Abstract
An effective immune response requires B cells to produce several classes of antibodies through the process of class switch recombination (CSR). Activation-induced cytidine deaminase initiates CSR by deaminating deoxycytidines at switch regions within the Ig locus. This activity leads to double-stranded DNA break formation at the donor and recipient switch regions that are subsequently synapsed and ligated in a 53BP1-dependent process that remains poorly understood. The DNA damage response E3 ubiquitin ligases RNF8 and RNF168 were recently shown to facilitate recruitment of 53BP1 to sites of DNA damage. Here we show that the ubiquitination pathway mediated by RNF8 and RNF168 plays an integral part in CSR. Using the CH12F3-2 mouse B cell line that undergoes CSR to IgA at high rates, we demonstrate that knockdown of RNF8, RNF168, and 53BP1 leads to a significant decrease in CSR. We also show that 53BP1-deficient CH12F3-2 cells are protected from apoptosis mediated by the MDM2 inhibitor Nutlin-3. In contrast, deficiency in either E3 ubiquitin ligase does not protect cells from Nutlin-3–mediated apoptosis, indicating that RNF8 and RNF168 do not regulate all functions of 53BP1.
Keywords: 53BP1, activation-induced cytidine deaminase , DNA damage response
Part of an effective immune response requires the production of antibodies of different classes, each of which mediates a different effector function. This process is initiated by activation-induced cytidine deaminase (AID), which induces class switch recombination (CSR) by deaminating deoxycytidines within Ig switch regions (1, 2). The recognition and subsequent processing of the mutated residues by base excision repair and/or mismatch repair machineries generates double-stranded DNA breaks (DSBs) at switch regions (3). In an attempt to mend the ensuing breaks, B cells mount a damage response similar to that signaled by irradiated cells (1). Ultimately, AID-induced DSBs are repaired predominantly by nonhomologous end-joining (4, 5) and to a lesser extent by an alternative end-joining pathway (6).
Generally, DSBs are readily recognized by sensor-kinase protein complexes, including the Mre11-Rad50-Nbs1 (MRN)/ataxia-telangiectasia mutated (ATM), Ku70/Ku80-DNA-PKcs, and ataxia-telangiectasia and Rad3-related (ATR)-ATR-interacting protein complexes (7). Upon binding to a DSB site, these factors are activated to trigger a cascade of events that culminates in cell cycle delay and/or DNA repair (7). During this process, sequential chromatin modifications around the break sites appear to be crucial for adequate resolution of the DNA breaks. Active chromatin modification is initiated by ATM-dependent phosphorylation of the histone variant H2AX to the γH2AX form (8–10). γ-H2AX then preferentially binds the tandem-BRCA1 C-terminal domain (BRCT) motifs of mediator of DNA damage checkpoint 1 (MDC1) (11, 12), which in turn recruits more MRN/ATM, thereby amplifying the γ-H2AX signal (11, 12). Recently, it was shown that ATM also phosphorylates MDC1 at a TQxF consensus, allowing for the recruitment of a novel E3 ubiquitin ligase, RNF8, via its phospho-binding forkhead-associated module (13–15). Once at the break, RNF8 mediates the monoubiquitination of H2A-type histones, among other potential proteins. Monoubiquitinated H2A-type histones subsequently signal the recruitment of another E3 ubiquitin ligase, RNF168 (16, 17), a mutation of which was recently reported in an immunodeficient and radiosensitive patient with RIDDLE syndrome (16, 18). RNF168 protects and amplifies the RNF8-mediated ubiquitination by catalyzing the addition of K63-linked polyubiquitin chains at DNA damage sites (16, 17). Although low levels of 53BP1 can transiently localize to DNA breaks via direct binding to dimethylated H4K20 (19), ubiquitination by RNF8 and RNF168 seem to be crucial for the accumulation and stabilization of 53BP1 at DSB sites.
