Abstract
Quantifying patterns of population structure in Africans and African Americans illuminates the history of human populations and is critical for undertaking medical genomic studies on a global scale. To obtain a fine-scale genome-wide perspective of ancestry, we analyze Affymetrix GeneChip 500K genotype data from African Americans (n = 365) and individuals with ancestry from West Africa (n = 203 from 12 populations) and Europe (n = 400 from 42 countries). We find that population structure within the West African sample reflects primarily language and secondarily geographical distance, echoing the Bantu expansion. Among African Americans, analysis of genomic admixture by a principal component-based approach indicates that the median proportion of European ancestry is 18.5% (25th–75th percentiles: 11.6–27.7%), with very large variation among individuals. In the African-American sample as a whole, few autosomal regions showed exceptionally high or low mean African ancestry, but the X chromosome showed elevated levels of African ancestry, consistent with a sex-biased pattern of gene flow with an excess of European male and African female ancestry. We also find that genomic profiles of individual African Americans afford personalized ancestry reconstructions differentiating ancient vs. recent European and African ancestry. Finally, patterns of genetic similarity among inferred African segments of African-American genomes and genomes of contemporary African populations included in this study suggest African ancestry is most similar to non-Bantu Niger-Kordofanian-speaking populations, consistent with historical documents of the African Diaspora and trans-Atlantic slave trade.
Keywords: Africa, human genomics, population genetics
Studies of African genetic diversity have greatly informed our understanding of human origins and history (1, 2), have identified genes under natural selection across evolutionary time (3), and hold great potential for elucidating the genetic bases of disease susceptibility and drug response among diverse human populations (4, 5). The study of African population structure is also critical for reconstructing patterns of African ancestry among African Americans and for enabling genome-wide association mapping of complex disease susceptibility and pharmacogenomic response in African-American populations (6–9).
Africa contains over 2,000 ethnolinguistic groups and harbors great genetic diversity (2, 10–17), but little is known about fine-scale population structure at a genome-wide level. This is, in part, because previous studies of high-density SNP and haplotype variation among global human populations (defined as studies with at least 100,000 SNP markers) have included few African populations (10, 12, 13, 18), whereas detailed studies of genetic structure among African populations have used a modest number of markers (2) (∼1,500 microsatellites and indels). Nonetheless, recent studies of microsatellite and DNA sequence variation suggest a significant population structure exists within sub-Saharan Africa, with geography, language, and mode of subsistence (e.g., hunter-gatherer, pastoralist, agriculturalist) as potential key factors (2, 12, 13, 19). Given that high-density genotype data have revealed discernible population structure within other continental populations (e.g., Europe, East Asia) and even among geographical regions within countries (e.g., Switzerland, Finland, United Kingdom) (20–24), there is strong reason to believe that high-density genotype data from African and African-American populations can elucidate patterns of genetic structure among these populations further.
We have thus genotyped on the Affymetrix GeneChip 500K array set 146 individuals from 11 populations in West and South Africa (Fig. S1 and Table S1) who speak Nilo-Saharan, Afro-Asiatic, and Niger-Kordofanian languages and integrated these data with our previous studies of human genomic diversity, including 57 Yorubans from Ibadan, Nigeria, genotyped as part of the International Haplotype Map project, 365 African Americans from throughout the United States, and 400 individuals of European ancestry (10, 25). Our study focuses on analysis of fine-scale population structure among the West African samples and its implication for high-resolution inference of admixture in African Americans. We use principal component analysis (PCA) to infer axes of genetic variation within Africa and examine individual and population clustering using the clustering algorithm FRAPPE (26). Next, we compare the West African, European, and African-American samples and seek to identify the set of West African populations closest to the ancestral population of African Americans. Finally, based on the results of the other two analyses, we evaluate individual patterns of European and African ancestry along each chromosome for each African-American subject in our dataset using a computationally efficient PCA-based method that infers admixture proportions based on high-density genome-wide data.
