Abstract
The remarkable fidelity of most DNA polymerases depends on a series of early steps in the reaction pathway which allow the selection of the correct nucleotide substrate, while excluding all incorrect ones, before the enzyme is committed to the chemical step of nucleotide incorporation. The conformational transitions that are involved in these early steps are detectable with a variety of fluorescence assays and include the fingers-closing transition that has been characterized in structural studies. Using DNA polymerase I (Klenow fragment) labeled with both donor and acceptor fluorophores, we have employed single-molecule fluorescence resonance energy transfer to study the polymerase conformational transitions that precede nucleotide addition. Our experiments clearly distinguish the open and closed conformations that predominate in Pol-DNA and Pol-DNA-dNTP complexes, respectively. By contrast, the unliganded polymerase shows a broad distribution of FRET values, indicating a high degree of conformational flexibility in the protein in the absence of its substrates; such flexibility was not anticipated on the basis of the available crystallographic structures. Real-time observation of conformational dynamics showed that most of the unliganded polymerase molecules sample the open and closed conformations in the millisecond timescale. Ternary complexes formed in the presence of mismatched dNTPs or complementary ribonucleotides show unique FRET species, which we suggest are relevant to kinetic checkpoints that discriminate against these incorrect substrates.
Keywords: alternating-laser excitation, conformational dynamics, fidelity checkpoints, Klenow fragment, fingers-closing
DNA polymerases copy a template sequence with extraordinary accuracy thanks to a series of noncovalent transitions that precede the chemical step of phosphoryl transfer and serve as kinetic checkpoints, rejecting inappropriate substrates early in the reaction pathway (1). An important example is the fingers-closing conformational change, inferred from cocrystal structures, in which the addition of the correct complementary deoxyribonucleotide (dNTP) to a polymerase-DNA (Pol-DNA) binary complex results in a transition from an open to a closed conformation, forming a snug binding pocket around the nascent base pair (Fig. 1A) (2–4). A variety of stopped-flow fluorescence studies, including two FRET-based studies focusing explicitly on the fingers-closing step, have provided information on prechemistry conformational transitions in DNA polymerases (5–8).
Fig. 1.
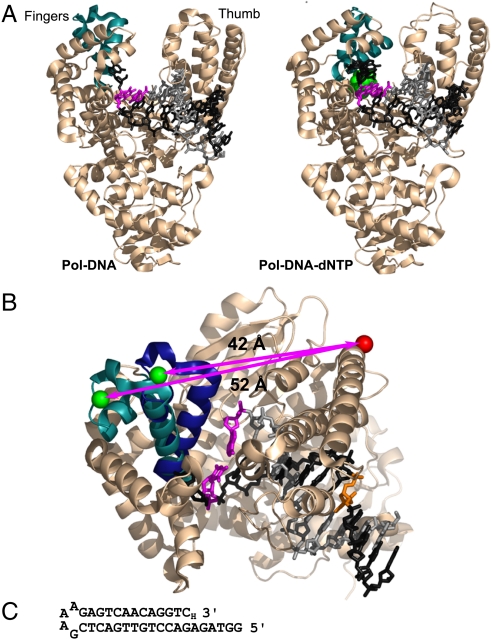
The fingers-closing conformational change in Pol I(KF). (A) The open Pol-DNA binary complex (PDB file 1L3U) and closed Pol-DNA-dNTP ternary complex (PDB file 1LV5) are illustrated using structural data from B. stearothermophilus (Bst) DNA polymerase (3), a close homologue of Pol I(KF). The α-carbon backbone of the protein is shown in Beige, except for the mobile segment of the fingers subdomain in Teal. The DNA template strand is in Dark Gray, the primer strand in Light Gray. The terminal base pair at the active site is Magenta, and the incoming complementary dNTP is Green. (B) Superposition of the open and closed structures shown in (A), viewed from above the polymerase active site. The mobile portion of the fingers subdomain (residues 680 to 714, equivalent to Pol I residues 732 to 766) is shown in Teal in the binary complex and Dark Blue in the ternary complex. The backbone structure of the rest of the protein, shown in Beige, is taken from the 1L3U PDB file but is essentially the same in both structures. The β carbons of the two side chains used as fluorophore attachment sites are shown in space-filling representation; residue 744 in Green, and residue 550 in red (Pol I residue numbers); the arrows indicate the distance between the Cβ positions in the open and closed conformations. The DNA primer-template is colored as in (A), except for the T(-8) position (Orange), which served as the attachment site for a dabcyl quencher in some experiments (see Fig. S1). The illustrations in (A) and (B) were made using PyMOL (DeLano Scientific). (C) DNA-hairpin oligonucleotide used in single-molecule FRET experiments. A dideoxy nucleotide (3′-H) prevents covalent addition to the 3′ end.
