Abstract
According to the “thermodynamic hypothesis”, the sequence of a biological macromolecule defines its folded, active structure as a global energy minimum on the folding landscape.1,2 But the enormous complexity of folding landscapes of large macromolecules raises a question: Is there indeed a unique global energy minimum corresponding to a unique native conformation, or are there deep local minima corresponding to alternative active conformations?3 Folding of many proteins is well described by two-state models, leading to highly simplified representations of protein folding landscapes with a single native conformation.4,5 Nevertheless, accumulating experimental evidence suggests a more complex topology of folding landscapes with multiple active conformations that can take seconds or longer to interconvert.6,7,8 Here we employ single molecule experiments to demonstrate that an RNA enzyme folds into multiple distinct native states that interconvert much slower than the time scale of catalysis. These data demonstrate that the severe ruggedness of RNA folding landscapes extends into conformational space occupied by native conformations.
Biopolymers face a challenge of folding a linear chain into complex three-dimensional conformations that have specific biological activities. According to the “thermodynamic hypothesis”, sequences of biological macromolecules have evolved to specify the active, or “native” conformation as a minimum in free energy, ensuring that native states are more stable than the sum of all possible inactive conformations.1 In the language of folding landscapes, the native conformation is the global energy minimum, separated from an ensemble of inactive conformations by a large energy gap.2–4 However, the enormous complexity of the conformational space of a typical macromolecule – at least 1030 conformations for a small protein containing 100 amino acids9 – raises the question of whether such energy minima are unique or whether there are multiple local energy minima that correspond to alternative active conformations. Observations of hysteresis in enzyme kinetics and of kinetic complexities in protein folding, unfolding and ligand binding provided early evidence that some protein enzymes can exist in several catalytically active forms.6,10, In the last decade, single molecule experiments have provided additional evidence for multiple active conformers that slowly interconvert.11,12,13
Functional RNAs represent a chemically distinct class of biopolymers that face a folding problem analogous to that of proteins. RNA folding landscapes have long been considered as more rugged than protein landscapes,14,15,16 due to the limited information content of RNA primary structure and the high stability of RNA secondary structures. Observations of very slow time scales for folding and the occurrence of multiple, long-lived intermediates appear to support these views. 15,17 Does ruggedness extend into the native state region, resulting in multiple active conformations? Experiments with a small catalytically active RNA –the hairpin ribozyme– revealed several distinct active forms of these molecules.18,19 While these results are suggestive of landscape ruggedness, there is insufficient evidence to demonstrate that distinct forms are interconverting conformations, and the results leave open the possibility of covalent differences between the molecules that exhibited different behavior.
We have used single molecule FRET experiments to test, whether the Tetrahymena group I ribozyme, a large, efficient RNA catalyst, folds into a unique native conformations or into multiple native conformations. Our results provide strong evidence for multiple conformations of the native state that interconvert but are separated by large energetic barriers.
The Tetrahymena group I ribozyme is a ~400 nucleotide RNA enzyme that folds in the presence of Mg2+ into an active form that catalyzes cleavage of an oligonucleotide substrate by an exogenous guanosine nucleophile. Binding of the oligonucleotide substrate to the folded ribozyme occurs in two steps (Fig. 1): in the first step, the substrate binds to a single-stranded 5′-end region forming the P1 duplex. In the second step, the P1 duplex docks into tertiary interactions in the active site.20, 21
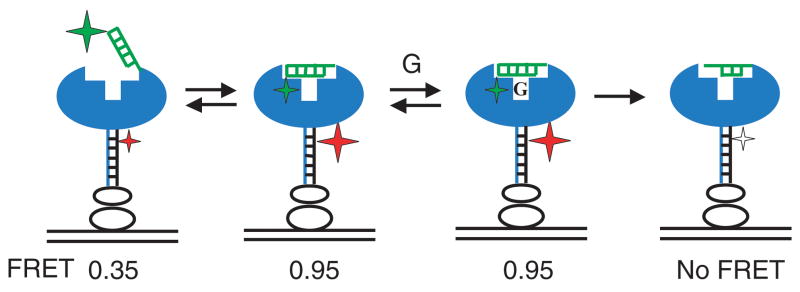
Figure 1.