Results
Genetic Structure of West African Populations.
Our study focused on West African populations, because previous genetic and historical studies suggest that region was the source for most of the ancestry of present-day African Americans (2, 27, 28). Among the sampled West African populations, Wright’s measure of population differentiation [autosomal FST (29)] was low (1.2%), suggesting quite recent common ancestry of all individuals in our sample or, alternatively, a large effective population size for the structured population from which the sample was drawn, with a large degree of gene flow among subpopulations. Nonetheless, we observed substantial variation in pairwise FST among sampled populations, suggesting genetic heterogeneity among the groups (Table 1). Differences in pairwise FST may reflect variation in effective population size or migration rates among the populations potentially attributable to isolation by distance or heterogeneity in geographical or cultural barriers to gene flow. For example, the Fulani appear to be genetically distinct from all other West African populations we sampled (average pairwise FST = 3.91%). Likewise, we found that the Bulala, Xhosa, and Mada populations consistently exhibited pairwise FST above 1% when compared with any other population, whereas the non-Bantu Niger-Kordofanian populations of the Igbo, Brong, and Yoruba exhibited little genetic differentiation from one another (average FST <0.4%). These results suggest that there are clear and discernible genetic differences among some of the West African populations, whereas others appear to be nearly indistinguishable even when comparing over 300,000 genetic markers.
Table 1.
FST distances between African populations
| Igbo | Brong | Yoruba | Kongo | Bamoun | Xhosa | Fang | Hausa | Kaba | Mada | Bulala | |
| Brong | 0.350% | — | |||||||||
| Yoruba | 0.084% | 0.200% | — | ||||||||
| Kongo | 0.282% | 0.425% | 0.291% | — | |||||||
| Bamoun | 0.293% | 0.448% | 0.318% | 0.175% | — | ||||||
| Xhosa | 1.448% | 1.636% | 1.251% | 1.106% | 1.277% | — | |||||
| Fang | 0.415% | 0.594% | 0.432% | 0.150% | 0.247% | 1.165% | — | ||||
| Hausa | 0.397% | 0.560% | 0.420% | 0.588% | 0.546% | 1.796% | 0.691% | — | |||
| Kaba | 0.516% | 0.510% | 0.471% | 0.501% | 0.484% | 1.498% | 0.567% | 0.619% | — | ||
| Mada | 1.296% | 1.336% | 1.300% | 1.282% | 1.276% | 2.319% | 1.380% | 1.299% | 0.968% | — | |
| Bulala | 1.862% | 1.905% | 1.879% | 1.736% | 1.806% | 2.646% | 1.929% | 1.773% | 1.280% | 0.931% | — |
| Fulani | 3.905% | 3.684% | 4.034% | 3.770% | 3.996% | 4.133% | 4.063% | 3.761% | 3.811% | 3.967% | 3.920% |
Bold face font is used to emphasize the genetic similarity of the Igbo, Brong and Yoruba.
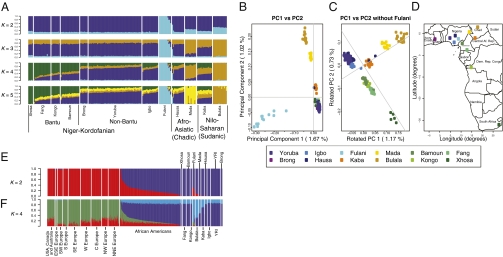
To investigate whether we could reliably distinguish ancestry among individuals from these populations, we used two approaches tailored for high-density genotype data. One, FRAPPE, implements a maximum likelihood method to infer genetic ancestry of each individual, wherein the individuals are assumed to have originated from K ancestral clusters (26). Fig. 1A and Fig. S2 summarize FRAPPE results when the number of clusters, K, is varied from K = 2 to K = 7. The small number of clusters was consistent with the small overall level of population differentiation among these populations. We next undertook PCA of the matrix of individual genotype values (i.e., the matrix with entries “0,” “1,” or “2” generated by tallying the number of copies of a given allele across all SNPs in a panel for all individuals genotyped) (30).