Most of the factors required for DNA damage response (DDR) also appear to contribute to the process of CSR (1). One important distinction between general DDR and CSR is the major reliance of the latter on 53BP1 (20–22). 53BP1 deficiency results in a drastic decrease in CSR. Whether this marked defect is due to a repair function that 53BP1 fulfills and/or because 53BP1 is required to bring distal switch regions into close proximity and assist in switch synapse formation is unclear (1). Recent data seem to focus on the latter aspect, especially because 53BP1-deficient B cells exhibit increased levels of intraswitch region recombination (23), which is consistent with 53BP1 promoting and/or stabilizing the synapsis of distant switch regions during CSR. Similarly, 53BP1 facilitates long-range end-joining during V(D)J recombination at the TCR locus (24) and also promotes end-joining of dysfunctional telomeres by increasing chromatin mobility (25). Historically, 53BP1 was first described as one of two proteins to interact with p53 (26). This aspect of 53BP1 function is generally considered to mediate apoptosis, which is seen as distinct from its ability to localize to DSBs.
We and others have previously shown that the ubiquitination of proliferating cell nuclear antigen (PCNA) is important for both somatic hypermutation and CSR (27, 28). In a continued effort to delineate the importance of the ubiquitin system in antibody diversification, we examined whether the RNF8 and RNF168 members of the E3 ubiquitin ligase cascade, which lie upstream of 53BP1 in the DDR pathway, also are required to mediate CSR.
Results
ShRNA-Mediated RNAi Can Establish Functional Knockdown in CH12F3-2 Cells.
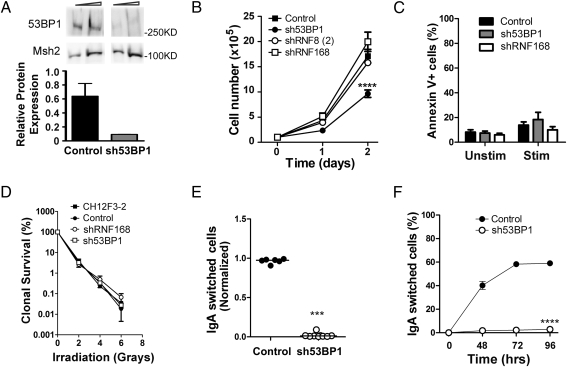
We first ascertained that the CH12F3-2 cell line is a faithful model of CSR with respect to the role of DDR factors. To do this, we assessed whether RNA interference (RNAi) by shRNA for 53BP1, which has been shown to be an essential factor in CSR (21, 22), can achieve functional knockdowns in CH12F3-2 cells. Cells were transduced with a lentiviral shRNA construct specific for 53BP1 (sh53BP1), and individual clones were isolated by puromycin selection. Compared with clones transduced with a scrambled, nonsilencing shRNA construct (control), 53BP1 knockdown cells showed an approximate five-fold reduction in 53BP1 transcript levels, as determined by RT-PCR analysis and Western blot analysis (Fig. 1A). These cells have a slightly lower growth rate compared with control cells (Fig. 1B) and demonstrate no major differences in apoptosis before or after stimulation (Fig. 1C). Although 53BP1-deficient cells have been shown to have a mild sensitivity to irradiation (29), 53BP1-depleted CH12F3-2 cells did not display increased sensitivity to irradiation compared with control cells (Fig. 1D).
Fig. 1.
shRNA-mediated RNAi knockdown of 53BP1 in CH12F3-2 cells abrogates CSR. (A) Western blot analysis of control (Left) and sh53BP1 (Right) CH12F3-2 cells quantifying 53BP1 and Msh2 (loading control). (B) Two clones of each indicated CH12F3-2 cell population were analyzed for growth in duplicate, and statistical significance was tested by two-way ANOVA. ****P <.0001. (C) Percentage of CH12F3-2 populations undergoing apoptosis (surface annexin V+) with and without stimulation for 2 days. The assay was carried out with at least two individual clones for each population. (D) Clonogenic survival assays with asynchronous CH12F3-2 populations exposed to varying x-ray doses. The assay was carried out with at least two individual clones for each population. (E) Six individual control and eight sh53BP1 CH12F3-2 clones were stimulated for 2 days and then analyzed for IgA expression. Values were normalized by dividing the % IgA-positive cells in the experimental group into the % IgA-positive cells in the stimulated parental CH12F3-2 cells. CSR assays were performed on each clone in duplicate, and statistical significance was tested using the one-tailed t test. (F) Three control and three sh53BP1 CH12F3-2 clones were stimulated for 2, 3, and 4 days, after which IgA expression was analyzed. CSR assays were performed in duplicate for each clone. Statistical significance tested by two-way ANOVA. ****P <.0001.
