Abstract
Transcription stimulates the genetic instability of trinucleotide repeat sequences. However, the mechanisms leading to transcription-dependent repeat length variation are unclear. We demonstrate, using biochemical and genetic approaches, that the formation of stable RNA·DNA hybrids enhances the instability of CTG·CAG repeat tracts. In vitro transcribed CG-rich repeating sequences, unlike AT-rich repeats and nonrepeating sequences, form stable, ribonuclease A-resistant structures. These RNA·DNA hybrids are eliminated by ribonuclease H treatment. Mutation in the rnhA1 gene that decreases the activity of ribonuclease HI stimulates the instability of CTG·CAG repeats in E. coli. Importantly, the effect of ribonuclease HI depletion on repeat instability requires active transcription. We also showed that transcription-dependent CTG·CAG repeat instability in human cells is stimulated by siRNA knockdown of RNase H1 and H2. In addition, we used bisulfite modification, which detects single-stranded DNA, to demonstrate that the nontemplate DNA strand at transcribed CTG·CAG repeats remains partially single-stranded in human genomic DNA, thus indicating that it is displaced by an RNA·DNA hybrid. These studies demonstrate that persistent hybrids between the nascent RNA transcript and the template DNA strand at CTG·CAG tracts promote instability of DNA trinucleotide repeats.
Keywords: RNA·DNA hybrids, transcription-induced instability, triplet repeats
Expansions of simple repeating sequences (microsatellites) are responsible for more than 20 human diseases (1). Moreover, instability of simple repeats (expansion as well as contraction) is observed throughout the genomes of all organisms studied, and it is considered an important source of genetic variation (2). The molecular basis of this unusual mutation mechanism has been studied extensively in the past 15 years using bacteria, yeast, flies, mice, and mammalian cells. Although these analyses uncovered several cis elements and trans-acting factors affecting repeat instability (3), a unifying, comprehensive model of the repeat expansion and contraction is lacking. Processes such as replication, recombination, and repair, which temporarily dissociate DNA complementary strands, strongly destabilize repeating sequences. Transient exposure of single-stranded DNA regions allows formation of non-B DNA structures in regions containing repeats. Virtually all current models of repeat instability incorporate non-B DNA structures as the proximate cause of instability (4, 5).
Recently, transcription through tandem repeat sequences has emerged as an important factor promoting instability of repeat sequences (6–12). Although the synthesis of RNA does not change the length of a DNA template, it can lead to the formation of non-B DNA structures, as shown in E. coli, yeast, and higher organisms (7–10, 13). These secondary structures may interfere with the progress of RNA polymerase, calling into play various DNA repair processes, whose action leads to repeat expansion and contraction. In human cells, transcription-induced repeat instability has been shown to involve mismatched repair components, transcription-coupled nucleotide excision repair, proteins that deal with stalled RNA polymerase complexes, and the proteasome (8, 9, 11).
The dependence of downstream events on non-B DNA secondary structures raises the question of how such unusual conformations are formed. Is the transient exposure of single strands by a passing polymerase sufficient for non-B DNA structure formation, or does the nascent RNA itself play a critical role? Extended stretches of RNA·DNA hybrids (so-called R loops) tend to form in GC-rich DNA because of the exceptional stability of rG·C base pairs (14–16). The high GC-content of most disease-causing repeat sequences can facilitate formation of R loops. RNA·DNA hybrids would likely extend the lifetime of single-stranded repeat DNA, thereby permitting increased formation of non-B DNA secondary structures and promoting repeat instability.
Cotranscriptional formation of RNA·DNA hybrids was first detected in prokaryotic cells (17). In yeast, formation of transcription-dependent R loops resulted in hyperrecombination and defects in transcription elongation, which were suppressed by overexpression of RNase H (18). In chicken DT40 cells, R loops accumulated after depletion of the ASF/SF2 splicing factor, leading to genomic instability (19). The R loops have also been identified in GC-rich, highly repetitive immunoglobulin class switch recombination sequences in mammalian cells (15, 20). More recently, RNA·DNA hybrids were detected in the GAA·TTC repeat region of a plasmid during transcription in bacteria (21). These examples suggest that RNA·DNA hybrids will ultimately be found to participate in many biological phenomena.
In this study, we demonstrate that stable RNA·DNA hybrids form at CTG·CAG repeat tracts in vitro and in vivo. We also show that these RNase H-sensitive structures stimulate CTG·CAG repeat instability in E. coli and in human cells. Hence, these studies provide evidence that transcription-induced RNA·DNA hybrids can cause genetic instability of DNA repeat sequences.
Results
In Vitro Formation of RNA·DNA Hybrids in Transcribed CTG·CAG Repeats.
