences (25) in the Ews intron 6 of the targeting vector (Supplement Fig. 1). This allows for mEF expression to be regulated by a Cre recombinase. As a control, we also inserted the Stop in the antisense direction (Supplement Fig. 1). The sense Stop-containing targeting vector was then used to screen ES cells for homologous recombination, but we again failed to obtain any positive clones (Supplement Fig. 1). This is probably due to the leaky expression of mEF even in the presence of Stop. Surprisingly, when we used the antisense Stop-containing targeting vector, we obtained 8 correctly targeted ES clones out of 170 screened (Supplement Fig. 1). However, the targeted ES cells expressed a read-through transcript which was slightly larger than the predicted Ews/FLI1 transcript as demonstrated by RT-PCR analysis (data not shown). Sequencing of the read-through transcript revealed that it contained an additional 147 nucleotides derived from the antisense Stop sequence resulting in an in-frame insertion of 49 aa between the Ews exons 6 and 7 (Supplement Fig. 2), likely due to the presence of cryptic splice acceptor and donor sites in the antisense Stop. We could not, however, detect a protein product generated from the read-through transcripts by Western blotting using polyclonal antibodies against the N-terminal region of Ews (data not shown) (24). However, we cannot exclude the possibility that a very low level of mEF containing the 49 aa might be expressed in these ES cells. Subsequently, we used the targeted ES cells to generate Ews/FLI1 knock-in mice harboring the conditional knock-in allele (EFStop) in their germline.
Expression of Ews/FLI1 causes apoptosis in mouse embryonic fibroblasts
We generated mouse embryonic fibroblasts (MEFs) expressing Ews/FLI1 (mEF) by crossing EFStop heterozygotes with a transgenic mouse constitutively expressing Cre recombinase fused to a mutant estrogen receptor ligand binding domain (CreER). CreER can be activated by a synthetic ligand 4-hydroxytamoxifen (4-HT) but not by the endogenous estrogen (28). As a control, we also generated MEFs expressing Ews/WT1(+KTS) (mEW), a translocation product found in DSRCT expressing EWS fused to the WT1(+KTS) isoform containing the three amino acids (Lys, Thr, and Ser, termed KTS), by crossing Ews/WT1(+KTS) knock-in heterozygotes (EWStop) (24) with CreER mouse.
Addition of 4-HT to the cultures of EFStop;CreER+ or EWStop;CreER+ MEFs led to the expression of mEF and mEW, respectively, as early as 4 hrs (Fig. 1B). After 24 hrs, cells expressing mEF (designated EF;CreER+) started to die in culture, and by 48–72 hrs, most cells were undergoing cell death (Fig. 1C). In contrast, 4-HT treatment had no effect on the cells expressing mEW (designated EW/CreER+) or wildtype MEFs expressing CreER (Wt;CreER+) (Fig. 1C). To further demonstrate that EF;CreER+ cells were undergoing apoptosis, we measured Caspase activity following the 4-HT treatment. We observed a 3-fold higher Caspase activity in EF;CreER+ cells as compared to the control cells at 24 hrs after the 4-HT treatment, and by 48–72 hrs, the Caspase activity increased dramatically (Fig. 1D). Taken together, our data showed that expression of mEF causes apoptosis in primary MEFs.
Caspase 3 mRNA is elevated following expression of Ews/FLI1
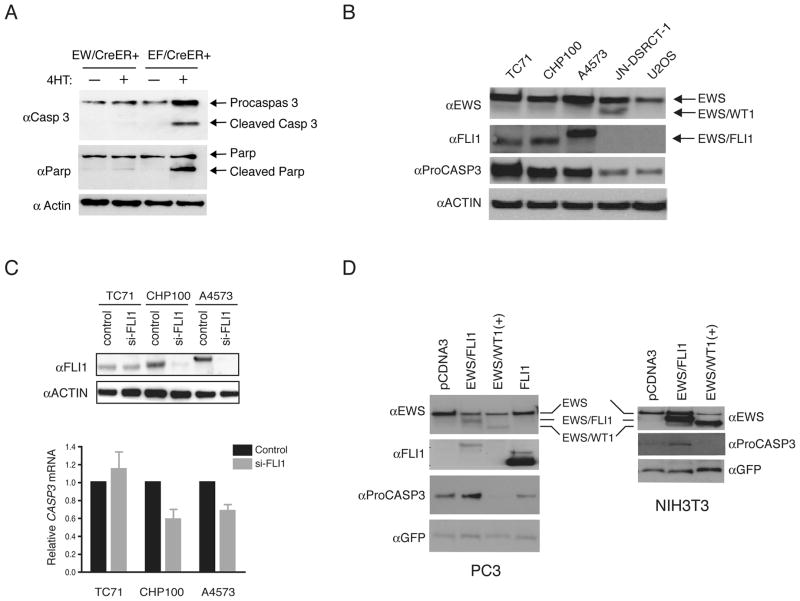
To determine the transcriptional changes induced by mEF which might contribute to apoptosis, we examined the expression levels of 84 apoptosis-related genes in EF/CreER+ MEFs using a Mouse Apoptosis RT2 Profiler PCR Array (SA Biosciences). We analyzed the expression levels of each gene in EF;CreER+ cells at 12 and 24 hrs after the 4-HT treatment in comparison to the levels observed without the treatment. Following the expression of mEF, a number of pro-apoptotic gene transcripts were increased compared to the uninduced cells, e.g., Death Associated Protein kinase 1 (Dapk1), Caspase 3 (Casp3), LIM homeobox protein 4 (Lhx4), and Transformation related protein 53 binding protein 2 (Trp53bp2), while several anti-apoptotic genes showed decreased expression (Supplement Table 1). We decided to further characterize Casp3 since it is directly involved in apoptosis and showed significant expression changes (p-value < 0.05) at both time-points (Supplement Table 1). We confirmed the induction of Casp3 following mEF expression in EF;CreER+ MEFs using a quantitative RT-PCR analysis (TaqMan, Applied Biosystems) (data not shown), as well as by immunoblotting (Fig. 2A). We also observed the cleavage of Casp3 and Parp (poly ADP-ribose polymerase) following mEF expression.