Schematic representation of docking and cleavage of the oligonucleotide substrate by the Tetrahymena ribozyme observed by single molecule FRET. The ribozyme is in blue, the P1 duplex is in green, and a DNA tether base-paired to a 3′-end extension of the ribozyme is in black. Streptavidin and biotinyilated bovine serum albumin used for surface attachment of the construct are shown as clear ovals. The substrate is labelled with the FRET donor (Cy3, green star), and the DNA tether is labelled with the FRET acceptor (Cy5, red & clear star). Donor fluorescence is excited with the 532 nm laser. FRET fluctuates between the levels of 0.3 in the open complex and 0.95 in the closed complex. After guanosine (G) binds, the substrate is cleaved and the Cy3-labelled 3′-end rapidly dissociates, leading to loss of Cy5 fluorescence, as indicated by the clear star, and the absence of FRET.27
We tested whether the ribozyme is folded into a single native conformation by measuring the thermodynamics and kinetics of docking for individual molecules by single molecule FRET. If all of the molecules were folded to a single native conformation, the docking behavior for each single molecule would be identical to the ensemble average behavior. However, in contrast to this simplest expectations, individual ribozyme molecules from a single sample that were folded together and observed side-by-side under identical conditions, displayed a broad distribution of docking behaviors (Fig. 2). For example, docking equilibrium constants for molecules A through D in Figure 2 vary by 300-fold.
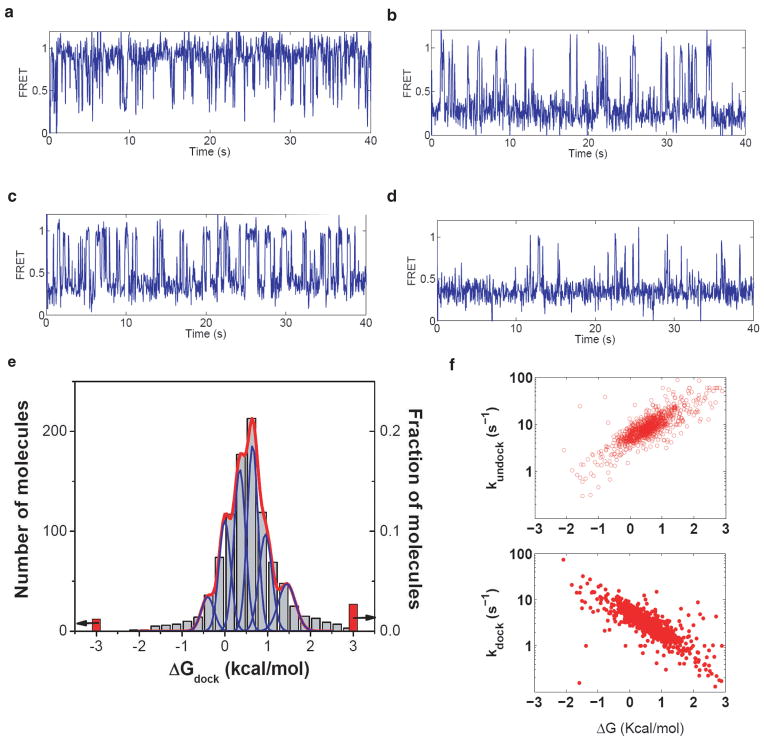
Figure 2.
Distribution of docking behaviors of individual ribozyme molecules. a–d, Representative examples of FRET traces, which give docking equilibria of Kdock = 8, 0.5, 0.08, and 0.03, respectively, corresponding to ΔGdock values of −1.4, 0.4, 1.4 and 2.0 kcal/mol. e, The distribution of docking behaviors of individual ribozyme molecules (grey bars). Red bars correspond to molecules that displayed no docking or no undocking transitions, for which only the limits of ΔGdock could be determined (as indicated by arrows). Blue peaks show the expected distribution widths for individual docking conformations (see Supplementary information). The red line shows the best fit of the distribution to a sum of distributions from six docking conformations. f, Docking (kdock) and undocking (kundock) rate constants of individual molecules. Experiments were performed in a standard buffer containing 10 mM MgCl2 at 22 °C. The substrate oligonucleotide was CCCUC(2′-methoxyU)AAACC-Cy3.