We next tested whether 53BP1 knockdown cells were deficient in CSR. Although CSR was not affected in control clones relative to the parental cells (Fig. 1E), eight individual 53BP1 knockdown clones demonstrated complete abrogation of CSR at 48 h after stimulation (Fig. 1E), even though both groups of cells expressed similar levels of AID (SI Fig. S1). This defect in CSR persisted even at 4 days poststimulation (Fig. 1F). Although the 53BP1-depleted cells showed a mild proliferation defect, previous findings have established a role for 53BP1 in CSR (21, 22), suggesting that the mild proliferative defect observed in these cells is not responsible for the dramatic lack of CSR. These results confirm the importance of 53BP1 for CSR in the CH12F3-2 cells used in these studies. They also indicate that lentiviral-mediated shRNA-mediated RNAi can achieve a functional knockdown in CH12F3-2 cells, thereby setting the stage for testing whether RNF8 and RNF168 are involved in CSR.
RNF8 Is Required for CSR in CH12F3-2 Cells.
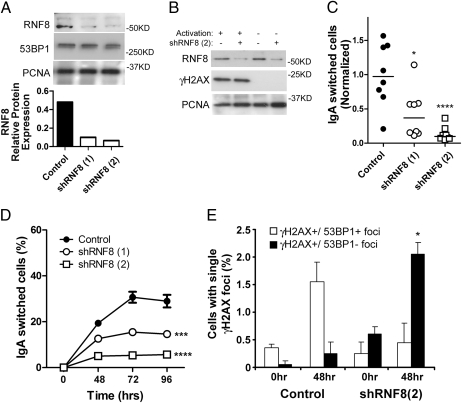
To achieve knockdown of RNF8, we tested five different shRNA lentiviral constructs specific for RNF8. Two of these constructs (designated 1 and 2) achieved strong depletion of RNF8 following puromycin selection in CH12F3-2 cells, as verified by Western blot analysis (Fig. 2A). We then assessed whether RNF8 depletion in those lines affected the response to DNA DSBs. As expected, activated cells had a considerably higher γH2AX signal compared with nonactivated samples (Fig. 2B). There was no difference in γH2AX protein levels in control and RNF8-depleted CH12F3-2 cells, however, indicating that both groups of cells responded similarly to AID-induced DNA DSBs.
Fig. 2.
CSR is impaired in RNF8-deficient CH12F3-2 cells. (A) Western blot analysis quantifying RNF8, 53BP1, and PCNA (loading control) in CH12F3-2 cells with control shRNA and two different lentiviral vectors targeting RNF8(1) and (2). (B) Western blot analysis of stimulated and unstimulated control or shRNF8-treated CH12F3-2 cells, quantifying RNF8 and γH2AX protein levels, with PCNA as the loading control. (C) Eight individual clones for each of control, shRNF8(1), shRNF8(2), and CH12F3-2 cells were stimulated for 3 days and then analyzed for IgA expression. Values were normalized by dividing the % IgA-positive cells in the experimental group to the % IgA-positive cells in the stimulated parental CH12F3-2 cells. Statistical significance was evaluated by the two-tailed t test. *P <.05; ****P <.0001. (D) Two control, two shRNF8(1)-treated, and two shRNF8(2)-treated CH12F3-2 clones were stimulated, and IgA expression was analyzed at 2, 3, and 4 days poststimulation. Statistical significance was tested by two-way ANOVA. ***P <.001; ****P <.0001. (E) Stimulated control and shRNF8 CH12F3-2 cells were collected at times 0 h and 48 h and then stained with antibodies specific to γH2AX and 53BP1. Cells from two separate experiments (each with n = 1,000), having single γH2AX foci with 53BP1 (γH2AX ± 53BP1+ foci) or without 53BP1 (γH2AX ± 53BP1- foci) colocalization were counted and analyzed as shown. More than 90% of the cells did not exhibit γH2AX foci after cytokine stimulation. Statistical significance was tested using the two-tailed t test between control 48 h and shRNF8(2) 48 h. *P = .014.