Figure 2.
EWS/FLI1 induces CASP3 expression. (A) MEFs cells were cultured with or without 1μM 4-HT and cell lysates were immunoblotted with α-CASP3, α-PARP and αACTIN antibodies. (B) Cell lysates were analyzed by immunoblotting with α-EWS, α-FLI1, α-CASP3, and α-ACTIN antibodies. (C) Cells transfected with si-FLI1 or scrambled siRNA were either analyzed by immunoblotting with α-FLI1 and α-ACTIN antibodies (upper panels) or total RNAs were analyzed for CASP3 by qRT-PCR (TaqMan). Data represent the mean ± SEM from three independent experiments. (D) Induction of Pro-CASP3 by EWS-FLI1. Empty vector (pcDNA3), pcDNA3-EWS/FLI1, or pcDNA3-EWS/WT1(+KTS) along with CMV-EGFP plasmids were transfected into NIH3T3 or into PC3 cells and immunoblotted as in (B).
EWS/FLI1 activates Caspase 3 transcription
To determine whether mEF-mediated induction of Casp3 also occurs in a relevant cell line, we examined the CASP3 transcript level in two Ewing sarcoma cell lines, A4573 and CHP100. As controls, we used U2OS, an osteosarcoma cell line, and JN-DSRCT-1, a cell line derived from DSRCT expressing EWS/WT1 (29). As compared to U2OS or JN-DSRCT-1 cells, CASP3 transcript levels were at least two-fold or higher in both Ewing sarcoma cells (Supplement Fig. 3A). We then examined the CASP3 at the protein level in three Ewing sarcoma cell lines: TC71 containing the type 1 EWS/FLI1 translocation (EWS exon 7 fused to FLI1 exon 6), CHP100 containing the type II fusion (EWS exon 7 fused to FLI1 exon 5, same as in EF;CreER+ MEFs), and A4573 harboring the infrequent type III fusion (EWS exon 10 fused to FLI1 exon 6). Western blot analysis with an anti-FLI1 antibody demonstrated the expression of different EWS/FLI1 fusion products in these cells (Fig. 2B), but not the endogenous FLI1 (Supplement Fig. 3B). Expression of EWS/WT1 in JN-DSRCT-1 cells was detected using rabbit polyclonal anti-EWS antibodies (24). In all three Ewing sarcoma cell lines, expression of Pro-CASP3 was much higher than U2OS or JN-DSRCT-1 cells (Fig. 2A). We note that the cleaved (activated) CASP3 was not observed in the any of the cell lines (data not shown).
We next used small interfering RNA (siRNA) to deplete EWS/FLI1 in Ewing sarcoma cells and measured CASP3 transcript levels. We employed the siRNA against the FLI1 DNA binding region (FLI-siRNA) that had been previously shown to silence EWS/FLI1 (27). Transfection of FLI-siRNA into CHP100 and A4573 cells reduced the expression of EWS/FLI1 efficiently, but not in TC71 cell (Fig. 2C, upper panel). The reason for this is not clear, but the inefficient knockdown in TC71 cells served as a negative control. Following the knockdown of EWS/FLI1, we observed approximately 30–40% reduction of CASP3 mRNA in A4573 and CHP100 cells, but not in TC71 cells (Fig. 2C, lower panel). To further demonstrate that EWS/FLI1 can induce CASP3 expression, we ectopically expressed EWS/FLI1 in mouse NIH3T3 or human prostate cancer cell line PC3. Compared to the empty vector-, EWS/WT1(+KTS)- or FLI1-transfected cells, EWS/FLI1 expression resulted in an increase expression of Pro-CASP3 in both NIH3T3 and PC3 cells (Fig. 2D). Western blotting of cotransfected green fluorescent protein (GFP) demonstrated similar transfection efficiency.