The accuracy of measuring docking equilibria from single molecule FRET traces is limited by the finite length of each trace caused by photobleaching. Therefore, a distribution of docking behaviors is expected even if all of the molecules were in the same conformation. Quantitative modelling of this effect (Supplementary Fig. S1, Fig. S2) demonstrated that the finite lengths can account for only a small fraction of the width of the observed distribution. More than six conformations with different docking thermodynamics are needed to account for this distribution (Fig. 2e and Supplementary Fig. S3). Docking and undocking rate constants determined for individual molecules also vary across a broad distribution (Fig. 2f) without discernable clustering into sub-groups that might indicate a small number of distinct conformations.
A simple model that could account for the multiple docking states suggests that some of the observed molecules represent only partially folded forms of the ribozyme that are capable of docking but have not reached the native, active state. To test this possibility we determined the catalytic activity of molecules across the broad range of docking behaviors. We first measured the docking equilibrium for each molecule, and then induced cleavage by adding guanosine to allow chemical catalysis to occur (Fig. 1). The number of molecules that have the substrate uncleaved decayed mono-exponentially with an endpoint of 6%, indicating that 94% of the molecules are catalytically active and only 6% are inactive or have much lower activity. Further, nearly all of the molecules throughout the distribution of docking conformations are catalytically active (Supplementary Table 1).
We next determined whether molecules in different docking conformations, as assessed by their distinct docking equilibria, exhibited the same or distinct cleavage kinetics. We binned molecules with similar docking behaviors (Fig. 3a,) and analysed cleavage kinetics within each bin (Fig. 3b & Supplementary Table 1). Both the cleavage rate constant kcleav and the endpoints were similar despite the >800-fold range docking equilibria. Thus, molecules that form docked states with the same catalytic activity exhibit distinct docking behaviors, providing direct evidence for a heterogeneous native state. Additional experiments suggest that the heterogeneity arises from different molecules forming different sub-sets of the tertiary docking interactions with the functional groups on the P1 duplex (Fig. S4).
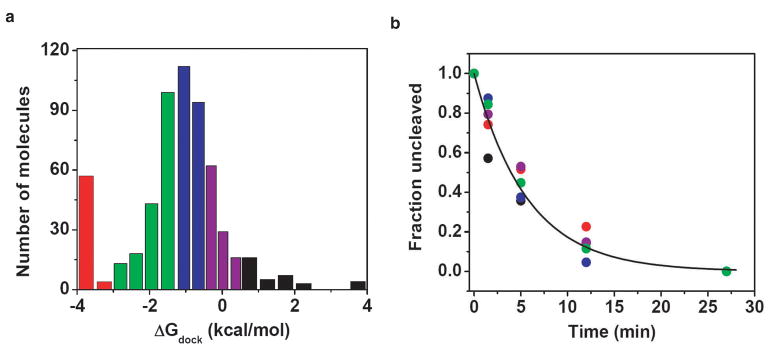
Figure 3.
Catalytic activity of molecules from different parts of docking distribution is the same. a, Distribution of docking behaviors was split into five bins marked by different colors. Cleavage experiments were performed with the CCCUC(2′-deoxyU)A-Cy3 substrate oligonucleotide that docks more stably than the CCCUC(2′-methoxyU)AAACC-Cy3 substrate in Fig. 2. b, Cleavage kinetics within each bin were the same as the global average (line indicates global fit), kinetic parameters for the global fit and fits individual bins are given in Supplementary Table 1.