Fig. 1.
Population structure within West Africa and relation to language and geography. (A) FRAPPE analysis of the West African populations. Individuals are represented as thin vertical lines partitioned into segments corresponding to the inferred membership in K = 2 through K = 5 genetic clusters as indicated by the colors (see Figs. S2–S5 for additional results). (B) Principal components 1 and 2 of the African individuals. (C) Principal components 1 and 2 of the African individuals, excluding the Fulani population, wherein the components have been rotated to emphasize further similarity with geography. (D) Approximate locations of sampled populations in Africa. (E and F) FRAPPE clustering of Europeans, African Americans, and West Africans. Individuals are represented as thin vertical lines partitioned into K segments corresponding to the inferred membership of the genetic clusters indicated by the colors. Values for K = 2 (E) and K = 4 (F) are shown for comparison between the two analyses.
Patterns of population structure were consistent between the two approaches (Figs. S2–S5). For example, in the FRAPPE analysis, the Fulani population was distinguished at K = 2, with Bulala, Mada, and Kaba populations showing some genetic similarity with the Fulani. PCA, likewise, separated the Fulani from other populations along the first principal component (PC1) (Fig. 1B). The two subsequent principal components, PC2 and PC3, reflect the geographical distribution of the populations. PC2 showed a Chadic and Nilo-Saharan dimension extending into inland Africa from the coast, distinguishing the Bulala, Mada, and Kaba populations. These populations belong to the Nilo-Saharan and Afro-Asiatic (Chadic) linguistic groups and live further inland. Analysis of the African populations, excluding the Fulani, gave a PC1 and PC2 that resemble the second and third principal components of the PCA with the Fulani (Fig. 1C). Rotating the PC1 and PC2 axes from the PCA without the Fulani reveals the similarity of the genetic and geographical maps (Fig. 1 C and D).
At K = 3, the FRAPPE algorithm clusters the Bulala into their own group and suggests genetic similarity of the Mada, Kaba, and Hausa, potentially indicating differentiation of Nilo-Saharan- and Afro-Asiatic-speaking populations from Niger-Kordofanian-speaking populations. At K = 4, all individuals from the Bantu-speaking Xhosa of South Africa cluster into a single group and individuals from the Bantu-speaking populations (Fang, Bamoun, and Kongo) exhibit considerable shared membership in this cluster. At K = 5, the Mada are distinguishable as a unique group, with modest genetic similarity with the Hausa and Kaba as well as with most of the Niger-Kordofanian populations. These results suggest that although these populations are quite closely related genetically, it is possible to detect meaningful population substructure given sufficient marker density [see also ref. (2)]. It is important to note that there is likely further substructure and diversity within these populations. Because we sample a modest number of individuals from each population (n = 13, on average, per population), we are not likely to have captured all the genetic variation within each population, region, or linguistic family. To compare patterns of haplotype structure and discern differences in demographic history among the African populations, we estimated linkage disequilibrium (LD) between all pairs of markers in the data for all populations (see SI Text and Fig. S6). All the African populations showed low levels of LD (even at closely linked sites) and a rapid decay of LD with distance genome-wide relative to populations of European ancestry.
Genome-Wide Patterns of Admixture in African Americans.