To test the effects of RNF8 depletion on CSR, we selected eight individual clones from each transduction for subsequent analysis. Although different switching efficiencies were achieved within each group of cells, CSR was significantly reduced in the RNF8-depleted clones (Fig. 2C). The extent of the switching defect was greater in the clones (2) that were more depleted for RNF8 (Fig. 2 A and C). In addition, we evaluated CSR at different time intervals in a representative sample of the clones and found a low CSR in RNF8-depleted cells relative to control cells (Fig. 2D). The RNF8-depleted cells exhibited no defects in proliferation (Fig. 1B). These data indicate that the reduced CSR observed in RNF8-depleted cells is not due to proliferation defects, and that RNF8 function is required for efficient CSR.
To gain insight into the role of RNF8 in CSR, we studied the effects of RNF8 knockdown by immunofluorescence. When unstimulated CH12F3-2 cells were fixed and stained with anti-γH2AX and anti-53BP1, almost all cells exhibited a diffuse 53BP1 nuclear pattern and a lack of γH2AX signal, as expected (Fig. S2, Top). On activation, control cells showed a marked increase in single γH2AX foci that colocalize with single 53BP1 foci, indicative of induced DSBs and hence active switching events (Fig. 2E, Fig. S2, Bottom). Importantly, compared with control cells, RNF8-depleted cells exhibited a significant increase in γH2AX foci with a diffused 53BP1 signal (Fig. 2E, Fig. S2). Of note, we observed a slight increase in γH2AX foci in nonactivated RNF8-depleted cells (Fig. 2E). likely due to an accumulation of spontaneously occurring breaks that could not be repaired efficiently following RNF8 knockdown. 53BP1 depletion has no effect on germline transcription (21) or AID expression (Fig. S1). In addition, we found that RNF8 depletion generated similar levels of AID-induced γH2AX as control cells when assayed by immunoblotting and immunofluorescence. This suggests that the cascade upstream of AID-induced breaks, which is dependent on germline transcription and AID, most likely was intact. Taken together, these findings suggest that the inability of RNF8-depleted cells to undergo efficient CSR is related to their inability to signal the accumulation and/or stabilization of 53BP1 at AID-induced breaks.
RNF168 Is Important for CSR in CH12F3-2 Cells.
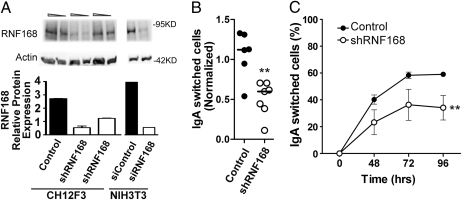
We next examined the contribution of RNF168 to CSR. To achieve knockdown of RNF168, we tested three different shRNA lentiviral constructs specific for RNF168, one of which led to an approximate two- to three-fold reduction in RNF168 protein levels (Fig. 3A). Akin to the shRNF8-transduced cells, the shRNF168-transduced cells and control cells displayed similar growth rates (Fig. 1B), similar apoptosis levels before and after stimulation (Fig. 1C), and similar sensitivity to irradiation (Fig. 1D). In contrast, the RNF168-knockdown cells had 46% lower CSR levels than the control cells (Fig. 3 B and C). As in the RNF8-depleted cells, this approximate two-fold reduction of CSR correlates with the level of knockdown achieved in the CH12F3-2 cells. Thus, the CSR defect observed in the absence of RNF8 can be recapitulated by depleting RNF168. Together, these findings indicate that the E3 ubiquitin ligase cascade composed of RNF8 and RNF168 plays a significant role in CSR.
Fig. 3.
CSR is impaired in RNF168-deficient CH12F3-2 cells (A) Western blot analysis of control or shRNF168 CH12F3-2 cells quantifying RNF168, with β-actin as the loading control. NIH 3T3 cells treated with control siRNA and siRNF168 were used as negative and positive controls for RNF168 protein expression, respectively. (B) Six individual control and seven individual shRNF168 CH12F3-2 clones were stimulated for 2 days and then analyzed for IgA expression. Values were normalized by dividing the % IgA-positive cells in the experimental group into the % IgA-positive cells in the stimulated parental CH12F3-2 cells. CSR assays were performed on each clone in triplicate, and statistical significance was evaluated by the two-tailed t test. **P = .0023 (C) Three control and three shRNF168 CH12F3-2 clones were stimulated for 2, 3, and 4 days, after which IgA expression was analyzed. CSR assays were performed in duplicate for each clone. Statistical significance was tested by two-way ANOVA. **P = .0034.