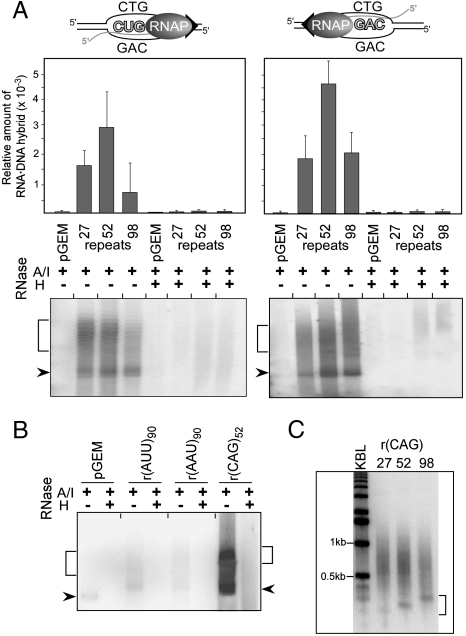
To determine whether transcription through a CTG·CAG repeat can generate RNA·DNA hybrids, we incubated supercoiled plasmids containing 0, 27, 52, or 98 CTG·CAG repeats with RNA polymerase. We used T7 RNA polymerase to transcribe the CAG template strand to generate potential rCUG·CAG hybrids, and SP6 RNA polymerase to transcribe the CTG template to generate potential rCAG·CTG hybrids. In all cases, the transcripts were tagged by including a radioactively labeled nucleotide in the reaction mixture. We used RNase A and RNase I (RNase A/I) treatment, which digests free RNA, to reveal putative hybrid RNA that remained associated with the plasmid DNA upon gel electrophoresis (Fig. 1A). RNase A/I-resistant RNA was demonstrated to be involved in RNA·DNA hybrids, because it was sensitive to RNase H, which cleaves RNA paired with DNA (Fig. 1A).
Fig. 1.
Stable RNA·DNA hybrids induced by in vitro transcription of CG-rich trinucleotide repeats. (A) RNA·DNA hybrids in transcribed CTG·CAG repeat tracts. Transcription reactions were treated with RNase A/I alone or in combination with RNase H prior to agarose electrophoresis. Plasmids containing 0, 27, 52, or 98 CTG·CAG repeats were all derivatives of pGEM3Zf(+). Arrowheads indicate migration of the supercoiled DNA; brackets indicate the position of relaxed plasmids. Plasmid-associated RNAs were quantified by scintillation counting and the efficiency of hybrid formation is expressed relative to the total amount of RNA prior to the RNase A/I/H treatment. (B) Lack of RNA·DNA hybrids in transcribed AT-rich (AAT·ATT)90 repeats. (C) Length of RNA·DNA hybrids formed at transcribed CTG·CAG repeat tracts of different lengths. Plasmid-associated RNAs were extracted from agarose gels, incubated with DNaseI and analyzed on a 1.2% agarose gel in denaturing conditions; KBL designates a 1 kb DNA ladder. The lengths of the shortest RNAs involved in RNA·DNA interactions, indicated by the bracket, correlate with the length of the CTG·CAG tracts.
Quantification of the hybrid RNA showed that it was specifically associated with templates that contained CTG·CAG repeat tracts. Only background levels of RNA·DNA hybrids were found in the control plasmid, which lacks a CTG·CAG tract (Fig. 1A). In the case of (CTG·CAG)52 and (CTG·CAG)98, approximately 2-fold more hybrid RNA was produced with SP6 RNA polymerase than with T7 RNA polymerase, likely due to the higher efficiency of formation or greater stability of rCAG·CTG hybrids relative to rCUG·CAG hybrids. Interestingly, although formation of hybrid RNA required CTG·CAG repeat tracts, the amount of hybrid RNA showed no significant dependence on the length of the repeat tract within the tested range. However, formation of hybrid RNA depended on GC content. Hybrid RNA was not detected when plasmids harboring 90 AAT·ATT repeats were transcribed with either T7 or SP6 RNA polymerase (Fig. 1B), but hybrids were detected for plasmids with 200 CCTG·CAGG repeats (Fig. S1A).
To analyze the effect of sequences capable of adopting non-B DNA structures on the formation of persistent RNA·DNA hybrids, we conducted in vitro transcription reactions using the (CTA·TAG)68 sequence (Table S1). These repeats have little or no capacity to form non-B DNA structures and are genetically stable (22). The r(GUA)68 sequences formed RNase H-resistant RNA·DNA interactions with an efficiency 5 times lower than r(CUG)52 repeats (Fig. S1). The complementary sequence r(CUA)68 did not form stable RNA·DNA hybrids in vitro (Fig. S1). These data strongly suggest that both the G content of the transcript and the propensity to adopt non-B DNA structures by unpaired nontemplate DNA strands influenced the formation of R loops at repeating sequences.
To determine the length of the RNA component of the RNA·DNA hybrids, RNase A/I-resistant hybrids formed during transcription of the plasmids carrying 27, 52, and 98 CTG·CAG repeats were extracted from the agarose gels. Next, we separated the radioactively labeled RNA from the DNA template using DNase I digestion and agarose gel electrophoresis under denaturing conditions. As shown in Fig. 1C, the length of the shortest RNAs involved in RNA·DNA interactions correlates with the length of the CTG·CAG tracts, suggesting that repeating sequence may serve as the R-loop initiation zone. Additionally, these data indicate that RNase H-resistant RNA·DNA interactions can extend beyond the repeats into the flanking sequences.