EWS/FLI1 activates the CASP3 promoter
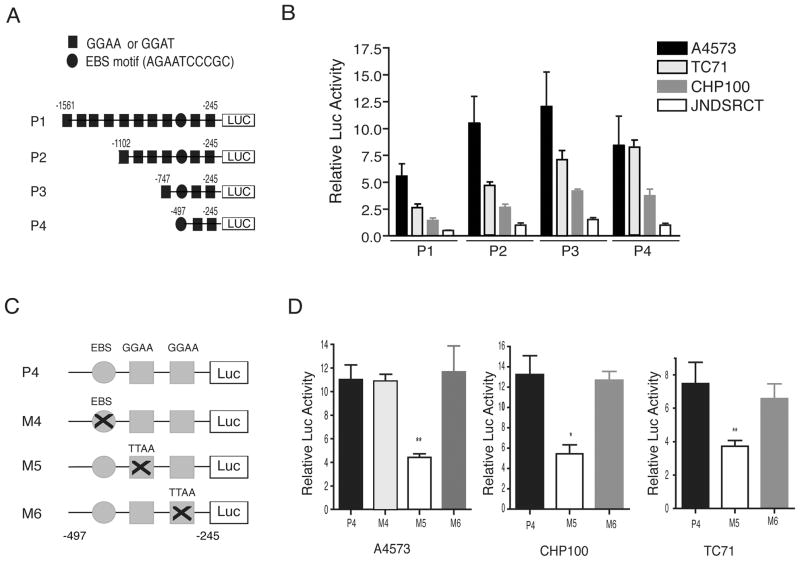
Analysis of the human CASP3 proximal promoter region (about 1.5 kb upstream of the transcription start, +1) revealed the presence of several ETS binding sites: one putative EBS (ETS binding site) motif (AGAATCCCGC) (30), and ten GGAA/T motifs (17) (Fig. 3A). To determine whether EWS/FLI1 can activate the CASP3 promoter, we subcloned a 1.3 kb fragment (from −1561 to −245) of the human CASP3 promoter (P1) into the promoterless luciferase plasmid, pGL3. We also generated a series of CASP3 promoter deletion constructs, P2, P3 and P4 (Fig. 3A). A promoter-reporter analysis of different CASP3 promoter constructs in Ewing sarcoma cell lines as compared to the JN-DSRCT-1 cells (control) revealed that the P4 promoter (−472 to −245) contained the minimal EWS/FLI1 responsive elements (Fig. 3B). The P4 promoter contains a single EBS and two GGAA sites as the potential EWS/FLI1 responsive elements (Fig. 3C). To determine whether these sites are involved, we mutated each site by site-directed mutagenesis to generate M4, M5, and M6 promoter constructs (Fig. 3C). Introduction of these constructs in Ewing sarcoma cells demonstrated that the mutation at the M5 site resulted in a significant reduction of luciferase activity compared to the P4 (wildtype) promoter (Fig. 3D). Mutations in the M4 or M6 site had no effect on the promoter activity.
Figure 3.
EWS/FLI1 activates CASP3 promoter. (A) P1–P4 CASP3 promoters. Putative ETS binding sites (GGAA and EBS motif) are indicated. The numbers are relative to the transcription start (+1). (B) pGL3-Basic or P1–P4 promoter constructs were co-transfected with renilla luciferase, and luciferase activities were measured and expressed as relative to the luciferase activities of pGL3-Basic (set to 1). Data represent the mean ± SEM from three independent experiments. (C) P4 and the mutated M4–M6 CASP3 promoters. Mutations in the EBS and the GGAA sites are indicated. (D) P4 or the mutated promoters (M4–M6) were introduced along with renilla luciferase and luciferase activities were measures as in (B). Data represent the mean ± SEM from three independent experiments (* p <0.01, ** p <0.05).
EWS/FLI1 binds to the GGAA site on the CASP3 promoter
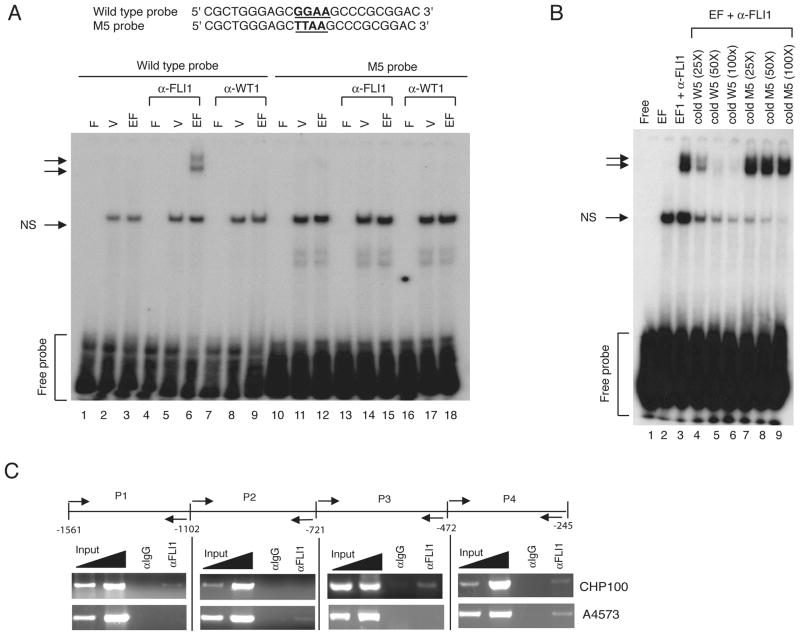
To determine whether EWS/FLI1 can bind directly to the M5 site on the P4 promoter, we synthesized EWS/FLI1 in vitro using a rabbit reticulocyte lysate and performed electro-mobility shift assay (EMSA) with P32-labeled oligonucleotides containing the wildtype GGAA or the mutated TTAA site (M5) (Fig. 4A). Incubation of EWS/FLI1-programmed lysate with the wildtype probe did not reveal any specific band as compared to the empty-vector programmed reticulolysate (Fig. 4A, lanes 2 and 3). However, when the antibody against FLI1 was added to the binding reaction, specific complexes appeared in the EMSA assay (Fig. 4A, arrows, lane 6). The specific bands were not observed when a control antibody (anti-WT1, lanes 7–9) nor when the mutated M5 probe was used (lanes 10–18), suggesting that the specific complexes were due to the stable interaction of EWS/FLI1, the wildtype probe, and the anti-FLI1 antibody. This was further confirmed by the addition of 25-fold excess cold wildtype probe to the binding reaction, which effectively reduced the formation of specific complexes (Fig. 4B, arrows, lane 4). Addition of 50X or 100X unlabeled wildtype probe (Fig. 4B, lanes 5 and 6), but not the mutant (M5) probe (Fig. 4B, lanes 7–9), completely abolished the formation of specific complexes. The reason for the absence of specific complexes when EWS/FLI1 was incubated with the wildtype probe could be due to a weak binding of EWS/FLI1 to a single GGAA site, which might be stabilized by the presence of anti-FLI1 antibody. In support of this view, recent reports demonstrated that EWS/FLI1 did not bind well to a single GGAA site but the binding increased dramatically when multiple GGAA sites were present (18, 19).