53BP1 Deficiency, But Not RNF8/168 Deficiency, Protects Cells From Nutlin-3–Induced Apoptosis.
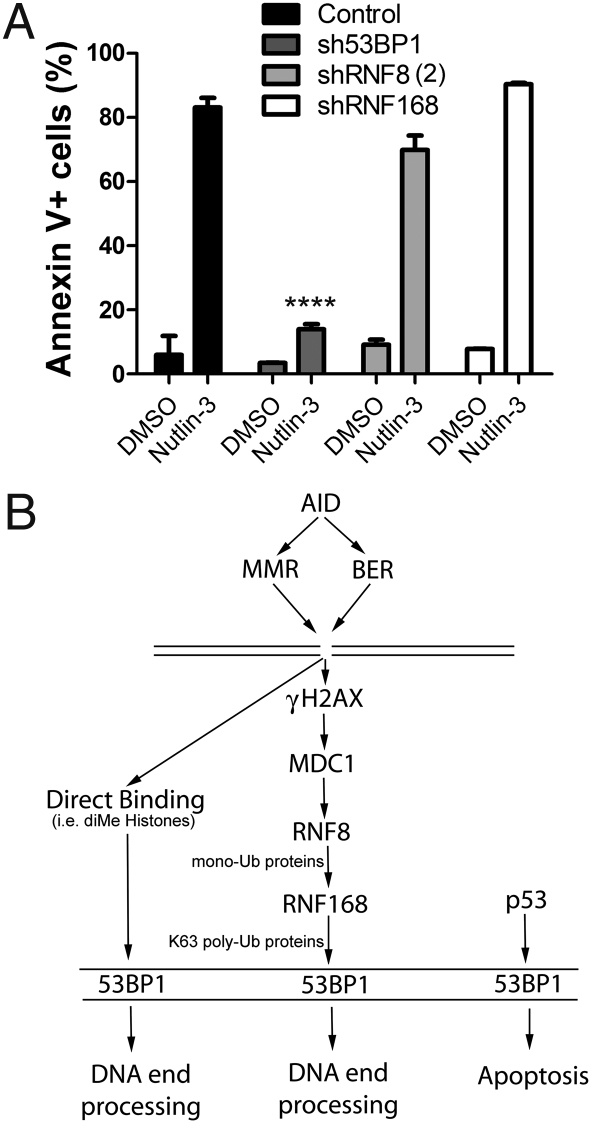
Nutlin-3 is a small-molecule inhibitor of MDM2 that induces activation of p53 and apoptosis of cancer cells in a process dependent on 53BP1 (30); that is, 53BP1-deficient cells are resistant to Nutlin-3–mediated apoptosis. To test whether this activity of 53BP1 is influenced by RNF8 or RNF168, we examined whether a deficiency of either protein protects CH12F3-2 cells from the effects of Nutlin-3. In contrast to control cells incubated with Nutlin-3, which exhibited dramatically increased apoptosis, Nutlin-3 did not induce high levels of apoptosis in 53BP1-depleted cells (Fig. 4A). Strikingly, RNF8-depleted cells and RNF168-knockdown cells remained sensitive to Nutlin-3–mediated apoptosis (Fig. 4A). These data indicate that 53BP1 has functions that are independent of RNF8 and RNF168 (Fig. 4B).
Fig. 4.
53BP1 knockdown cells, but not RNF8/168 knockdown cells, are resistant to Nutlin-3–mediated apoptosis. (A) Percentage of control, sh53BP1, shRNF8(2), and shRNF168 CH12F3-2 cells undergoing apoptosis (i.e., surface annexin V+) after treatment with Nutlin-3 or DMSO for 1 day. Each assay was carried out with two different clones of each population. Statistical significance was tested by two-way ANOVA. ****P <.0001. (B) Schematic illustrating two different roles for 53BP1. AID indirectly induces a DSB by the actions of the mismatch repair (MMR) and base excision repair (BER) pathways. 53BP1 is recruited to the DNA DSB site either by direct binding (via the MRN complex or dimethylated histones) or through the γH2AX-RNF8-RNF168 amplification pathway, the latter of which leads to the accumulation and stabilization of 53BP1 at the break site. These pathways ultimately lead to end-processing and resolution of the DNA break. Independent of the RNF8/168 cascade, 53BP1 could lead to p53 stabilization or enhanced transcriptional activity in response to apoptosis signaling.