Reduced Activity of RNase HI Stimulates Transcription-Dependent Instability in E. coli.
One strategy for demonstrating that downstream effects are caused by stable RNA·DNA hybrids is to change the expression or activity of RNase H (19–21). In E. coli, RNase H activity is encoded by two genes: rnha (RNase HI) and rnhb (RNase HII) (23). The activity of the RNase HII is severalfold lower than RNAse HI, and its effects on RNA·DNA hybrids are minimal (24). Additionally, prior studies on R-loop formation in E. coli revealed that active RNase HII alone does not efficiently prevent formation of RNA·DNA hybrids (25). Therefore, we focused our studies on RNase HI.
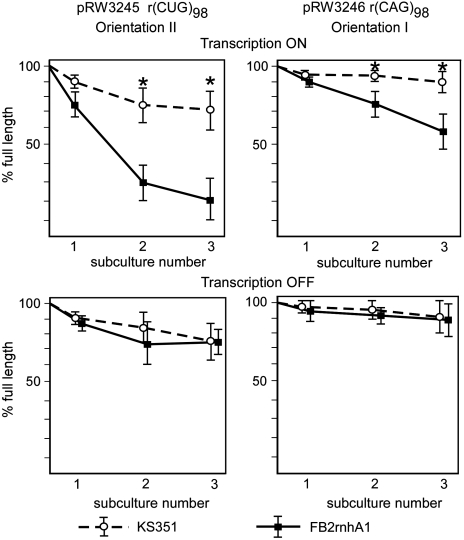
To determine whether RNA·DNA hybrids influence CTG·CAG repeat instability, we used two isogenic strains of E. coli: the parental strain, KS351, and a derivative, FB2, which harbors the rnhA1 mutation that decreases the activity of RNase HI by 70% (26). We introduced CTG·CAG repeats into these strains by transformation with plasmids pRW3245 and pRW3246, each of which harbors a tract of 98 CTG·CAG repeats (Table S1). These plasmids differ in the orientation of the CTG·CAG repeat tracts relative to the origin of replication. In orientation I, the CTG repeats are located on the template strand for leading strand synthesis; in orientation II, the CTG repeats are on the lagging strand template (27). In both cases, the lacZ promoter controls transcription through the repeats, so that transcription can be stimulated by addition of IPTG or inhibited by expression of an exogenous lacIq repressor. The pRW3245 and pRW3246 serve as templates for rCUG and rCAG transcripts, respectively. These bacterial strains and plasmids allow us to measure repeat instability in the presence and absence of transcription in cells with normal or deficient RNase HI activity.
We introduced plasmids into cells and subcultured them through three rounds of growth (≈60 generations). Repeat tracts were then excised from the isolated plasmids and analyzed by polyacrylamide gel electrophoresis. In the cells with active transcription through the CTG·CAG tract, the amount of full-length repeat decreased in the RNase HI-deficient strain, generating deleted CTG·CAG tracts. These results confirm that CTG·CAG repeats are substantially more unstable in the RNase HI-deficient strain than in the parental strain (Fig. 2 Upper). Repeat deletions were especially apparent in orientation II, where only about 20% of full-length (CTG·CAG)98 remained after the third subcultivation (Fig. 2). By contrast, episomal overexpression of RNase HI reduced the frequency of both deletions and expansions of the (CTG·CAG)98 tract (Fig. S2).
Fig. 2.
Transcription-induced CTG·CAG repeat deletions in RNase HI-deficient E. coli. Quantitative analysis of repeat instability in the presence (Upper) and absence (Lower) of transcription in RNase HI-proficient and -deficient strains of E. coli. Transcription through the CTG·CAG repeats in pRW3246 and pRW3245 in the parental (KS351) and RNase HI-deficient (FB2) strains was induced by 1 mM IPTG; transcription was blocked by expression of the lacIq repressor (7). The amount of full-length CTG·CAG inserts relative to the contraction products was quantified after each of three subcultivations by phorophorimager. Error bars represent standard deviations, and the asterisks indicate statistically significant differences (p < 0.05) between the two strains.
RNase HI could potentially influence repeat instability by affecting initiation of ColE1 plasmid replication. To determine whether RNase HI influences CTG·CAG repeat deletions during replication or during transcription, we inhibited transcription from the lacZ promoter, thus separating these two functions of RNase HI. As shown in Fig. 2 Lower, when transcription was shut off, the repeat tract was substantially more stable and there was no significant difference between the parental strain and the RNase HI-deficient strain.
Together, these results demonstrate that transcription-induced RNA·DNA hybrids stimulate the genetic instability of CTG·CAG repeats in vivo.
RNase H1 and H2 Knockdowns Destabilize CTG·CAG Tracts in Human Cells.