Our studies, on the Klenow fragment of E. coli DNA polymerase I (Pol I(KF)), have identified two potentially interesting conformational transitions, whose rapid rate (≈200 s-1) places them ahead of the slow step that is rate-limiting for nucleotide addition (6, 8). One is the fingers-closing step, which does not take place normally with either mispaired dNTPs or ribonucleotides (rNTPs). The other step is assigned as a DNA rearrangement; this step is blocked by mispaired nucleotides but takes place with complementary nucleotides regardless of whether they contain deoxyribo or ribo sugars. This observation places the DNA conformational transition ahead of the fingers-closing step and suggests a sequence of events in which a DNA polymerase checks the incoming nucleotide first for complementarity to the templating base and subsequently for the correct sugar structure.
A drawback to ensemble studies of the type discussed above is that the measured fluorescence signal is a population average. Therefore one may fail to detect dynamic behavior within the population or the existence of subpopulations of molecules with interesting properties. We have therefore used single-molecule FRET (smFRET) to learn more about the conformational states that precede phosphoryl transfer. A further improvement over our previous experimental design is the attachment of both the donor and acceptor fluorophores to the polymerase, instead of having one partner on the DNA. This expanded the scope of our experiments to include studies of the polymerase in its unliganded form and, by doing so, revealed its highly dynamic state. Thus, Pol I joins a small but growing group of enzymes that, in the absence of substrate, have been shown to explore the conformational states that predominate in enzyme-substrate complexes during the catalytic process (9–12).
Results
Protein Labeling.
To detect the fingers-closing conformational transition in Pol I(KF), we placed one fluorophore on the mobile segment of the fingers subdomain (residue 744) and a complementary fluorophore at the base of the thumb subdomain (residue 550); the latter position is essentially unchanged during fingers-closing as inferred from cocrystal structures (Fig. 1B). This labeling scheme maximizes the interfluorophore distance change upon fingers-closing, since the fluorophore on the fingers moves along the vector defined by the initial interprobe separation. The Cβ positions of the labeled side chains are ≈52 Å apart in the open conformation and ≈42 Å apart in the closed conformation; such a large distance change can be detected easily using FRET. We chose Cy3B as the FRET donor (abbreviated as “G” for emitting upon green-laser excitation) and ATTO647N as the acceptor (abbreviated as “R” for emitting upon red-laser excitation); this pair has a Förster distance, Ro, of 67 Å (determined as in ref. 13), which is consistent with the FRET changes described below, assuming a contribution of 15–20 Å from the fluorophore linkers. By exploiting differences in the reactivity of Cys side chains at 550 and 744, we obtained two doubly labeled Pol I (KF) preparations (SI Materials and Methods and Figs. S1 and Fig. S2). In one, the donor-acceptor population was almost exclusively (≥97%) 550-Cy3B,744-ATTO647N (abbreviated as G550R744); in the other, the donor-acceptor population was heavily biased (≈88%) towards 550-ATTO647N,744-Cy3B (abbreviated as R550G744). In smFRET experiments, the G550R744 protein behaved as if stuck in the closed conformation (Fig. S3), and, unlike the R550G744 derivative, did not show a fingers-closing transition in ensemble FRET measurements (Fig. S2). We attribute this aberrant behavior to the ATTO647N probe at position 744 which, in doubly or singly labeled Pol I(KF) derivatives, resulted in lower polymerase activity (Fig. S2), perhaps related to the high anisotropy of this probe (Fig. S1E). Consequently, the experiments described below were carried out using the R550G744 protein, and, when appropriate, the calculated amounts of various species were corrected for the presence of 12% G550R744 in the closed conformation.
smFRET Demonstrates Open and Closed Conformations.