To understand the genetic structure of the African-American population better and to determine African-American ancestry, we used FRAPPE to evaluate African Americans together with European and African individuals genotyped on the same marker set. At K = 2, African populations (blue) were distinguished from European populations (red), with African Americans showing highly variable levels of European and West African ancestry (Fig. 1 E and F). For the African Americans, estimated mean West African ancestry was 77%, consistent with prior studies (2, 28, 31–34). Analysis at K = 4 revealed additional substructure in a North-South cline within Europe and clusters coinciding with the linguistic and geographical substructure within Africa (see SI Text, Tables S2–S4, and Figs. S7 and S8 for additional FRAPPE and population genetic analyses). PCA of the genotype value matrix of the European, West African, and African-American samples revealed the primary axis of variation (PC1) to correspond with “European” vs. “West African” ancestry (see Fig. 2A) and explained ∼ 9.8% of the genetic variance. Specifically, we observed two centroids in the data, with all the individuals of European ancestry exhibiting negative loadings along PC1, whereas all the West African individuals exhibited positive loadings. African Americans exhibited a wide range of loadings along PC1, presumably attributable to differences in European vs. West African ancestry. PC2 corresponds to population substructure within West Africa and largely mirrors the patterns discussed above.
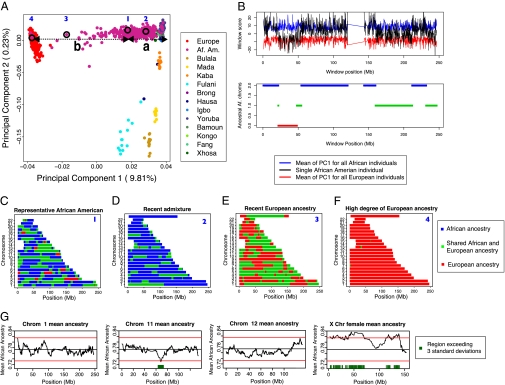
Fig. 2.
Results of our PCA-based ancestry estimation method. (A) Graphical illustration of approach: Euclidean distances from a given individual's coordinates in PCA space (i.e., “loadings”) and the West African centroid (“a”) and the European centroid (“b”) along PC1 for PCA space that includes Europeans, African Americans, and West Africans. (B) Local ancestry estimation using the PCA sliding window approach and associated HMM for number of chromosomes for a given individual (i.e., “0,” “1,” or “2”) with African ancestry. (C–F) Individual ancestry estimates of 4 representative African-American individuals (denoted 1, 2, 3, and 4 in Fig. 2A) in our dataset of 365 individuals. The colors represent two chromosomes of West African ancestry (blue), two chromosomes of European ancestry (red), or one chromosome of West African and one chromosome of European ancestry (green). (G) Mean ancestry of 365 African-American individuals at each window across chromosome (chrom) 1, chrom 11, chrom 12, and chromosome X (X Chr). The black line shows the overall mean estimated ancestry. Red bands indicate +3 and −3 SDs from the mean ancestry. (All chromosomes are reported in Fig. S10).
Estimation of Admixture in Local Genomic Regions.
We reconstructed estimated European or West African ancestry for every African American in our dataset at every position in the genome using a PCA-based algorithm (Fig. 2A). Our method is a generalization of the approach of Paschou et al. (35) and estimates genome-wide proportion of West African ancestry for a given individual as P = b/(a + b), where b and a are the chord distances from the European and West African centroids, respectively, for the given individual along PC1. Our generalization involves undertaking the PC1 distance analysis on a grid of points along the genome (as opposed to genome-wide) centered on 15 SNP windows and using a Hidden Markov Model (HMM) for inference of ancestry state (i.e., having “0,” “1,” or “2” chromosomes of recent African origin; see SI Text, Fig. 2B, and Fig. S9). An ancestry plot summarizing the number of segments of European (i.e., “0”), West African (i.e., “2”), or admixed (i.e., “1”) ancestry for a representative African-American individual with 73.5% West African ancestry is illustrated in Fig. 2C. There is a great deal of variation among the ancestry plots of the 365 self-identified African Americans in the study, ranging from an estimate of over 99% West African ancestry to an estimate of less than 1% West African ancestry (Fig. 2F). Some patterns reflected a high level of West African ancestry and only one or two ancestry-switching events per chromosome, suggesting very recent direct African ancestry (Fig. 2D). Other patterns reflected only European and admixed ancestry throughout the genome, suggesting one parent of European ancestry and one parent of African-American ancestry (Fig. 2E).