Multiple native conformations of the ribozyme observed in our experiments could represent multiple distinct folds of the same sequence, as implicitly assumed above. Alternatively, they could instead represent similar folded states of RNA molecules that have different sequences because of errors in synthesis, unintended chemical modifications or degradation. Covalent heterogeneity of RNA has been considered as a possible origin of multiple conformations of the hairpin ribozyme, although no direct evidence for or against this proposal has been accumulated, and it may be difficult to decisively rule out covalent heterogeneity by assays such as mass-spectrometry because modifications can be mass-neutral and the formation of multiple modified species each in low-abundance can be difficult to detect.19,22
Stable covalent versus interconvertible conformational differences can be distinguished if interconversion between different conformations can be observed under some conditions. If each detected ‘conformation’, or behavior, can interconvert to other conformations (behaviors), conformational heterogeneity is strongly suggested, whereas the absence of extensive interconversion under any condition leaves open the possibilities of covalent heterogeneity or extremely stable and long-lived RNA species.23,24
We determined if molecules can change their docking behavior by comparing docking of individual molecules before and after unfolding under mild denaturing conditions. Removing Mg2+ by EDTA disrupts tertiary structure of the ribozyme but leaves most of the secondary structure intact and allows the molecules to remain tethered to the surface. As illustrated in Fig. 4a, if all molecules quickly interconvert in the unfolded state, each molecule could then re-fold to any of the docking conformations, independent of which conformation it populated before unfolding. We grouped molecules into three non-overlapping sub-populations based on their docking equilibrium constant before unfolding (Fig. 4b) and compared the distributions of conformations that each molecule populated after refolding. As observed in Fig. 4c, molecules from each of the three original sub-populations displayed docking behaviors across the entire distribution. These observations indicate that in the unfolded state all or nearly all of the conformational states can interconvert to any other conformation. Incomplete overlap of the distributions suggests that either some interconversions are slow on the minute timescale even in the unfolded state or that covalent differences are present in a small subset of the molecules.
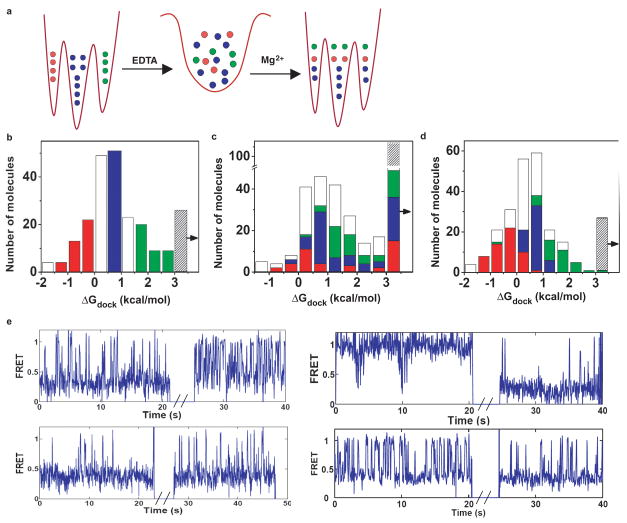
Figure 4.
Interconversion of docking behaviors. a, Scheme illustrating changes in docking following fast interconversion in the unfolded state. If interconversions were very slow, each molecule would display the same docking behavior before and after unfolding. b, Distribution of ΔGdock before unfolding. Red, blue and green bars indicate three selected “conformations”. Clear bars indicate molecules not included in the analysis to ensure nonoverlap of selected “conformations”. c, Distribution of ΔGdock after refolding. The rightmost bin marked by the arrow indicates misfolded molecules (Supplementary information). 26,29 Colored bars indicate the number of molecules in the correspondingly colored bins before unfolding (in Fig. 4b). d, Distribution of ΔGdock after 40 min in the folded state. Colored bars indicate the number of molecules in the correspondingly colored bins at the zero timepoint. e, Example traces showing interconversions after refolding (see also Supplementary Fig. 7). Breaks in FRET traces correspond to the time unfolded. ΔGdock before and after refolding for molecules 1–4 were 1.2 → 0.1, −1.8 → 2.1, 2.0 → 1.2, and 1.5 → 0.4 kcal/mol, and the p-values indicating the significance level for interconversion were 0.01, 10−15, 0.1, and 0.1, respectively.
The original observation of multiple docking behaviors of active molecules (Fig. 2 and Fig. 3) indicated that in the folded state the interconversions must be slow relative to the minute timescale of the observations. Are interconversions in the folded state slower than in the unfolded? Or, in other words, does the energy landscape become more “rugged” going from the unfolded state towards the native state? To address this question we evaluated the extent of interconversion in the folded state. The data in Fig. 4d show that there was little interconversion between docking behaviors even after 40 minutes – i.e., most of the molecules ‘remembered’ their initial docking equilibrium. Nevertheless, some of the molecules changed their docking behavior during this time period (9% of all molecules; Fig. 4d & Supplemental information). A simple explanation that accounts for much smaller extent of interconversion in the folded state than in the unfolded state is that the energy barriers separating different conformations are higher in the folded state so that the rate of interconversion is lower (~ 100 fold less). The data on interconversion in different conditions suggest a view of the ribozyme folding landscape that is moderately rugged in the region occupied by unfolded conformations and becomes more rugged towards the region occupied by the native conformations.