Single-molecule FRET studies of Pol I(KF), unliganded and in complexes with DNA and nucleotides, were performed on molecules diffusing freely through a femtoliter observation volume defined by a focused laser beam and confocal optics. Our experiments were performed using alternating-laser excitation (ALEX), which reports on the status of both donor and acceptor fluorophores by sorting molecules on the basis of relative donor:acceptor stoichiometry (S) and apparent FRET efficiency (E∗) (14, 15). The DNA substrate was a hairpin template-primer with A as the templating base (Fig. 1C). The 3′ terminus was a dideoxynucleotide, allowing formation of a ternary complex with an incoming dNTP, but preventing subsequent phosphoryl transfer and therefore restricting our observations to prechemistry species.
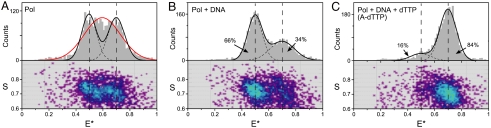
The FRET distributions obtained for Pol-DNA and Pol-DNA-dNTP complexes each revealed a biased bimodal distribution of molecular species with the majority species in each case having a FRET value consistent with the structurally defined open and closed complexes, respectively (Fig. 2B and C). In the Pol-DNA binary complex, the predominant species (66%) peaked at mean E∗ = 0.5, corresponding to the open conformation; nevertheless, the closed conformation was significantly represented even at a DNA concentration that should drive > 95% of Pol I(KF) into binary complexes. Addition of dTTP, complementary to the templating base, gave the closed ternary complex with mean E∗ = 0.7. The fraction of molecules in the closed conformation increased as a function of dNTP concentration, reflecting a Kd app of 0.2 μM (Fig. S4A and B). At saturating dTTP concentration, 84% of the molecules were in the closed conformation. This 5∶1 ratio of closed to open complexes considerably strengthens our previous estimate of the equilibrium across the fingers-closing step (6). Similar FRET species were observed using a DNA substrate with a template T (Fig. S4C and D), indicating that the FRET characteristics of Pol I(KF) complexes are independent of the identity of the templating base and incoming complementary dNTP.
Fig. 2.
FRET-related histograms for Pol I(KF) complexes. (A–C) FRET-related histograms for (A) unliganded R550G744 Pol I(KF), (B) the Pol-DNA binary complex, and (C) the Pol-DNA-dNTP ternary complex (with a correct dNTP, forming an A-dTTP pair). The numbers of bursts in A, B, and C are comparable (2731, 2908, and 2598 events, respectively). The binary complex was formed by adding 100 nM DNA to Pol I(KF); the ternary complex was formed by adding 100 nM DNA and 10 μM dTTP to Pol I(KF). Each sample is described by two histograms: The lower shows a two-dimensional histogram of probe stoichiometry, S, vs. apparent FRET efficiency, E∗, for each diffusing molecule that contains both G and R fluorophores (i.e., molecules with 0.6 < S < 0.9; see SI Materials and Methods and refs. 14, 31). The upper panel shows a one-dimensional E∗ histogram (80 bins) of the molecules in the lower panel. In (B) and (C), the E∗ distributions were fitted to double-Gaussian distributions (black solid lines, sum of Gaussians; dashed lines, individual Gaussians) using an iterative process (see SI Materials and Methods). The percentage of the population in each Gaussian (after correction for the contaminating G550R744 protein) is indicated. The two dashed vertical lines mark the mean E∗ values of the main subpopulations in the binary (open) and ternary (closed) complexes. For the unliganded Pol I(KF) in (A), the red line shows the best fit by a single Gaussian; the black dashed lines show a fit to a constrained double-Gaussian distribution with the means and standard-deviations of the Gaussian peaks set to equal those of the open (〈E∗〉 = 0.5, σSN = 0.054) and closed (〈E∗〉 = 0.7, σSN = 0.060) complexes determined from the data in (B) and (C); neither of these strategies provide a good fit to the experimental data (see text). The residuals of all fits are in Fig. S5.