An interesting question one can address with these kinds of data is whether regions of the genome show substantially high European or West African ancestry across all individuals in the sample [e.g., as may be the case if a particular allele from one of the ancestral populations was under strong selection (36–39)]. For our analysis, we considered genomic regions as potential candidates for increased European or West African ancestry if the mean ancestry for the region across the 365 African-American individuals was 3 SDs above or below the genome-wide average of West African ancestry (78.1%). Using this approach, we found that several genomic regions of autosomal chromosomes 5, 6, and 11 could be considered outliers from the genome-wide distribution of ancestry, although these differences were not significant after correction for multiple tests. In Fig. 2G, we show mean ancestry across two example chromosomes that do not show any outlier regions (chromosomes 1 and 12) and one chromosome showing a region falling outside the 3 SD criteria (chromosome 11). Mean ancestry estimates for all chromosomes can be found in Fig. S10, and a precise listing of molecular regions for the three outlier regions may be found in Table S4. In contrast to the autosomes, the X chromosome shows significantly high West African ancestry along the majority of the chromosome, consistent with a gender-biased model of admixture with excess European male and West African female ancestry (Fig. 2G).
Discussion
The Bantu expansion occurred ∼4,000 years ago, originating in Cameroon or Nigeria and expanding throughout sub-Saharan Africa (40, 41). The clustering of the Xhosa, Fang, Bamoun, and Kongo populations, all of which are Bantu Niger-Kordofanian-speaking populations, likely reflects a Bantu migration from Nigeria/Cameroon expanding toward the south. Although we have limited sample sizes (with three of our populations having sample sizes of less than 10), the relative order of clustering (the East-West axis, followed by the North-South axis) suggests that the strongest differentiating axis among the African populations is linguistic classification corresponding to Chadic and Nilo-Saharan vs. Niger-Kordofanian ancestry. The relatively weaker North-South axis may result from the genetic similarity among the Niger-Kordofanian linguistic groups because of their recent common ancestry. Although sampled in Nigeria, the very distinct Fulani are part of a nomadic pastoralist population that occupies a broad geographical range across Central and Western Africa. Analyses of microsatellite and insertion/deletion polymorphisms indicate that they share ancestry with Niger-Kordofanian, North African, and Central African Nilo-Saharan populations, as well as low levels of European and/or Middle Eastern ancestry (2). Exempting the Fulani, our LD analyses show no large differences in rates of LD decay among our sampled African populations, with all populations exhibiting a faster decay of LD (i.e., larger inferred effective population size) than previously characterized populations of European ancestry (see SI Text).
Interestingly, the Kongo population does not follow the overall trend of East-West and North-South clustering. The Kongo population’s genetic proximity to geographically distant Bantu populations from Cameroon could be explained by the genetic similarity of Bantu-speaking populations in the region, as seen in the FRAPPE analyses (Fig. 1). Alternatively, although these individuals self-identified as Kongo and were refugees from locations within the Democratic Republic of Congo, the samples were collected in Cameroon; therefore, self-identified ancestry might poorly represent the long-term geographical origins or may reflect recent admixture.