Figure 4.
EWS/FLI1 binds to the GGAA. (A) Sequences of the wildtype and the mutated (M5) probes are shown. P32-labled wildtype (lanes 1–9) or M5 probes (lanes 10–18) were incubated with the indicated reticulocyte lysates (F: free probe, V: empty vector, EF: EWS/FLI1) containing either α-FLI1 (C19, Santa Cruz) or α-WT1 (C19, Santa Cruz) antibodies. The arrows indicate specific complexes. NS denotes a nonspecific band. (B) Competitive EMSA assay was performed as in (A) with either 25-, 50-, 100-fold molar excess of unlabeled wildtype (lanes 4–6) or M5 probe (lane 7–9). (C) ChIP assay. Chromatin from CHP100 and A4573 cells immunoprecipitated with either rabbit IgG or α-FLI1 (C19, Santa Cruz) was amplified by PCR with the indicated primers.
To determine whether EWS/FLI1 is present on the human CASP3 promoter in vivo, we performed chromatin immunoprecipitation (ChIP) experiment using the anti-FLI1 antibody (C-19, Santa Cruz), which has been previously used for ChIP assay (18, 19). In both CHP100 and A4573 cells, the ChIP analysis demonstrated that EWS/FLI1 is consistently recruited to the P4 region of the CASP3 promoter (Fig. 4C). We also observed the recruitment of EWS/FLI1 to the P1 (−1102 to −1561) and the P3 regions (−472 to −721) of the CASP3 promoter in CHP100 cells but not in A4573 cells. Recruitment of EWS/FLI1 to the CASP3 P4 promoter region in CHP100 and A4573 cells was further confirmed by ChIP assay performed with the N-terminal EWS-specific antibody (24), but not with the C-terminal EWS-specific antibody (Supplement Fig. 3C). Together, these results suggest that EWS/FLI1 binds directly to the P4 region of CASP3 promoter.
EWS/FLI1 is efficiently expressed upon tamoxifen treatment in vivo
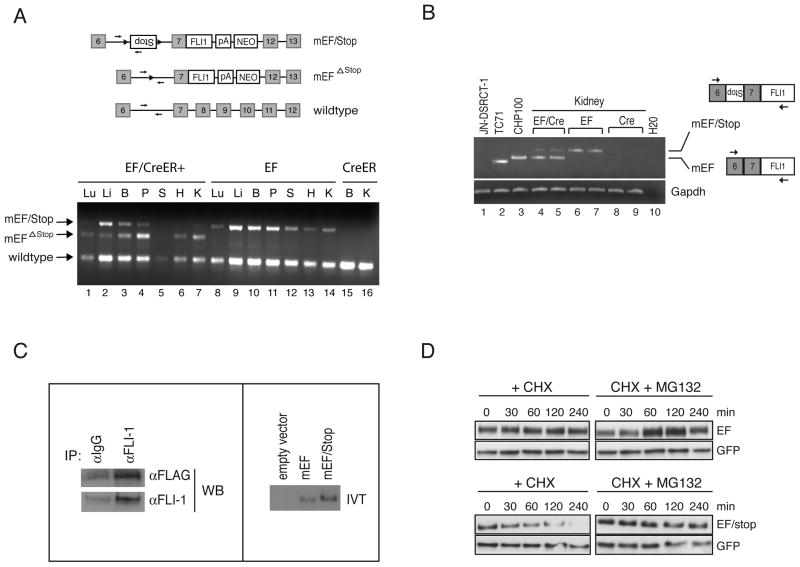
We next examined the effect of mEF expression in the mouse. To detect the expression of mEF in vivo, we administered three daily injections (i.p.) of tamoxifen to 4–8 weeks old EFStop;CreER+ and EFStop;CreER− mice. Following the last injection, tissues were harvested and genomic DNAs were analyzed for the Cre-mediated recombination by PCR (Fig. 5A). In most tissues examined, three daily administration of tamoxifen resulted in the Cre-mediated excision of Stop (Fig. 5A). However, the recombination efficiency varied in different tissues: kidney, heart, lung and pancreas showed the highest level of recombination, while the recombination was less efficient in the other tissues.