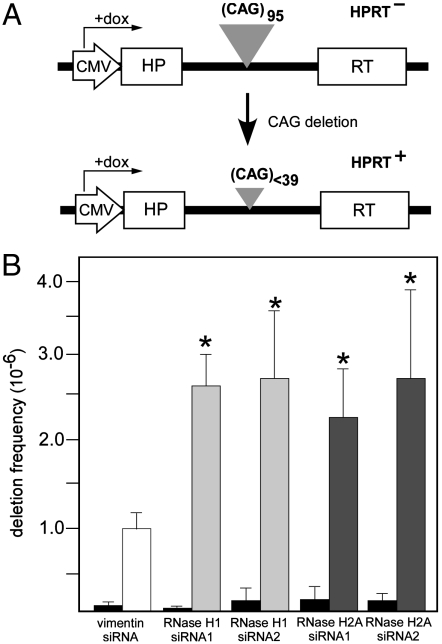
To investigate the potential role of the RNase H activity in transcription-induced CTG·CAG repeat contraction in human cells, we used a well-established genetic assay based on the activity of the hypoxanthine phosphoribosyltransferase (HPRT) minigene in FLAH25 cells (8, 9). In these cells, the HPRT minigene is regulated by the Tet-ON promoter and contains an intron harboring 95 CTG·CAG repeats, which is oriented so that an rCAG transcript is synthesized. Long CTG·CAG repeat tracts inactivate the HPRT minigene by interfering with normal splicing and rendering the cells HPRT-. Contraction of the CTG·CAG tract to less than 39 repeats permits expression of the HPRT minigene, allowing cells with contractions to grow in hypoxanthine/aminopterin/thymidine selection media (Fig. 3A).
Fig. 3.
Effects of RNase H activity on transcription-induced CTG·CAG repeat instability in human cells. (A) HPRT selection system for analyzing the frequency of (CTG·CAG)95 repeat contractions (see text for details). (B) Effects of siRNA knockdown of RNase H1 and RNase H2A on transcription-induced (CTG·CAG)95 repeat contraction. Frequencies of contractions were determined using the HPRT selection assay. Six independent experiments were conducted, using two different siRNAs for RNase H1 and for RNase H2A. Gray bars indicate results when transcription was turned on (plus doxycycline); black bars indicate results with transcription turned off (minus doxycycline). Asterisks indicate statistically significant differences (t test, p < 0.01) between treatments with RNase H1- or RNase H2A-specific siRNAs and treatment with a control vimentin siRNA.
Mammalian cells express two different RNase H enzymes: RNase H1 and RNase H2. Each enzyme is potentially capable of removing RNA·DNA hybrids (28). To determine whether the activity of either of these enzymes affects CTG·CAG repeat instability, we used siRNAs to knock down expression of RNase H1 and RNase H2A, which is the catalytic subunit of RNase H2. For each RNase H, we analyzed two different siRNAs. The siRNAs against RNase H1 lowered its expression by 64% and 71%; the two RNase H2A siRNAs reduced its expression by 66% and 72%. Knockdown of either RNase H1 or H2A significantly increased the frequency of CTG·CAG repeat contractions when the minigene was actively transcribed as compared to siRNA against vimentin (Fig. 3B). By contrast, in the absence of transcription, no statistically significant changes in contraction frequencies occurred (Fig. 3B). These results indicate that the ability of the cells to remove RNA·DNA hybrids affects transcription-induced repeat instability.
Detection of RNase H-Sensitive RNA·DNA Hybrids in Human Cells.
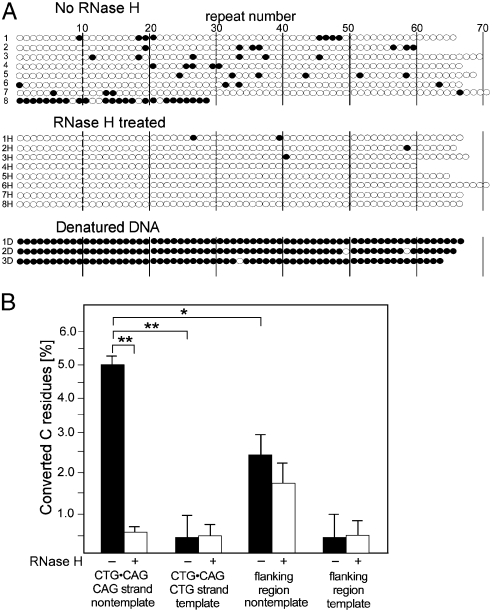
If R loops exist at transcribed CTG·CAG repeats, as suggested by our previous results, we should be able to detect the displaced nontemplate DNA strand by bisulfite modification, which converts Cs in single-stranded DNA to Us. For these experiments, we used a derivative of HEK293 cells in which we had site-specifically integrated a (CTG·CAG)67 tract so that it was transcribed to generate an rCAG transcript. We isolated genomic DNA under nondenaturing conditions to preserve potential R loops (20), and then incubated it with recombinant RNase H to cleave the RNA·DNA hybrids and collapse the R loop or in the absence of RNase H (an incubation control). Subsequently, these two genomic DNA samples were subjected to bisulfite modification under nondenaturing conditions (20). As a positive control to determine if all Cs in the CTG·CAG tract could be converted to Us, we treated the genomic DNA with bisulfite under denaturing conditions. After these treatments, we PCR amplified and cloned the CTG·CAG tract and then sequenced individual clones to detect the bisulfate modifications (Fig. 4).