A concern in estimating admixture is the effect of choice of ancestral populations. Often, the true ancestral population is no longer available for sampling; thus, using a proxy may introduce bias when evaluating the admixed population. For example, individual admixture estimates in Latin Americans have been shown to depend on the ancestral populations evaluated (42). Some studies estimating admixture proportions in African Americans have used a single ancestral African population, the Yoruba (39), and our data provide an effective means of testing whether other populations may serve as better proxies for the ancestral population of African Americans and whether using the Yoruba biases inferences. Comparison of the inferred West African segments of African-American genomes with contemporary West African populations (Table S3) reveals that the ancestry of the West African component of African Americans is most similar to the profile from non-Bantu Niger-Kordofanian-speaking populations, which include the Igbo, Brong, and Yoruba, with FST values to African segments of the African Americans ranging from 0.074 to 0.089%. That these FST values are all nearly identical (and quite small), coupled with the small pairwise FST values of the Igbo, Yoruba, and Brong populations (Table 1), suggests that considering the set of West African populations sampled, any of these three populations may serve as a proxy for the ancestral population of the African Americans and that, in fact, all three are likely to have contributed ancestry to present-day African Americans (43). This is wholly in line with historical documents showing that the Igbo and Yoruba are 2 of the 10 most frequent ethnicities in slave trade records, although it is important to note that other African populations not sampled, including those from Sierra Leone, Senegal, Guinea Bissau, and Angola, may also serve as good (or potentially even better) proxies for the ancestral population of some African Americans (44).
That some individuals who self-identify as African American show almost no West African ancestry and others show almost complete West African ancestry has implications for pharmacogenomics studies and assessment of disease risk. Although individuals with very low West African or very low European ancestry may be expected by chance after several generations of admixture, these individuals are most likely descendants of individuals of European ancestry or recent African immigrants, respectively. Assuming these individuals are not simply mislabeled, it appears that the range of genetic ancestry captured under the term African American is extremely diverse, which suggests caution should be used in prescribing treatment based on differential guidelines for African Americans (45).
We found regions on chromosomes 5, 6, and 11 that show deviations from the overall mean West African ancestry. These regions do not overlap with those previously suggested to be under selection (39), and about a dozen genes are found across these regions. Whether these genes or regions are potentially under selection in African Americans merits further investigation.
In conclusion, we believe the data presented here speak to several important points. First, patterns of genomic diversity within Africa are complex and reflect deep historical, cultural, and linguistic impacts on gene flow among populations. These patterns are discernible using high-density genotype data and allow us to differentiate closely related populations along linguistic and geographical axes, even with limited sample sizes from many of our populations. Second, admixture can be reconstructed for local genomic regions efficiently at a high density of genetic markers. For this study, we tailored the method to admixed populations with two ancestral source populations, but the approach is generalizable to multiple populations. Application of the method to genome-wide patterns of genomic variation in African Americans reveals the rich mosaic structure of admixture in this population. We find that we can distinguish African ancestry among West African populations to a large degree (e.g., Bantu from non-Bantu Niger-Kordofanian populations) but that some populations (e.g., Igbo, Yoruba, and, to a lesser extent, Brong) are so closely related genetically that their contribution to patterns of African ancestry in African Americans is not reliably distinguishable. We believe that increasing the density of markers and, more importantly, sequencing directly in these populations to identify ancestry-informative markers may make this possible in the future.
Materials and Methods
Datasets.
We genotyped 225 individuals from 11 African populations [see the article by Tishkoff et al. (2) for sampling locations] on the Affymetrix GeneChip 500K array set and incorporated data from the Yoruban population of Ibadan, Nigeria, from the HapMap project, thinned to the same SNP set (10). European samples were from the GlaxoSmithKline Population Reference Sample (POPRES) project, a resource of nearly 6,000 control individuals from North America, Europe, and Asia (25) genotyped on the Affymetrix GeneChip 500K array set. For our analyses, we extracted a subset of 400 individuals from Europe, randomly sampling 15 individuals from each European country represented in POPRES when possible and 15 individuals each from the United States, Canada, and Australia. We include 365 African Americans from this dataset (see SI Text and ref. 25). Written informed consent was provided by the study participants and approved by the proper institutional review boards, and permits were obtained for collection of African populations as described by Tishkoff et al. (2).
Population Structure Analyses.