Figure 5.
In vivo expression of mEF. (A) A schema of the wildtype and the mEF alleles before and after Cre recombination. The arrows indicate the three primers used in the genomic PCR analysis. Genomic DNAs from tamoxifen-treated EF;CreER+ (EF/Cre), EFStop;CreER− (EF) and CreER tissues were analyzed by PCR (lower panels). Lu: lung, Li: liver, B: brain, P: pancreas, S: spleen, H: heart, K: kidney. (B) Total RNAs from the kidneys of tamoxifen-treated EF;CreER+ (EF/Cre), EFStop;CreER− (EF) or CreER+ (Cre) mice were analyzed by RT-PCR. Gapdh was amplified as a control. The arrows indicate the primers used in RT-PCR. (C) mEF expression analysis. Left panel: Tamoxifen-treated EF;CreER+ kidney lysate was immunoprecipitated with α-FLI (C19) and immunoblotted with α-FLAG or α-FLI1 antibodies. Right panel: In vitro translated mEF and mEF/Stop cDNAs were immunoblotted with α-FLI1. (D) U2OS cells co-transfected with pCMV-SPORT6-mEF or -mEF/Stop along with CMV-EGFP vector, treated with cycloheximide (CHX, 10μg/ml) or CHX plus MG132 (10μM/ml) were immunobloted with α-FLI1.
To detect the expression of mEF transcripts in different tissues, we performed RT-PCR analysis (Fig. 5B). As positive controls, we used total RNAs isolated from TC71 and CHP100 cells expressing either the type I or the type II EWS/FLI1, respectively. Total RNA from JN-DSRCT-1 cells was used as a negative control. In the kidneys of tamoxifen-treated EF;CreER+ mice, mEF fusion transcripts (type II) were readily detected along with a residual amount of the larger transcripts containing the Stop (mEF/Stop) (Fig. 5B, lanes 4 and 5). mEF transcripts were also detected in other tissues of EF;CreER+ mice (Supplement Fig. 4A). In the kidneys of tamoxifen treated EFStop;CreER− mouse, only the larger mEF/Stop transcript was detected (Fig. 5B, lanes 6 and 7). No product was amplified in the Wt;CreER+ kidneys (Fig. 5B, lanes 8 and 9). To demonstrate the expression of mEF at the protein level, we performed immunoprecipitation of kidney lysates from the tamoxifen-treated EF;CreER+ mouse with the anti-FLI1 antibody followed by immunoblotting with either anti-FLAG or anti-FLI1 antibody. The results demonstrated that mEF is expressed in the kidneys of EF;CreER+ mice (Fig. 5C, left panel).
The read-through transcript containing the Stop produces an unstable protein
Despite the presence of the mEF/Stop transcripts in the EFStop;CreER− mouse (Fig. 5B, lanes 6 and 7), we could not detect a protein product generated from the read-through transcripts in the MEFs nor in the kidneys of these mice (data not shown), indicating that the protein generated from the mEF/Stop transcript might be unstable. To test this, we cloned the full-length mEF/Stop and mEF cDNAs by RT-PCR. Using an in vitro transcription-translation system, mEF/Stop produced a higher migrating protein compared to the mEF (Fig. 5C, right panel), demonstrating that a protein can be synthesized from mEF/Stop transcript. However, when transiently-transfected into U2OS cells, expression of mEF/Stop was markedly less than mEF (Supplement Fig. 4B), consistent with the notion that mEF/Stop encoded an unstable protein.
Next, we transfected mEF/Stop or mEF cDNAs together with a small amount of CMV-GFP into U2OS cells and examined the protein half-life. We used cycloheximide to block de novo protein synthesis and examined the steady-state levels of mEF/Stop and mEF. The results demonstrated that mEF/Stop encoded an unstable protein with a half-life of 30–60 min, while the half-life of mEF was more than 240 min (Fig. 5D). The addition of a proteasome inhibitor, MG132, prolonged the half-life of mEF/Stop in U2OS (Fig. 5D), suggesting that mEF/Stop might be degraded via proteasome pathway.
To determine whether a short-lived mEF/Stop might still retain its transcriptional activity, we performed a promoter-reporter assay. Cotransfection of mEF along with the CASP3 P4 promoter resulted in a 2.5-fold activation of the promoter (Supplement Fig. 4C), but mEF/Stop failed to activate the CASP3 P4 promoter. A similar 2-fold activation of the CASP3 promoter was observed when the human EWS/FLI1 type II cDNA (hEF) was used. Similar results were obtained with the c-MYC promoter (Supplement Fig. 4C), which has been shown to be regulated by EWS/FLI1 (31).