Fig. 4.
Detection of stable RNA·DNA hybrids in human cells. (A) Sequence analysis of clones of bisulfite modified genomic DNA isolated from HEK293_5150 cells, which contain a (CTG·CAG)67 tract oriented to give an rCAG transcript. Genomic DNA was extracted in nondenaturing conditions and subjected to the bisulfite treatment, PCR amplification, cloning, and sequencing. Clones 1–8 were obtained after bisulfite modification of genomic DNA under nondenaturing conditions. Clones 1H–8H were obtained after bisulfite modification under nondenaturing conditions after the genomic DNA had been treated with RNase H. Clones 1D–3D were isolated after bisulfite treatment genomic DNA in denaturing conditions. Each horizontal line of circles represents a single clone. The differences in the lengths of individual clones reflect repeat instability introduced during PCR amplification and cloning. Each open circle represents the C of a CTG·CAG triplet; each filled circle represents a C that was converted to a U. Modifications of the nontemplate strand are shown. A total of 26 clones (corresponding to 1,710 CTG·CAG repeats) without RNase H treatment and 25 clones (corresponding to 1,681 CTG·CAG repeats) after the RNase H treatment were analyzed in two independent experiments. (B) Effect of RNase H treatment on the frequency of C to U conversion in the CTG·CAG and repeat flanking regions. Flanking sequences, bordered by primers used for PCR amplification, consist of 178 bp including 49 Cs on the template strand and 59 Cs on nontemplate strand for the transcription. C to U conversion events were scored on both the template and nontemplate strands for transcription. Asterisks indicate statistically significant differences (** p < 0.01, * p < 0.05).
Because the displaced CAG strand in the R loops can form double-stranded hairpins, we expected that only some Cs would be available for bisulfite modification under nondenaturing conditions, namely, those in hairpin loops and those between adjacent hairpins. We observed just such an interspersed pattern of modified Cs in 20 out of the 26 clones that were sequenced (Fig. 4A, clones 1–8). A total of 58% of clones carried three or more modified C residues. This pattern of C modification was virtually eliminated by pretreatment with RNase H (only a single clone with three C residues modified), which would be expected to collapse R loops (Fig. 4A, clones 1H–8H). Bisulfite treatment of DNA subsequent to denaturation, which would eliminate R loops and intrastrand secondary structures, showed that > 98% of Cs in the CTG·CAG tracts were capable of modification (Fig. 4A, clones 1D–3D). These results demonstrate that single-stranded DNA exists at transcribed CTG·CAG repeat tracts in human cells.
DNA sequence analyses of the clones of bisulfite treated samples also reveal which parental strand of the CTG·CAG repeat was originally modified. Modification of the CTG strand generates a TTG·CAA triplet, whereas modification of the CAG strand generates a TAG·CTA triplet (Fig. S3). As expected for an R loop formed by an RNA·DNA hybrid between the nascent RNA (rCAG) and the template strand for transcription (CTG), the displaced CAG strand was preferentially modified by bisulfite treatment (Fig. 4B). Moreover, it is these modifications that are specifically eliminated by RNase H digestion (Fig. 4B). No significant strand-specific RNase H sensitivity is apparent for the rare modifications of the CTG strand. Additionally, bisulfite modification of C residues within the CTG strand is two times more frequent than C nucleotides in the regions flanking the repeat tract (Fig. 4B). It should be noted that 178-bp region flanking the repeats and the CTG·CAG tract exhibit similar GC content (61% and 66%, respectively). These results confirm the existence of single-stranded segments of the nontemplate CAG strand, which is consistent with the formation of R loops at transcribed CTG·CAG repeats in human cells.
Discussion
Transcription stimulates expansions and contractions of tracts of DNA triplet repeats, as demonstrated in bacteria, flies, and human cells (7–9, 12, 29, 30). In terminally differentiated cells, which no longer replicate their DNA, transcription may be the major factor affecting somatic instability (8, 9). It has been postulated that transient dissociation of DNA strands upon transcription enhances formation of non-B DNA structures and, consequently, the instability of repeating sequences (10). Herein, we investigated whether newly synthesized, nascent RNA strands play a role in stimulating CTG·CAG repeat instability via formation of stable R loops. Using a combination of biochemical and cellular assays, we demonstrated that the transcribed CTG·CAG repeat sequences can exist in human cells as R loops. Bisulfite treatment preferentially modified the displaced nontemplate DNA strand, indicating its partial single-stranded character, and this preference was eliminated by RNase H treatment. Finally, we used genetic approaches in E. coli and human cells to show that these structures promoted the instability of CTG·CAG repeats. These results demonstrate the involvement of R loops in the generation of trinucleotide repeat instability.
R loops tend to form in GC-rich regions due to the exceptional stability of the rG·C base pairs that are generated when the template strand is C rich (14, 31). Substitution of GTP with ITP or other analogs during transcription dramatically reduced the propensity for R-loop formation (31). Because most tri- and tetranucleotide sequences associated with repeat expansion disorders are highly GC rich (1), it may be that R loops play a general role in repeat instability. Our results demonstrated that R loops formed in CTG·CAG repeat tracts (as well as in CCTG·CAGG repeats), whereas they were undetectable in a plasmid containing (AAT·ATT)90 and in a control plasmid lacking repeats.