Dynamic Behavior of the Unliganded Polymerase.
In contrast to the binary and ternary complexes, the unliganded Pol I(KF) exhibited a highly heterogeneous FRET distribution, suggestive of interconversions between open and closed conformations occurring at the millisecond timescale. In the smFRET experiments (Fig. 2A), unliganded Pol I(KF) gave a broad, flat FRET distribution with mean E∗ ≈ 0.6. If this distribution is fitted by a single Gaussian function, the fit is poor (red line; reduced χ2 = 125) and the distribution has a width (σdist = 0.134) that exceeds by > 2-fold the width obtained for the open and closed states. Static or dynamic heterogeneity of the unliganded Pol I(KF) could account for the excessive width of its E∗ distribution.
In the case of static heterogeneity, the broad distribution might be due to molecules with noninterconverting conformations corresponding to closely spaced E∗ values. However, fitting with a double-Gaussian function, with the means and widths of the individual Gaussians fixed to those of the open and the closed conformations (Fig. 2A, black solid line; reduced χ2 = 198), fails to account for the experimental data (see Fig. S5 for residuals); it underestimates the frequency of molecules with E∗ values in the middle part of the distribution and slightly overestimates the frequency of molecules with E∗ values on either side of the distribution. Thus static heterogeneity could explain the experimental data only if there was a third, and unique, species with E∗ intermediate between the open and closed complexes (and not resolvable given our current FRET resolution), or if the two noninterconverting species had E∗ values distinct from those of the open and closed complexes.
Dynamic heterogeneity can cause a broad E∗ distribution if each molecule interconverts between different E∗ states on a timescale close to its observation time (16). Monte-Carlo simulations of ALEX experiments (Fig. S6 and SI Materials and Methods) showed that the flat broad shape of the E∗ distribution in unliganded Pol I(KF) can be recapitulated if transitions between open and closed states equivalent to those seen in the binary and ternary complexes occur on a timescale similar to that of diffusion (with a diffusion time of 3 ms, as measured using fluorescence correlation spectroscopy [FCS]). In contrast, faster dynamics (e.g. 10-fold faster than diffusion through the confocal volume) result in Gaussian-like distributions with narrow widths, whereas slower dynamics (e.g. 10-fold slower than diffusion through the confocal volume) result in two distinct, well-resolved Gaussian distributions.
It is technically difficult to observe single-molecule dynamics around the 3 ms timescale: detection of diffusing single molecules in solution is generally limited to short observation times, whereas observations of surface-immobilized proteins are often complicated by surface-induced artifacts (17). By recording long confocal datasets in solution, we collected a population of long (≥8 ms) bursts corresponding to the small fraction of molecules that remained longer than average in the confocal volume. In the case of the unliganded Pol I(KF), many of these long bursts showed FRET fluctuations consistent with transitions between the open and closed states (Figs. 3A and S7), whereas the binary and ternary complexes remained primarily in a single conformational state (open for binary complexes, closed for ternary complexes) with occasional sampling of the less populated state (Fig. S7).
Fig. 3.