Discussion
Role of RNF8 and RNF168 in CSR and Its Relation to 53BP1 Localization.
Although the necessity of 53BP1 for CSR has been reported previously (21, 22, 31), the exact function of the 53BP1-dependent pathway in CSR remains elusive. Recent findings demonstrate that RNF8 and RNF168 play essential roles in the recruitment and stabilization of 53BP1 at DSB sites during irradiation-induced DDR (13–17). In support of a role for 53BP1 localization to sites of AID-induced DSB, our data indicate that knockdown of either RNF8 or RNF168 causes a significant decrease in CSR. These findings support the notion that the accumulation of 53BP1 at sites of AID-induced DSB in switch regions is dependent on RNF8/RNF168. Because we also used the knockdown of RNF8/RNF168 to separate the roles of 53BP1 in CSR and apoptosis, this indirectly supports the idea that 53BP1 is required for the efficient bridging of the two distal DNA ends (1, 24, 25).
Because the recruitment of RNF8 and subsequent recruitment of RNF168 to DSB sites requires ATM/MRN, γH2AX, and MDC1, it would be reasonable to assume that all of these factors have similar effects on CSR (Fig. 4B). In reality, however, the loss of these DDR factors seems to have different effects on CSR. Whereas ATM, MRN, or H2AX deficiency leads to a ∼70% reduction in CSR (32–36), MDC1 deficiency has a milder effect (25%–50% reduction (11)), and 53BP1 deficiency has the most profound effect (∼90% reduction; this study and refs. 21 and 22). A lesser dependence of CSR on MDC1 may be due to either (i) a sufficient residual level of MDC1 protein in the MDC1 gene-trap knockout mice to amplify the ATM/MRN-mediated γH2AX signal (11) or (ii) a redundant MDC1-independent mechanism that allows for recruitment of low levels of RNF8 and subsequently 53BP1, allowing for intermediate levels of CSR. On the other hand, the difference between the low levels of CSR observed in ATM, MRN, and H2AX deficiency and the nearly complete lack of CSR in 53BP1-deficient cells might be explained by the potential transient recruitment of 53BP1 to switch regions via either direct binding to dimethylated H4K20 exposed at the DSB site (19) or other, as-yet undetermined, factors. As a result, this transient recruitment of 53BP1 could be sufficient for the resolution of a subset of DNA breaks mediating low levels of CSR in the absence of the upstream proteins required for 53BP1 accumulation and stabilization (Fig. 4B). Whether a loss of RNF8 or RNF168 affects CSR in a manner akin to the loss of 53BP1 or γH2AX remains to be seen. Our data suggest that RNF8/RNF168 have an incomplete effect on CSR; however, our shRNA constructs cannot completely deplete these factors. The finding that serum IgG is reduced by ∼10-fold in a patient with RIDDLE syndrome deficient in RNF168 (18) suggests that complete ablation of RNF8 or RNF168 would produce a pronounced phenotype.
53BP1 Has a Unique Function Independent of RNF8 and RNF168.
53BP1 has been shown to facilitate several interrelated cellular functions, including CSR (21, 22), cell cycle checkpoint, DNA repair (31, 37), and apoptosis (30, 38). It is not yet clear whether these different pathways always act in unison or whether they could act independently. Using Nutlin-3, a small-molecule inhibitor of MDM2, we tested whether RNF8 and RNF168 could differentially affect some functions of 53BP1, such as DNA end-processing and p53-dependent apoptosis.