Prior work suggested that expansions of CTG·CAG repeat tracts strongly correlate with the GC content of the neighboring flanking sequences (32). For example, the highly expandable CTG·CAG repeat associated with myotonic dystrophy type 1, spinocerebellar ataxias type 1, 2, and 7, Huntington disease, and Kennedy’s disease are located in regions of high GC content, ranging from 59% to 79% in the 500 bp flanking the repeats (32), whereas the average genome GC content is 41% (33). As with these natural repeats, the flanking sequences in our experiments contained 61% GC base pairs. Perhaps the DNA sequences flanking transcribed trinucleotide repeats affect their capability to adopt R-loop conformations, hence influencing the susceptibility of the repeat tracts to contraction or expansion.
Depending on the direction of transcription, the RNA·DNA hybrids formed during in vitro transcription are composed of rCAG·CTG or rCUG·CAG units. We demonstrated that rCAG transcripts were present in R loops more frequently than rCUG transcripts. Because both RNA·DNA hybrids form identical rG·C and rC·G interactions, differences in their stability may depend on the relative stability of rA·T versus rU·A base pairs. UV melting and NMR spectroscopy studies of RNA·DNA hybrids have demonstrated that rA·T base pairs are more thermodynamically stable than rU·A base pairs (16, 34). In the case of CTG·CAG repeats, the contribution of a single base pair is multiplied by the number of repeats, which may account for the observed greater propensity for hybrid formation by rCAG transcripts.
Recent studies suggest that the nascent RNA and the nontemplate DNA strand compete for interactions with the transcription template strand, thereby influencing the efficiency of RNA·DNA hybrid formation (35). Our results are consistent with this idea. We find that displaced nontemplate strands that are capable of forming non-B DNA structures (e.g., CTG·CAG or CCTG·CAGG), and are therefore less available for re-pairing with the DNA template strand, allow efficient formation of RNA·DNA hybrids. By contrast, nontemplate strands with structurally inert (CTA·TAG)68 repeats form R loops inefficiently, if at all. Thus, structure-forming repeat tracts may contribute to R-loop formation (and repeat instability) in two ways: by the effect of CG content on strength of pairing with RNA and by the effect of structure on the rate of re-pairing separated strands.
Our experiments in bacteria and in human cells demonstrate that decreasing the activity of RNase H increases repeat instability in a transcription-dependent manner. In contrast, overexpression of the enzyme in E. coli led to the stabilization of the CTG·CAG repeats. In the absence of transcription, changes in RNase H activity have no effect on CTG·CAG tract length. In agreement with these results, abolishing the activity of RNase HI in yeast had no effect on the instability of nontranscribed CTG·CAG tracts (36). Our results and those in yeast indicate that the “replicative” functions of RNase H (i.e., Okazaki fragment processing) are not directly relevant to repeat instability. Interestingly, our data suggest that both the level of transcription through a repeat-containing gene and the activity of RNase H can affect the instability of the repeat tracts. Differential expression of the enzymes that remove RNA·DNA hybrids could contribute to the tissue-specific pattern of repeat instability observed in repeat expansion diseases (1).
Bisulfite modification of nondenatured samples provides critical evidence for the existence of R loops in transcribed CTG·CAG tracts in human cells. The frequency of converted C residues was ≈10-fold higher in the nontemplate CAG strand than in the template CTG strand. Elimination of this strong strand bias by pretreatment with RNase H demonstrates that it depends on the RNA generated by transcription. These data show that both the movement of the RNA polymerase machinery and the presence of the nascent RNA in R loops affect the lifetime of the displaced nontemplate DNA strand.
Strikingly, the main pattern of bisulfite modification—individual Cs and clusters of two to three Cs—indicates that only a fraction of the Cs in the nontemplate strand are unpaired, consistent with formation of multiple stem-loop structures in the displaced strand (Fig. 5). These structures are reminiscent of the multiple stem loops that form in slipped-strand duplexes, which arise when complementary repeats pair out of register (37). Indeed, the presence of a multiloop structure in the nontemplate strand may enhance formation of slipped-strand duplexes when the nascent RNA is removed from the DNA template and the DNA strands rehybridize.
Fig. 5.