Real-time conformational dynamics of unliganded Pol I(KF). (A) Examples of timetraces of long fluorescence bursts (≥8 ms; for burst selection, see SI Materials and Methods) for a static-DNA control and unliganded Pol I(KF) molecules diffusing in solution (for a large gallery of bursts, see Fig. S7). The upper panel shows photon counts upon donor excitation in the donor (Green) and acceptor (Red) emission channels, as well as photon counts upon acceptor excitation in the acceptor emission channel (Gray), all partitioned in 0.5-ms time bins; the lower panel shows the corresponding E∗ traces. For the static-DNA burst shown, E∗ fluctuations display a standard-deviation (σE∗) approaching the shot-noise-limited standard-deviation of E∗ [σE∗SN; calculated as in (16)]; for the Pol I(KF) burst shown, E∗ fluctuations significantly exceed the shot-noise limit. (B) Standard-deviation analysis: simulations. Plots of E∗ and σE∗ for bursts of > 4 ms taken from simulated timetraces, selected based on FAex,Aem photons and using L = 30, M = 6, T = 0.5 ms; see SI Materials and Methods). Dotted parabola: shot-noise-limited σE∗ at different E∗ values for a 20-photon segment. A static sample displays a distribution with a mean σE∗ value matching the shot-noise limit. A two-state system (state one with E∗ = 0.5; state two with E∗ = 0.7) that fluctuates at a timescale (k1 = k-1 = 17 s-1) much slower than diffusion (τD = 3 ms) displays two shot-noise-limited distributions (Red contours); when the same system fluctuates around the timescale of diffusion (k1 = k-1 = 166 s-1), it displays an additional, wide population (centered at E∗ ≈ 0.6) with σE∗ that exceeds the shot-noise limit by 5–15%. A further 10-fold increase in the interconversion rate leads to a single, narrow distribution (centered at E∗ ∼ 0.6) with σE∗ that exceeds the shot-noise limit by ≈20%. (C) Standard-deviation analysis experiments. Burst analysis and display as in panel B. Unliganded Pol I(KF) displays a profile similar to the one predicted for interconversions occurring around the timescale of diffusion, whereas the rest of the samples appear static on that timescale. (D) Determination of interconversion rates using FCS on samples in polyacrylamide gels. (Top): donor autocorrelation [GDD(τ), Black line] and donor-acceptor cross correlation [GDA(τ), Red line] curves for unliganded Pol normalized such that the GDD(τ) curve falls between 0 and 1. The amplitude of the cross correlation curve is proportional to the relative concentration of doubly labeled species; thus, samples with different donor-acceptor concentrations have different maximum correlation amplitudes. (Bottom): ratios of GDD(τ) and GDA(τ) for unliganded Pol (Dark Gray) and a static-DNA control (Light Gray). The curve segments above 10 ms are dominated by noise (due to the low amplitude of the correlation curves) and were not fitted. While the control showed no dynamics, the unliganded Pol exhibited a large fluctuation that was fitted (Blue line) with a stretched exponential function with mean relaxation time of ≈3 ms (see SI Materials and Methods). (E) Same as in D, Bottom, but for binary and ternary complexes. No significant fluctuations were observed during the probed timescale.
A statistical approach to analyze FRET fluctuations within single bursts relies on calculating the standard-deviation of E∗ (σE∗) for each burst (18). Molecules that interconvert many times between conformational states during their confocal-volume transit produce highly time-varying E∗ values and, in turn, σE∗ values higher than those of static or slowly fluctuating samples (see Fig. 3B for simulations). Our experimental results for unliganded Pol I(KF) revealed a large fraction of molecules that exceed the static limit of σE∗ (“shot-noise limit”; dotted parabola, Fig. 3C and SI Materials and Methods), in qualitative agreement with simulations of a two-state system interconverting around the diffusion timescale (Fig. 3B; k1 = k-1 = 166 s-1). In contrast, the binary and ternary complexes (Fig. 3C) appear largely static within the observation timescale.
Additional support for millisecond-timescale dynamics in the unliganded Pol came from FCS studies in 5% nondenaturing polyacrylamide gels. These gels increased the mean diffusion time by ≈3-fold (to ≈8 ms), providing a longer time-window for observing dynamics by FCS in a nonperturbing fashion, even for large conformational changes (e.g., the folding/unfolding of a DNA hairpin, ref. 18). Using published procedures (19), we were able to separate the timescale of FRET fluctuations from the timescale of diffusion (a major complication in earlier FCS approaches) by fitting the ratio of correlation functions GDD(τ) and GDA(τ) to an exponential function that represents the FRET fluctuations (see SI Materials and Methods and ref. 18). Applying this method to unliganded Pol I(KF) showed a clear, large-amplitude FRET-fluctuation process that relaxes at ≈3 ms (Fig. 3D bottom, dark gray line). In contrast, the amplitude of FRET fluctuations for the binary and ternary complexes (Fig. 3E) was small, similar to that of a static-DNA control (Fig. 3D bottom, light gray line), reflecting the absence of dynamics within the probed timescale (1 μs–10 ms).