Nutlin-3 has been shown to disrupt p53–MDM2 interaction, inducing a p53-mediated apoptosis in a 53BP1-dependent manner (30). Consistent with this finding, our results indicate that 53BP1-deficient CH12F3-2 cells are resistant to Nutlin-3–induced apoptosis. Of note, the C terminus of AID interacts with MDM2 (39), which potentially could interfere with the analysis of our Nutlin-3 treatment. But because our experiments were conducted in nonactivated CH12F3-2 cells, in which the activation of AID protein is not yet initiated (Fig. S1), it is safe to assume that any such interaction would not interfere with our analysis. 53BP1 interacts with p53 through its C-terminal tandem-BRCT motif in a phosphorylation-independent manner. This interaction is thought to stabilize p53 and stimulate p53-mediated transcriptional activation (40). It is possible that p53, on release from MDM2-mediated inhibition, is degraded in 53BP1-deficient cells, and thus apoptosis is not induced, or that 53BP1 functions as a cofactor of p53 to induce apoptosis. Whether this function of 53BP1 is related to its role in localization at sites of DSB remains unclear. But because the p53–53BP1 interaction is phosphorylation-independent (40), this plausibly might be a function of 53BP1 distinct from foci formation at DSB sites. In support of this notion, we found that RNF8- and RNF168-depleted CH12F3-2 cells remained sensitive to the apoptotic effects of Nutlin-3, indicating that RNF8 and RNF168 do not regulate all functions of 53BP1 signaling. Thus, 53BP1 seems to have distinct and independent roles in response to genotoxic stress (Fig. 4B).
Methods
In Vitro Cell Culture.
CH12F3-2 cells were maintained and CSR assays performed as described previously (41). In brief, cells were stimulated with 1 ng/mL of recombinant human TGF-β1 (R&D Systems), 10 ng/mL of recombinant mouse IL-4 (R&D Systems) and 2 μg/mL of functional-grade purified anti-mouse CD40 (eBiosciences) and then analyzed by flow cytometry as described below. Lentiviral shRNA constructs and protocols for 53BP1 (V2LMM 83391), RNF8 (AAF59E8 and AAF59E12), RNF168 (V2MM 16141), and nonsilencing negative control (RHS4346 and SHC002) shRNA were purchased from Open Biosystems. CH12F3-2 cells were transduced with lentivirus by centrifugation at 800 × g for 90 min at room temperature in the presence of 5 μg/mL of polybrene. Cells were then incubated for 3 days, after which positively transduced clones were obtained by limiting dilution cloning and puromycin selection. For growth curve analysis, CH12F3-2 cells were diluted to a concentration of 1 × 105 cells/mL and aliquoted in duplicate on a 96-well plate. At various time points, the numbers of live trypan blue–excluded cells were counted with a hemocytometer. CH12F3-2 cells were treated with 25 μM Nutlin-3 (Sigma) or DMSO as a negative control. NIH 3T3 cells were treated with an siRNA SMARTpool (ThermoFisher) targeting murine RNF168. siRNA transfections were performed using Dharmafect 1 (ThermoFisher) in the forward transfection mode.
Flow Cytometric Analyses.
CH12F3-2 cells were analyzed by intracellular staining with PE-conjugated anti-mouse IgA clone 11–44-2 (eBiosciences), using Cytofix/Cytoperm and Perm/Wash buffers (BD Biosciences). The cells were surface-stained for annexin V using the Annexin V–APC Apoptosis Detection Kit (eBiosciences). Stained cells were analyzed by FACSCalibur (BD Biosciences) and FlowJo software (Truestar).
Irradiation Sensitivity Assays.
CH12F3-2 cell lines were irradiated with various x-ray doses and then plated at various dilutions in duplicate 96-well plates. Survival was determined by counting the number of expanding clones normalized to plating efficiency.
Western Blot Analyses.
53BP1 (Alexis Biochemicals and Novus), RNF8 (Abcam), γH2AX (Upstate Biotechnology), PCNA (Santa Cruz Biotechnology), β-actin (Abcam), and Msh2 (BD Pharmingen) antibodies were used in accordance with the manufacturers’ protocols. The RNF168 polyclonal antibody was raised against a murine GST-RNF168381-567 fusion protein and affinity-purified using a murine HIS6-RNF168381-567 Sepharose column.
Immunofluorescence.