Induction of Ews/FLI1 expression in EF mouse leads to rapid death
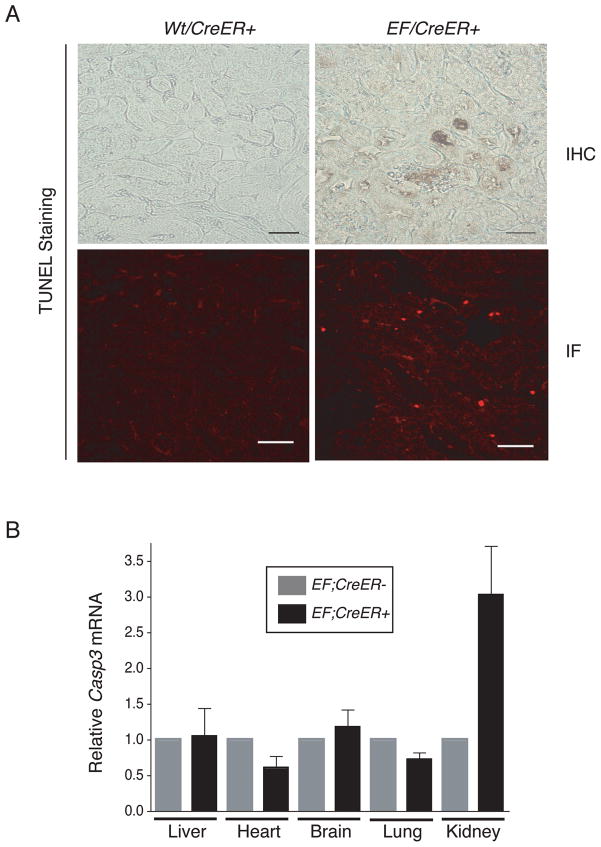
Two studies have recently shown lethal effects of EWS/FLI1 expression in the mouse (32, 33). Consistent with these reports, we observed a similar lethal effect of mEF expression in the tamoxifen injected EF;CreER+ mice. Upon tamoxifen injection, all EF;CreER+ mice quickly became moribund and died within 4–6 days. The lethality was dose-dependent as the animals injected with lower doses of taxomifen survived longer but eventually, all EF;CreER+ animals died even at the lowest dose of tamoxifen (0.25mg/40g b.w.) (data not shown). Tamoxifen had no discernible effects on EFStop;CreER− or Wt;CreER+ mice even at the highest dose (3mg/40g b.w.) up to 6 months following tamoxifen injection (data not shown). Histological analysis of tamoxifen injected EF;CreER+ tissues showed no gross abnormalities, except in the kidneys where the dying cells were readily observed in the epithelia of renal tubules as confirmed by TUNEL staining (Fig. 6A).
Figure 6.
mEF triggers apoptosis in the kidney. (A) Kidneys of tamoxifen-treated Wt/CreER+ and EF;CreER+ were examined for TUNEL assays. Bright field images (IHC) were taken under Leica DMLB microscope (scale bar: 100 μm). Immunofluorescent images (IF) were captured by confocal microscopy (scale bar: 50μm). (n=3 for each genotype). (B) Total RNAs from tissues of tamoxifen-treated EFStop;CreER− and EF;CreER+ mice were analyzed for Casp3 and Gapdh levels by qRT-PCR (TaqMan). The Casp3 levels in the EFStop;CreER− tissues were used as a control. (n=3 for each genotype).
To determine whether mEF was activating Casp3 expression in EF;CreER+ mouse, we isolated various tissues following tamoxifen treatment (3mg/40g b.w.) and examined the expression of Casp3 by qRT-PCR analysis. Of the tissues examined, only the kidneys showed 3-fold higher level of Casp3 mRNA compared to the control mice (EFStop;CreER−) (Fig. 6B).
Discussion
The apoptotic effects of EWS/FLI1 in primary cells have been well documented in the literature (15, 22). However, the molecular mechanism(s) underlying the cellular toxicity has not been fully elucidated. A previous study has shown that loss of p16INK4A, p19ARF or p53 pathway is sufficient to attenuate the toxic effects of EWS/FLI1 (22), but how these pathways are involved remains unknown. In the present study, we demonstrated that EWS/FLI1 directly activates CASP3 transcription and causes apoptosis in the MEFs and in the kidneys, which is the likely cause of death in the EF;CreER+ mouse. CASP3 is one of the central Caspases essential for the execution of apoptosis (34), and overexpression of Pro-CASP3 alone was sufficient to trigger apoptosis in primary MEFs (Supplement Fig. 5A and B). Thus, our findings suggest that the induction of CASP3 might be one of the mechanisms by which EWS/FLI1 triggers apoptosis. Interestingly, expression of EWS/FLI1 in a transgenic mouse also resulted in apoptosis in liver and spleen, and an increase in CASP3 transcripts (33). At the cellular level, the induction of CASP3 by EWS/FLI1 was also observed in human mesenchymal progenitor cells following EWS/FLI1 expression (35). Collectively, these results provide strong evidence for CASP3 as a bona-fide target of EWS/FLI1.