The ≈3 ms timescale obtained for unliganded Pol I(KF) using FCS is in excellent agreement with our simple two-state simulations (Figs. 3B and S6), in which an ≈3 ms relaxation time (equivalent to opening and closing rates of ≈166 s-1) gave the best approximation of the experimental results. Taken together, simulations, the standard-deviation analysis, and the FCS analysis, supported by time-trace data, all point to the conclusion that unliganded Pol I(KF) fluctuates between open and closed conformations on a timescale of ≈3 ms. By contrast, the apparent absence of dynamics in the ternary complex is consistent with the opening and closing rates provided by our ensemble stopped-flow experiments, which showed that the rate of fingers-closing at saturating dNTP is 140 s-1 (6). Using this rate, and the observed 5∶1 equilibrium in favor of the closed conformation (Fig. 2C), we calculate an opening rate of ≈30 s-1.
Complexes with Mispaired dNTPs and with Ribonucleotides.
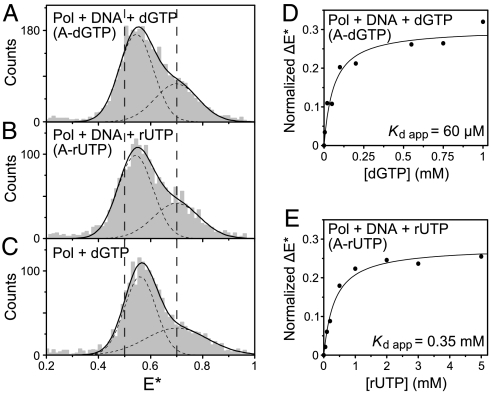
An essential characteristic of DNA polymerases is their ability to reject mismatched dNTPs. Our smFRET experiments suggest that discrimination against mismatches involves a unique ternary complex species, illustrated by the complex containing an A-dGTP mispair (Fig. 4A). The mismatched species had a mean E∗ of ≈0.55 (Fig. 4A), a value only slightly greater than that of the open binary complex (0.50; Fig. 2B) so that there was no clear separation between the mispair ternary complex and the open binary complex in the FRET histograms. Instead, there was a gradual shift in the mean E∗ of the lower-FRET peak, which showed saturation behavior as a function of nucleotide concentration (Fig. 4D and Fig. S8A). The saturation position of the peak was at ≈25% of the FRET difference between the open and closed complexes, indicating that the mispair ternary complex is distinct from the open and closed complexes described earlier. We observed similar FRET changes with other mispairs, including a T-dTTP mispair (Fig. S8C) where the steric constraints on formation of the closed complex should be less severe than for A-dGTP. Interestingly, Pol I(KF) complexed with a dNTP in the absence of DNA gave a species with similar E∗ to that of the mispair ternary complex (Fig. 4C), implying that Pol-DNA-dNTP(mis) complexes might resemble the open Pol-dNTP complex that has been observed in cocrystals (20, 21).
Fig. 4.
Novel FRET species observed for Pol I(KF) ternary complexes with incorrect substrates. (A–C) E∗ histograms of the doubly labeled molecules (0.6 < S < 0.9, as in Fig. 2) in (A) a Pol-DNA-dNTP ternary complex with mispaired A-dGTP (1 mM dGTP), (B) a Pol-DNA-rNTP ternary complex with complementary A-rUTP (1 mM UTP), and (C) a binary Pol-dNTP complex (lacking DNA) with 1 mM dGTP. In (A) and (B), the DNA (Fig. 1C) was present at 100 nM; control experiments (Fig. S8D) established that DNA was present in these complexes. The vertical dashed lines correspond to the mean E∗ for the open (E∗ = 0.5) and closed (E∗ = 0.7) conformations that predominate in the binary and matched ternary complexes, respectively (Fig. 2). The E∗ histograms were fitted to double-Gaussian distributions (solid black lines, sum of Gaussians; dashed lines, individual Gaussians). In all cases, the fit of the lower-E∗ subpopulation was unconstrained, whereas the mean of the higher-E∗ subpopulation (which gives rise to a shoulder) was fixed at E∗ = 0.7, corresponding to the closed conformation. At high nucleotide concentration (1 mM), all three experiments showed a shift of the mean of the lower-E∗ peak to a position at ≈25% of the difference between the mean E∗ values of the open and closed conformations. (D) Nucleotide dependence of the E∗ shift for the A-dGTP mispaired ternary complex. See Fig. S8A for the E∗ histograms at each dGTP concentration. The shift of the mean of the lower-E∗ peak was normalized relative to the E∗ difference between the means of the open and closed conformations. Normalized ΔE∗ was plotted as a function of nucleotide concentration, and an apparent equilibrium dissociation constant Kd app was obtained by fitting to a hyperbolic function. (E) As in (D), but for the A-rUTP ternary complex. See Fig. S8B for the E∗ histograms at each rUTP concentration.