FRAPPE implements an efficient maximum likelihood version of the Bayesian clustering algorithm, STRUCTURE (26, 46, 47). After thinning markers to have Pearson product-moment correlation of allele frequency, r2, less than 0.5 in 50 SNP windows, shifted and recalculated every 5 SNPs, we ran FRAPPE on all 204,457 remaining markers for 5,000 iterations. Clusters at K = 6 and higher did not correspond to known linguistic or population substructures (Fig. S2). We ran PCA using the program smartpca from the package eigenstrat (30) on a reduced dataset of 251,253 SNPs, where r2 < 0.8 in 50 SNP windows. FST was calculated using a C++ implementation of Weir and Cockerham’s weighted equations (29). Minor allele frequency (MAF) was thresholded at >0.1 in the populations being compared for all comparisons, except when calculating distances between African Americans and each of the African populations. To reduce the SNP ascertainment biases associated with SNP discovery in the Yoruba, we used only markers with a MAF >0.1 in Europeans for the FST estimates.
Admixture Analysis.
Our local genomic PCA admixture method normalizes the genotype matrix of all individuals using the procedure as in eigenstrat (30). Each chromosome is divided into 15 SNP nonoverlapping windows. The score for an individual for a given window is the product of an individual’s normalized and scaled genotypes across this window with the corresponding segment of the PC1 eigenvector (see SI Text for more details of the procedure). Windows that have one or more missing genotypes for an individual are not given a score and are omitted by the HMM. This gives a vector of scores for each individual across all chromosomes. We assume that ancestral population scores are drawn from a normal distribution and use the ancestral population sample means and variances as the estimated parameters for the distribution (see SI Text for mathematical details of the model and validation).
Supplementary Material
Acknowledgments
We thank K. King for her work in managing and preparing the POPRES data. We thank J.D. Degenhardt for helpful discussions and suggestions throughout the project, and K.E. Lohmueller for discussion, LD scripts, and constructive comments on the manuscript. This work was supported by the National Institutes of Health (Grant 1R01GM83606). S.A.T. additionally acknowledges support by the National Institutes of Health (Grant R01GM076637), National Science Foundation (Grants BCS-0196183, BSC-0552486, and BCS-0827436), and David and Lucile Packard and Burroughs Wellcome Foundation Career Awards.
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909559107/DCSupplemental.
References
- 1.Reed FA, Tishkoff SA. African human diversity, origins and migrations. Curr Opin Genet Dev. 2006;16:597–605. doi: 10.1016/j.gde.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sirugo G, et al. Genetic studies of African populations: An overview on disease susceptibility and response to vaccines and therapeutics. Hum Genet. 2008;123:557–598. doi: 10.1007/s00439-008-0511-y. [DOI] [PubMed] [Google Scholar]
- 5.Campbell MC, Tishkoff SA. African genetic diversity: Implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma L, et al. Distribution of CCR2-64I and SDF1-3′A alleles and HIV status in 7 ethnic populations of Cameroon. J Acquir Immune Defic Syndr. 2005;40:89–95. doi: 10.1097/01.qai.0000157008.66584.d6. [DOI] [PubMed] [Google Scholar]
- 7.Williamson C, et al. Allelic frequencies of host genetic variants influencing susceptibility to HIV-1 infection and disease in South African populations. AIDS. 2000;14:449–451. doi: 10.1097/00002030-200003100-00020. [DOI] [PubMed] [Google Scholar]
- 8.Reich D, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JA. Ethnic differences in cardiovascular drug response: Potential contribution of pharmacogenetics. Circulation. 2008;118:1383–1393. doi: 10.1161/CIRCULATIONAHA.107.704023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazer KA, et al. International HapMap Consortium (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature. 449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrigan D, et al. Inferring human population sizes, divergence times and rates of gene flow from mitochondrial, X and Y chromosome resequencing data. Genetics. 2007;177:2195–2207. doi: 10.1534/genetics.107.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 13.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 14.Tishkoff SA, et al. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 15.Tishkoff SA, Kidd KK. Implications of biogeography of human populations for ‘race’ and medicine. Nat Genet. 2004;36(11 suppl):S21–27. doi: 10.1038/ng1438. [DOI] [PubMed] [Google Scholar]