Multiple active conformations of the Tetrahymena ribozyme offer a glimpse of the full complexity of the folding landscape of a functional RNA. Taken together with the prior evidence for proteins, the results presented herein suggest that the complexity of native folded state may be an inherent property of all biopolymers. These observations prompt us to revisit the Levinthal paradox in a different context. Levinthal noted that proteins cannot sample all of conformational space on a biological timescale, leading to the notion that there must be a preferred pathway to the native state.25 More recent work has emphasized the likelihood of multiple pathways to the folded state of proteins and RNAs. Multiple pathways, combined with limitations to full sampling may lead to multiple ‘endpoints’ or multiple native state local minima. This scenario, while counter to traditional viewpoints, has some support in the protein folding literature and is strongly supported for the Tetrahymena group I RNA by this and prior26 folding studies. We further speculate that perhaps folded RNAs or proteins in different conformations may respond to cellular or environmental cues in different ways and contribute to stochastic diversity.
Methods Summary
The L-16 form of the Tetrahymena group I ribozyme, extended at the 3′-end by a 26 nucleotide T2 tail, was synthesized by in vitro transcription and purified by denaturing PAGE. Oligonucleotides were obtained commercially (IDT and Dharmacon), fluorescently labelled, and purified by denaturing PAGE. The DNA tether was bound to the ribozyme by annealing (95 °C heating for 3 min, cooling to 50 °C at 0.1 °C/s), followed by folding in 10 mM was MgCl2 (30 min at 50 °C).
The oligonucleotide substrates were bound to the tether-bound, pre-folded ribozyme at 1 μM concentration (10 min at 22 °C). The ribozyme was then diluted to 30–100 pM final concentration for deposition on BSA-biotin/streptavidin coated slides, as described.27 Single molecule FRET experiments were carried out on a home-built prism-based total internal reflection microscope, similar to one previously described.28 Data collection and analysis were performed using home-written programs in C++ and Matlab (Mathworks Co.). Details of the methods, including sample preparation, data collection and analysis, and numerical simulations, are presented as the online Methods.
Supplementary Material
Acknowledgments
We thank T.H. Lee, B. Cui, H. Kim, W. Zhao and other current and former members of the Chu lab, and the Mabuchi lab, for technical assistance. We thank members of the Herschlag lab for discussions and comments on the manuscript. Financial support for this work was provided by an NIH program project grant P01-GM-66275 and an NIH grant GM49243 to D.H. We thank Stanford Bio-X Foundation for the fellowship support to S.V.S.
Footnotes
Supplementary information is linked to the on-line version of the paper at www.nature.com/nature
Author Contributions All authors contributed to the experimental design and writing of the manuscript; S.V.S. performed the experiments; M.G. and S.V.S. carried out data analysis.
Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
dU stands for 2′-deoxyuridine
mU stands for 2′-methoxyuridine
References
- 1.Anfinsen CB. Principles that Govern the Folding of Protein Chains. Science. 1973;181:223. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways and the energy landscape of protein folding: a synthesis. Protiens Struct Funct Genet. 1995;21:167. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 3.James LC, Tawfik DS. Conformational diversity and protein evolution - a 60-year-old hypothesis revisited. Trends Biochem Sci. 2003;28:361. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 4.Zwanzig R. Two-state models of protein folding kinetics. Proc Natl Acad Sci U S A. 1997;94:148. doi: 10.1073/pnas.94.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 6.Schmid FX, Blaschek H. A Native-Like Intermediate on the Ribonuclease A Folding Pathway. Eur J Biochem. 1981;114:111. doi: 10.1111/j.1432-1033.1981.tb06180.x. [DOI] [PubMed] [Google Scholar]
- 7.Jennings PA, Finn BE, Jones BE, Matthews CR. A reexamination of the folding mechanism of dihydrofolate reductase from Escherichia coli: Verification and refinement of a four-channel model. Biochemistry. 1993;32:3783. doi: 10.1021/bi00065a034. [DOI] [PubMed] [Google Scholar]
- 8.Kamagata K, Sawano Y, Tanokura M, Kuwajima K. Multiple Parallel-pathway Folding of Proline-free Staphylococcal Nuclease. Journal of Molecular Biology. 2003;332:1143. doi: 10.1016/j.jmb.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Dinner AR, Sali A, Smith LJ, Dobson CM, Karplus M. Understanding protein folding via free-energy surfaces from theory and experiment. Trends Biochem Sci. 2000;25:331. doi: 10.1016/s0968-0004(00)01610-8. [DOI] [PubMed] [Google Scholar]
- 10.Frieden C. Slow Transitions and Hysteretic Behavior in Enzymes. Ann Rev Biochem. 1979;48:471. doi: 10.1146/annurev.bi.48.070179.002351. [DOI] [PubMed] [Google Scholar]
- 11.Flomenbom O, et al. Stretched exponential decay and correlations in the catalytic activity of fluctuating single lipase molecules. Proc Natl Acad Sci U S A. 2005;102:2368. doi: 10.1073/pnas.0409039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu HP, Xun L, Xie XS. Single-Molecule Enzymatic Dynamics. Science. 1998;282:1877. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 13.English BP, et al. Ever-fluctuating single enzyme molecules: Michaelis-Menten equation revisited. Nat Chem Biol. 2006;2:87. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 14.Herschlag D. RNA Chaperones and the RNA Folding Problem. J Biol Chem. 1995;270:20871. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 15.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Current Opinion in Structural Biology. 1999;9:339. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 16.Chen SJ, Dill KA. RNA folding energy landscapes. Proc Natl Acad Sci U S A. 2000;97:646. doi: 10.1073/pnas.97.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan J, Thirumalai D, Woodson SA. Folding of RNA involves parallel pathways. J Mol Biol. 1997;273:7. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 18.Zhuang X, et al. Correlating structural dynamics and function in single ribozyme molecules. Science (Washington, D C, 1883-) 2002;296:1473. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 19.Tan E, et al. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc Natl Acad Sci U S A. 2003;100:9308. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herschlag D. Evidence for processivity and two-step binding of the RNA substrate from studies of J1/2 mutants of the Tetrahymena ribozyme. Biochemistry. 1992;31:1386. doi: 10.1021/bi00120a015. [DOI] [PubMed] [Google Scholar]
- 21.Bevilacqua PC, Kierzek R, Johnson KA, Turner DH. Dynamics of ribozyme binding of substrate revealed by fluorescence-detected stopped-flow methods. Science. 1992;258:1355. doi: 10.1126/science.1455230. [DOI] [PubMed] [Google Scholar]
- 22.Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG. A rugged free energy landscape separates multiple functional RNA folds throughout denaturation. Nucleic Acids Res. 2008;36:7088. doi: 10.1093/nar/gkn871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindahl T, Adams A, Fresco JR. Renaturation of transfer ribonucleic acids through site binding of magnesium. Proc Natl Acad Sci U S A. 1966;55:941. doi: 10.1073/pnas.55.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korennykh AV, Plantinga MJ, Correll CC, Piccirilli JA. Linkage between Substrate Recognition and Catalysis during Cleavage of Sarcin/Ricin Loop RNA by Restrictocin. Biochemistry. 2007;46:12744. doi: 10.1021/bi700931y. [DOI] [PubMed] [Google Scholar]
- 25.Levinthal C. Are there pathways for protein folding? J Chim Phys. 1968;65:44. [Google Scholar]
- 26.Russell R, et al. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci U S A. 2002;99:155. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuang X, et al. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 28.Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. Direct measurement of tertiary contact cooperativity in RNA folding. J Am Chem Soc. 2008;130:6085. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway. J Mol Biol. 2001;308:839. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 30.Narlikar GJ, Bartley LE, Khosla M, Herschlag D. Characterization of a local folding event of the Tetrahymena group I ribozyme: effects of oligonucleotide substrate length, pH, and temperature on the two substrate binding steps. Biochemistry. 1999;38:14192. doi: 10.1021/bi9914309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.