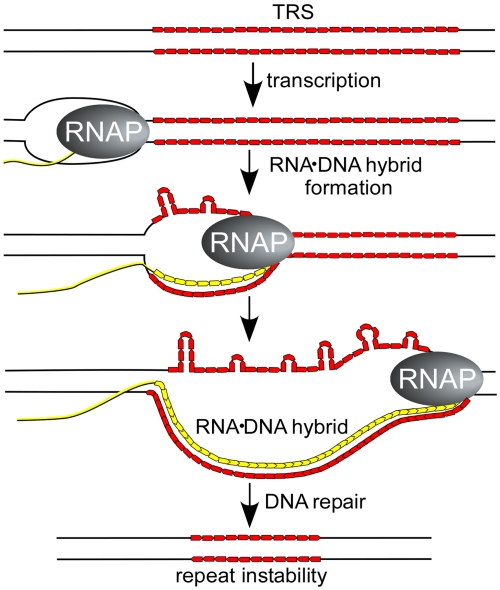
Proposed mechanism for stable RNA·DNA hybrids stimulating repeat instability. Transcription of DNA regions containing a CG-rich trinucleotide repeat (shown in red) favors formation of stable RNA·DNA hybrids. The nontemplate DNA strand, which is displaced and rendered single-stranded (unpaired), can adopt non-B DNA structures such as CTG or CAG hairpins. The unpaired regions of the nontemplate strand are susceptible to bisulfite modification. These non-B DNA structures may be recognized directly by cellular DNA repair processes such as transcription-coupled repair, nucleotide excision repair, mismatch repair, or they may lead to formation of double-stranded DNA breaks. Alternatively, if the RNA (shown in yellow) is ultimately removed from the R loop, the template strand could re-pair with the nontemplate strand to generate a slipped duplex (37), which then engages a DNA repair process. We propose that repeat instability is a consequence of the repair process that are called into play by the non-B DNA structures whose formation is enhanced by transcriptional R loops.
Hairpin loops, unpaired nucleotides, slipped-strand structures, or other aberrant conformations may be recognized as damage signals by one or more DNA repair systems such as mismatch repair and nucleotide excision repair (38), which are ultimately responsible for changing the length of the repeat sequence (Fig. 5). Alternatively, both RNA·DNA hybrids and non-B DNA conformations have been shown to pause the progression of the RNA polymerases (39). Stalled transcription elongation complexes may block the progression of the DNA replication fork leading to fork collapse, formation of double-strand breaks, and stimulation of repeat instability (39, 40).
Recurrent interactions between RNA and DNA are unavoidable during the expression of genetic information. More than 60% of the human genome undergoes transcription, and it frequently occurs in the sense and antisense directions, using both DNA strands as transcription templates (41). Hundreds of microsatellite sequences, potentially capable of forming transcription-dependent R loops, exist in the genomes of most organisms. Thus R loops represent a significant threat not only to the repeats in identified disease genes, but also to overall genome stability. Indeed, the formation of the RNA·DNA hybrids at simple repeat sequences may represent a global mechanism of mutagenesis.
Materials and Methods
Transcription In Vitro and RNA·DNA Hybrid Detection.
pGEMZf(+) and its derivatives were transcribed in vitro using 10 units of T7 or SP6 RNA polymerase (New England Biolabs, Inc.). Transcription was carried out in the presence of 1 μg of supercoiled plasmid (greater than 95% supercoiled) and 0.5 mM of each of four rNTP in a 30 μl volume for 30 min at 37 °C. Transcripts were labeled using 10 μCi of [α32P]rCTP for rCAG and rCUG transcripts and [α32P]rATP for rAAU and rAUU transcripts. R loops were detected using RNase A/I and/or RNase H digestion as described in SI Materials and Methods.
Assay for Genetic Instability in E. coli.
The plasmids pRW3245 and pRW3246 were transformed into the appropriate E. coli strains and grown for a number of generations, as described previously (27) (SI Materials and Methods). E. coli KS351 (Hfr, thi, lacY482, rha, relA1, spoT1) was used as a parental strain for the RNase HI mutant FB2 (KS351 rnhA1).
Detection of RNA·DNA Hybrids in Genomic DNA.
The procedure described by Yu et al. (20), with some modifications, was used to analyze for the presence of RNA·DNA hybrids in the transcribed (CTG·CAG)67 tract of HEK293_5150. See SI Materials and Methods for details.
Construction of the HEK293-5150 Cell Line.
pRW5150, a derivative of pcDNA5/FRT/TO (Invitrogen), was constructed by cloning the (CTG·CAG)67 insert from pRW4026 (42). This plasmid was site-specifically integrated into the genome of HEK293Flp-InT-Rex cells creating the HEK293_5150 cell line as described previously (30).
Frequency of CTG·CAG Contractions in Human Cells.
The human-cell system used to analyze the frequency of the CTG·CAG repeat contractions was described previously (8, 9). Briefly, HPRT- FLAH25 cells contain an HPRT minigene that is inactivated by an intronic insertion of a (CTG·CAG)95 repeat oriented to give an rCAG transcript. Contraction of the repeat tract to fewer than 39 repeats leads to the HPRT+ phenotype (8, 9). For detailed culture conditions and methods of the contraction frequency calculation see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Richard Bowater for plasmid expressing E. coli RNase HI. We thank Elizabeth McIvor for technical assistance. This work was supported by grants from the National Institutes of Health [ES11347], Friedreich’s Ataxia Research Alliance, Seek a Miracle, and the Robert A. Welch Foundation (R.D.W.), and by National Institutes of Health Grant GM38219 (to J.H.W). M.N. was supported by the Friedreich’s Ataxia Research Alliance and the National Ataxia Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909740107/DCSupplemental.