- 16.Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: Implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- 17.Tishkoff SA, Williams SM. Genetic analysis of African populations: Human evolution and complex disease. Nat Rev Genet. 2002;3:611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- 18.Adeyemo AA, Chen G, Chen Y, Rotimi C. Genetic structure in four West African population groups. BMC Genet. 2005;6:38. doi: 10.1186/1471-2156-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patin E, et al. Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set. PLoS Genet. 2009;5:e1000448. doi: 10.1371/journal.pgen.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lao O, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 21.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xing J, et al. Fine-scaled human genetic structure revealed by SNP microarrays. Genome Res. 2009;19:815–825. doi: 10.1101/gr.085589.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McEvoy BP, et al. Geographical structure and differential natural selection among North European populations. Genome Res. 2009;19:804–814. doi: 10.1101/gr.083394.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelis M, et al. Genetic structure of Europeans: A view from the North-East. PLoS One. 2009;4(5):e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson MR, et al. The Population Reference Sample, POPRES: A resource for population, disease, and pharmacological genetics research. Am J Hum Genet. 2008;83:347–358. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: Analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 27.Lovejoy PE. Transformations in Slavery. New York: Cambridge Univ Press; 2000. [Google Scholar]
- 28.Salas A, et al. Shipwrecks and founder effects: Divergent demographic histories reflected in Caribbean mtDNA. Am J Phys Anthropol. 2005;128:855–860. doi: 10.1002/ajpa.20117. [DOI] [PubMed] [Google Scholar]
- 29.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 30.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parra EJ, et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Lind JM, et al. Elevated male European and female African contributions to the genomes of African American individuals. Hum Genet. 2007;120:713–722. doi: 10.1007/s00439-006-0261-7. [DOI] [PubMed] [Google Scholar]
- 33.Smith MW, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parra EJ, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paschou P, et al. PCA-correlated SNPs for structure identification in worldwide human populations. PLoS Genet. 2007;3:1672–1686. doi: 10.1371/journal.pgen.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Workman PL, Blumberg BS, Cooper AJ. Selection, gene migration and polymorphic stability in a U.S. white and Negro population. Am J Hum Genet. 1963;15:429–437. [PMC free article] [PubMed] [Google Scholar]
- 37.Reed TE. Caucasian genes in American Negroes. Science. 1969;165:762–768. doi: 10.1126/science.165.3895.762. [DOI] [PubMed] [Google Scholar]
- 38.Cavalli-Sforza L, Bodmer W. The Genetics of Human Populations. San Francisco: Freeman; 1971. [Google Scholar]
- 39.Tang H, et al. Recent genetic selection in the ancestral admixture of Puerto Ricans. Am J Hum Genet . 2007;81:626–633. doi: 10.1086/520769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehret C. Bantu expansions: Re-envisioning a central problem of early African history. Int J Afr Hist Stud. 2001;34:5–40. [Google Scholar]
- 41.Klieman KA. The Pygmies Were Our Compass. 2003 (Heinemann, Portsmouth, NH) [Google Scholar]
- 42.Tian C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 4(1):e4. Am J Hum Genet. 2008;81(1):626–633. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 44.Hall GM. Slavery and African Ethnicities in the Americas: Restoring the Links. Chapel Hill, NC: Univ North Carolina Press; 2005. [Google Scholar]
- 45.Reiner AP, et al. Population structure, admixture, and aging-related phenotypes in African American adults: The Cardiovascular Health Study. Am J Hum Genet. 2005;76:463–477. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchard JK, Donnelly P. Case-control studies of association in structured or admixed populations. Theor Popul Biol. 2001;60:227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- 47.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.