Cells were pelleted at 1,000 rpm for 3 min in Eppendorf tubes, then fixed with 4% paraformaldehyde (Sigma), permeabilized with 0.3% Triton X (Roche), and blocked with 10% FCS (HyClone) and 0.01% saponin (Sigma) in PBS. The following antibodies were used: mouse FITC anti-γH2AX (Millipore), rabbit anti-53BP1 (Novus), and anti-rabbit Alexa Fluor 568 (Invitrogen). Cells were stained overnight at 4°C, washed with blocking buffer, and then pelleted on slides using a Shandon cytospin machine at 400 rpm for 2 min. DAPI stain in mounting dye (Vectashield) was used to detect DNA.
Statistical Analysis.
Analyses were performed using GraphPad Prism. For the Student t test, two-way ANOVA, and Mann-Whitney U test, a P value of ≤.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Jason Moffat and the Martin Laboratory for helpful discussions and Tasuku Honjo and Frederick Alt for the CH12F3-2 cells. This research is supported by grants from the National Cancer Institute (RO1CA72649 and R01CA102705, to M.D.S.) and the Canadian Institutes of Health Research (MOP10703115, to D.D., and MOP66965, to A.M.). A.M. is supported by a Canada Research Chair award.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913790107/DCSupplemental.
References
- 1.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 3.Saribasak H, Rajagopal D, Maul RW, Gearhart PJ. Hijacked DNA repair proteins and unchained DNA polymerases. Philos Trans R Soc Lond B Biol Sci. 2009;364:605–611. doi: 10.1098/rstb.2008.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan-Hammarström Q, et al. Impact of DNA ligase IV on nonhomologous end-joining pathways during class switch recombination in human cells. J Exp Med. 2005;201:189–194. doi: 10.1084/jem.20040772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franco S, et al. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy-chain class switch recombination. J Exp Med. 2008;205:557–564. doi: 10.1084/jem.20080044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan CT, et al. IgH class switching and translocations use a robust nonclassical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 7.Harper JW, Elledge SJ. The DNA damage response: Ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: The histone guardian of the genome. DNA Repair (Amst) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 10.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 11.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 12.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 13.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 16.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 17.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 18.Stewart GS, et al. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci USA. 2007;104:16910–16915. doi: 10.1073/pnas.0708408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward IM, Minn K, van Deursen J, Chen J. p53-binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy-chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 22.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1-/- B cells. Eur J Immunol. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- 24.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes nonhomologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S. Two cellular proteins that bind to wild-type, but not mutant, p53. Proc Natl Acad Sci USA. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roa S, et al. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc Natl Acad Sci USA. 2008;105:16248–16253. doi: 10.1073/pnas.0808182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langerak P, Nygren AO, Krijger PH, van den Berk PC, Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J Exp Med. 2007;204:1989–1998. doi: 10.1084/jem.20070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura K, et al. Genetic dissection of vertebrate 53BP1: A major role in nonhomologous end-joining of DNA double-strand breaks. DNA Repair (Amst) 2006;5:741–749. doi: 10.1016/j.dnarep.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Brummelkamp TR, et al. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- 31.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 32.Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reina-San-Martin B, et al. H2AX is required for recombination between immunoglobulin switch regions, but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lähdesmäki A, Taylor AM, Chrzanowska KH, Pan-Hammarström Q. Delineation of the role of the Mre11 complex in class switch recombination. J Biol Chem. 2004;279:16479–16487. doi: 10.1074/jbc.M312796200. [DOI] [PubMed] [Google Scholar]
- 35.Dinkelmann M, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16:808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reina-San-Martin B, Nussenzweig MC, Nussenzweig A, Difilippantonio S. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc Natl Acad Sci USA. 2005;102:1590–1595. doi: 10.1073/pnas.0406289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53-binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao L, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by BRCA1 deficiency. Mol Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacDuff DA, Neuberger MS, Harris RS. MDM2 can interact with the C-terminus of AID but it is inessential for antibody diversification in DT40 B cells. Mol Immunol. 2006;43:1099–1108. doi: 10.1016/j.molimm.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 40.Iwabuchi K, et al. Stimulation of p53-mediated transcriptional activation by the p53-binding proteins, 53BP1 and 53BP2. J Biol Chem. 1998;273:26061–26068. doi: 10.1074/jbc.273.40.26061. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura M, et al. High-frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.