Examination of the expression profiling data from Khan et al (36) as well as the data from Kauer et al (37) revealed that CASP3 expression is moderately elevated in primary Ewing sarcoma (data not shown), while no evidence of elevated CASP3 expression was found in other datasets (38, 39). We have thus analyzed CASP3 expression in 9 primary Ewing sarcomas and 9 osteosarcomas (control), and the results showed moderate but consistent increase in CASP3 transcripts in Ewing sarcoma as compared to the control (Supplement Fig. 6). In addition to CASP3, expression of other genes involved in apoptosis was altered following mEF expression (Supplement Table 1), which could also contribute to apoptosis. Among them, Dapk1 and Lhx4 were highly induced by mEF. Interestingly, induction of Dapk1 was also documented following EWS/FLI1 expression in human mesenchymal stem cells (13) as well as in primary Ewing sarcoma (37–39). However, it remains to be determined whether these genes are directly regulated by EWS/FLI1. Increased expression of CASP3 and other pro-apoptotic genes by EWS/FLI1 raises an interesting hypothesis about the tumor cell of origin. Since the only cell type which tolerates EWS/FLI1 expression is the mesenchymal stem cells (13–15), this suggests that the mesenchymal stem cells have a mechasnism(s) to oppose EWS/FLI1-mediated apoptosis. Thus, identifying the anti-apoptotic mechanism(s) of the mesenchymal stem cells could have therapeutic implications for Ewing sarcoma.
Generation of EWS/FLI1 transgenic mouse has been difficult due to its toxic effects. Previously, a conditional Ews/ERG knock-in mouse was generated using an invertor approach (40), which caused T cell leukemia (41). Recently, a conditional EWS/FLI1 transgenic mouse was used to express EWS/FLI1 in the hematopoietic compartment, which resulted in a rapid onset of myeloid leukemia (33). A second transgenic mouse with EWS/FLI1 expression in the mesenchymal cells of the limbs caused limb deformities, but no tumors (32). However, with a concomitant loss of p53, EWS/FLI1 expression led to the development of poorly differentiated sarcoma (32). Thus, our conditional Ews/FLI1 knock-in mouse, which allows the expression of the fusion product from the native Ews locus, represents an important first step toward generating an animal model of Ewing sarcoma. The obvious challenge will be to circumvent the lethality of EWS/FLI1. One approach might be to use a transgenic mouse expressing CreER under a tissue-specific promoter, in which EWS/FLI1 expression is tolerated. In this regard, both human and mouse mesenchymal stem cells from the bone-marrow have been shown to not only tolerate EWS/FLI1 expression, but also form tumors in nude mice (13–15). Thus, limiting the expression of EWS/FLI1 to the mesenchymal stem cell lineage in our Ews/FLI1 knock-in mouse will be the next step towards the generation of Ewing sarcoma mouse model.
Supplementary Material
Acknowledgments
We thank Jun Nishio (Fukuoka University, Fukuoka, Japan) and Crystal Mackall (NCI, NIH) for providing the cell lines, Cuiling Li and Chuxia Deng (NIDDK Knockout Mouse Core) for the generation of Ews/FLI1 knock-in mouse, Su Young Kim (NCI, NIH) for providing primary Ewing sarcoma and osteosarcoma samples and Yun-Ping Wu (NIDDK, NIH) for the confocal microscopy.
This research was supported by the Intramural Research Program of the NIH, NIDDK (S.B.L.).
Footnotes
The authors declare no competing interests.
References
- 1.Burchill SA. Molecular abnormalities in Ewing’s sarcoma. Expert Rev Anticancer Ther. 2008;8:1675–87. doi: 10.1586/14737140.8.10.1675. [DOI] [PubMed] [Google Scholar]
- 2.Arvand A, Denny CT. Biology of EWS/ETS fusions in Ewing’s family tumors. Oncogene. 2001;20:5747–54. doi: 10.1038/sj.onc.1204598. [DOI] [PubMed] [Google Scholar]
- 3.Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–5. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 4.Janknecht R. EWS-ETS oncoproteins: the linchpins of Ewing tumors. Gene. 2005;363:1–14. doi: 10.1016/j.gene.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Kovar H. Ewing’s sarcoma and peripheral primitive neuroectodermal tumors after their genetic union. Curr Opin Oncol. 1998;10:334–42. doi: 10.1097/00001622-199807000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Ladanyi M, Gerald W. Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res. 1994;54:2837–40. [PubMed] [Google Scholar]
- 7.Panagopoulos I, Hoglund M, Mertens F, Mandahl N, Mitelman F, Aman P. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene. 1996;12:489–94. [PubMed] [Google Scholar]
- 8.Labelle Y, Zucman J, Stenman G, et al. Oncogenic conversion of a novel orphan nuclear receptor by chromosome translocation. Hum Mol Genet. 1995;4:2219–26. doi: 10.1093/hmg/4.12.2219. [DOI] [PubMed] [Google Scholar]
- 9.Zucman J, Delattre O, Desmaze C, et al. EWS and ATF-1 gene fusion induced by t(12;22) translocation in malignant melanoma of soft parts. Nat Genet. 1993;4:341–5. doi: 10.1038/ng0893-341. [DOI] [PubMed] [Google Scholar]
- 10.Lessnick SL, Braun BS, Denny CT, May WA. Multiple domains mediate transformation by the Ewing’s sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10:423–31. [PubMed] [Google Scholar]