References
- 1.Wells RD, Ashizawa T. Genetic Instabilities and Neurological Diseases. 2nd Ed. San Diego, CA: Elsevier Academic; 2006. [Google Scholar]
- 2.Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 4.Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 6.Mangiarini L, et al. Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nat Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- 7.Bowater RP, Jaworski A, Larson JE, Parniewski P, Wells RD. Transcription increases the deletion frequency of long CTG·CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 1997;25:2861–2868. doi: 10.1093/nar/25.14.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 10.Lin Y, Hubert L, Jr, Wilson JH. Transcription destabilizes triplet repeats. Mol Carcinog. 2009;48:350–361. doi: 10.1002/mc.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Y, Wilson JH. Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells. DNA Repair. 2009;8:878–885. doi: 10.1016/j.dnarep.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ditch S, Sammarco MC, Banerjee A, Grabczyk E. Progressive GAA·TTC repeat expansion in human cell lines. PLoS Genet. 2009;5(10):e1000704. doi: 10.1371/journal.pgen.1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belotserkovskii BP, et al. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J Biol Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 14.Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol. 2009;29:3124–3133. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tracy RB, Hsieh C-L, Lieber MR. Stable RNA/DNA hybrids in the mammalian genome: Inducible intermediates in immunoglobulin class switch recombination. Science. 2000;288:1058–1061. doi: 10.1126/science.288.5468.1058. [DOI] [PubMed] [Google Scholar]
- 16.Sugimoto N, et al. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 17.Drolet M, et al. Over expression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci USA. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huertas P, Aguilera A. Cotranscriptionally formed DNA: RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Yu K, Roy D, Huang FT, Lieber MR. Detection and structural analysis of R-loops. Methods Enzymol. 2006;409:316–329. doi: 10.1016/S0076-6879(05)09018-X. [DOI] [PubMed] [Google Scholar]
- 21.Grabczyk E, Mancuso M, Sammarco MC. A persistent RNA·DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Razidlo DF, Lahue RS. Mrc1, Tof1 and Csm3 inhibit CAG·CTG repeat instability by at least two mechanisms. DNA Repair. 2008;7:633–640. doi: 10.1016/j.dnarep.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tadokoro T, Kanaya S. Ribonuclease H: Molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 24.Itaya M. Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc Natl Acad Sci USA. 1990;87:8587–8591. doi: 10.1073/pnas.87.21.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Usongo V, et al. Depletion of RNase HI activity in Escherichia coli lacking DNA topoisomerase I leads to defects in DNA supercoiling and segregation. Mol Microbiol. 2008;69:968–981. doi: 10.1111/j.1365-2958.2008.06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carl PL, Bloom L, Crouch RJ. Isolation and mapping of a mutation in Escherichia coli with altered levels of ribonuclease H. J Bacteriol. 1980;144:28–35. doi: 10.1128/jb.144.1.28-35.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 28.Chon H, et al. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 2009;37:96–110. doi: 10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 30.Soragni E, et al. Long intronic GAA ∗ TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res. 2008;36:6056–6065. doi: 10.1093/nar/gkn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizuta R, Mizuta M, Kitamura D. Guanine is indispensable for immunoglobulin switch region RNA-DNA hybrid formation. J Electron Microsc. 2005;54:403–408. doi: 10.1093/jmicro/dfi058. [DOI] [PubMed] [Google Scholar]
- 32.Brock GJ, Anderson NH, Monckton DG. Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: Associations with flanking GC content and proximity to CpG islands. Hum Mol Genet. 1999;8:1061–1067. doi: 10.1093/hmg/8.6.1061. [DOI] [PubMed] [Google Scholar]
- 33.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 34.Huang Y, Chen C, Russu IM. Dynamics and stability of individual base pairs in two homologous RNA-DNA hybrids. Biochemistry. 2009;48:3988–3997. doi: 10.1021/bi900070f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy D, Zhang Z, Lu Z, Hsieh CL, Lieber MR. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: A nick can serve as a strong R-loop initiation site. Mol Cell Biol. 2009 doi: 10.1128/MCB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Callahan JL, Andrews KJ, Zakian VA, Freudenreich CH. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol Cell Biol. 2003;23:7849–7860. doi: 10.1128/MCB.23.21.7849-7860.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson CE, et al. Slipped-strand DNAs formed by long (CAG·CTG) repeats: Slipped-out repeats and slip-out junctions. Nucleic Acids Res. 30:4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G, Vasquez KM. Models for chromosomal replication-independent non-B DNA structure-induced genetic instability. Mol Carcinog. 2009;48:286–298. doi: 10.1002/mc.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- 40.Krasilnikova MM, Samadashwily GM, Krasilnikov AS, Mirkin SM. Transcription through a simple DNA repeat blocks replication elongation. EMBO J. 1998;17:5095–5102. doi: 10.1093/emboj/17.17.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Napierala M, Parniewski PP, Pluciennik A, Wells RD. Long CTG·CAG repeat sequences markedly stimulate intramolecular recombination. J Biol Chem. 2002;277:34087–34100. doi: 10.1074/jbc.M202128200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.