- 11.May WA, Gishizky ML, Lessnick SL, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci U S A. 1993;90:5752–6. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.May WA, Lessnick SL, Braun BS, et al. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13:7393–8. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riggi N, Suva ML, Suva D, et al. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 2008;68:2176–85. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 14.Castillero-Trejo Y, Eliazer S, Xiang L, Richardson JA, Ilaria RL., Jr Expression of the EWS/FLI-1 oncogene in murine primary bone-derived cells Results in EWS/FLI-1-dependent, ewing sarcoma-like tumors. Cancer Res. 2005;65:8698–705. doi: 10.1158/0008-5472.CAN-05-1704. [DOI] [PubMed] [Google Scholar]
- 15.Riggi N, Cironi L, Provero P, et al. Development of Ewing’s sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res. 2005;65:11459–68. doi: 10.1158/0008-5472.CAN-05-1696. [DOI] [PubMed] [Google Scholar]
- 16.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–37. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 17.Mao X, Miesfeldt S, Yang H, Leiden JM, Thompson CB. The FLI-1 and chimeric EWS-FLI-1 oncoproteins display similar DNA binding specificities. J Biol Chem. 1994;269:18216–22. [PubMed] [Google Scholar]
- 18.Gangwal K, Sankar S, Hollenhorst PC, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc Natl Acad Sci U S A. 2008;105:10149–54. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillon N, Tirode F, Boeva V, Zynovyev A, Barillot E, Delattre O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS ONE. 2009;4:e4932. doi: 10.1371/journal.pone.0004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owen LA, Lessnick SL. Identification of target genes in their native cellular context: an analysis of EWS/FLI in Ewing’s sarcoma. Cell Cycle. 2006;5:2049–53. doi: 10.4161/cc.5.18.3213. [DOI] [PubMed] [Google Scholar]
- 21.Riggi N, Stamenkovic I. The Biology of Ewing sarcoma. Cancer Lett. 2007;254:1–10. doi: 10.1016/j.canlet.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Deneen B, Denny CT. Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene. 2001;20:6731–41. doi: 10.1038/sj.onc.1204875. [DOI] [PubMed] [Google Scholar]
- 23.Lessnick SL, Dacwag CS, Golub TR. The Ewing’s sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1:393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Watford W, Li C, et al. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Invest. 2007;117:1314–23. doi: 10.1172/JCI31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Smolen GA, Beers LF, et al. Adenosine transporter ENT4 is a direct target of EWS/WT1 translocation product and is highly expressed in desmoplastic small round cell tumor. PLoS ONE. 2008;3:e2353. doi: 10.1371/journal.pone.0002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chansky HA, Barahmand-Pour F, Mei Q, et al. Targeting of EWS/FLI-1 by RNA interference attenuates the tumor phenotype of Ewing’s sarcoma cells in vitro. J Orthop Res. 2004;22:910–7. doi: 10.1016/j.orthres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci U S A. 1995;92:6991–5. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishio J, Iwasaki H, Ishiguro M, et al. Establishment and characterization of a novel human desmoplastic small round cell tumor cell line, JN-DSRCT-1. Lab Invest. 2002;82:1175–82. doi: 10.1097/01.lab.0000028059.92642.03. [DOI] [PubMed] [Google Scholar]
- 30.Kikuchi R, Murakami M, Sobue S, et al. Ewing’s sarcoma fusion protein, EWS/Fli-1 and Fli-1 protein induce PLD2 but not PLD1 gene expression by binding to an ETS domain of 5′ promoter. Oncogene. 2007;26:1802–10. doi: 10.1038/sj.onc.1209973. [DOI] [PubMed] [Google Scholar]
- 31.Dauphinot L, De Oliveira C, Melot T, et al. Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2and c-Myc expression. Oncogene. 2001;20:3258–65. doi: 10.1038/sj.onc.1204437. [DOI] [PubMed] [Google Scholar]
- 32.Lin PP, Pandey MK, Jin F, et al. EWS-FLI1 induces developmental abnormalities and accelerates sarcoma formation in a transgenic mouse model. Cancer Res. 2008;68:8968–75. doi: 10.1158/0008-5472.CAN-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torchia EC, Boyd K, Rehg JE, Qu C, Baker SJ. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol Cell Biol. 2007;27:7918–34. doi: 10.1128/MCB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degterev A, Yuan J. Expansion and evolution of cell death programmes. Nat Rev Mol Cell Biol. 2008;9:378–90. doi: 10.1038/nrm2393. [DOI] [PubMed] [Google Scholar]
- 35.Miyagawa Y, Okita H, Nakaijima H, et al. Inducible expression of chimeric EWS/ETS proteins confers Ewing’s family tumor-like phenotypes to human mesenchymal progenitor cells. Mol Cell Biol. 2008;28:2125–37. doi: 10.1128/MCB.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan J, Wei JS, Ringner M, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–9. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kauer M, Ban J, Kofler R, et al. A molecular function map of Ewing’s sarcoma. PLoS One. 2009;4:e5415. doi: 10.1371/journal.pone.0005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baird K, Davis S, Antonescu CR, et al. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res. 2005;65:9226–35. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- 39.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–9. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 40.Forster A, Pannell R, Drynan L, et al. Chromosomal translocation engineering to recapitulate primary events of human cancer. Cold Spring Harb Symp Quant Biol. 2005;70:275–82. doi: 10.1101/sqb.2005.70.008. [DOI] [PubMed] [Google Scholar]
- 41.Codrington R, Pannell R, Forster A, et al. The Ews-ERG fusion protein can initiate neoplasia from lineage-committed haematopoietic cells. PLoS Biol. 2005;3:e242. doi: 10.1371/journal.pbio.0030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.