In addition to discriminating against mismatches, DNA polymerases also select for the correct sugar structure, choosing dNTPs over rNTPs by a factor of ≈104 (22). Like the A-dGTP ternary complex described above, an A-rUTP complex, containing a complementary ribonucleotide, showed the presence of a unique FRET species distinct from the open and closed conformations (Fig. 4B). The E∗ value of the A-rUTP species, and its saturation behavior as a function of UTP concentration (Fig. 4E and Fig. S8B) resembled that of the mismatched ternary complex.
Despite the similar E∗ values, we suspect that the mispair and ribonucleotide ternary complexes are distinct from one another, based on our ensemble stopped-flow studies showing that rNTPs and mispairs are detected at different steps of the reaction pathway. Incorporation of complementary rNTPs by Pol I(KF) is blocked by steric interference in the closed complex between the ribose 2′-OH and a highly conserved side chain (Glu710) (23). This steric clash should develop only as fingers-closing proceeds, consistent with the stopped-flow data showing that an rNTP substrate diminished both the rate and the extent of fingers-closing (6). Thus, it is likely that the Pol-DNA-rNTP complex corresponds to a partially closed conformation limited by steric constraints, and the inability of this complex to proceed further along the reaction pathway results in rejection of rNTPs. By contrast, ternary complexes containing mispaired dNTPs are blocked at or before the DNA rearrangement step that precedes fingers-closing (6, 8). This implies that Pol I(KF) can detect a mispair without accessing the fully closed conformation and is consistent with the absence of a FRET change in the mispair ternary complexes that we have examined using an AEDANS-Dabcyl FRET pair in our ensemble stopped-flow assay for fingers-closing (6). We suggest, therefore, that the FRET pair used in the current study is able to detect a rearrangement, occurring on binding a mispaired dNTP, that is distinct from fingers-closing.
Discussion
The smFRET technique has provided a unique window on the DNA polymerase reaction pathway, revealing molecular species and conformational dynamics that would be difficult to detect by other means. Crystallographic data for A-family and other high-fidelity DNA polymerases fostered the expectation that Pol-DNA binary complexes would populate exclusively the open conformation and Pol-DNA-dNTP ternary complexes the closed conformation. By measuring the population of FRET states in solution, unconstrained by the requirement for molecular uniformity in a crystal, smFRET demonstrates that both binary and ternary complexes populate the less favored conformation to a significant extent. Our data imply that the binary complex favors the open conformation by ≈0.4 kcal mol-1, and the ternary complex favors the closed conformation by ≈1 kcal mol-1. Rather more unexpected were the conformational heterogeneity and rapid dynamics seen in the unliganded polymerase, contrasting with crystal structures which showed unliganded DNA polymerases in the open conformation characteristic of the corresponding Pol-DNA complexes (24–28). Our data strongly suggest that unliganded Pol I(KF) can access the full range of motion required for its reaction mechanism, consistent with the hypothesis, developed from observations on several different enzymes, that conformational motions employed in enzyme catalysis are intrinsic to the unliganded enzyme (9–12). The unliganded motions of Pol I(KF) appear to be faster than the prechemistry rate-limiting step (6, 29); this may play an important role in the fast rejection of the large number of incorrect nucleotide substrates encountered by a DNA polymerase in vivo.
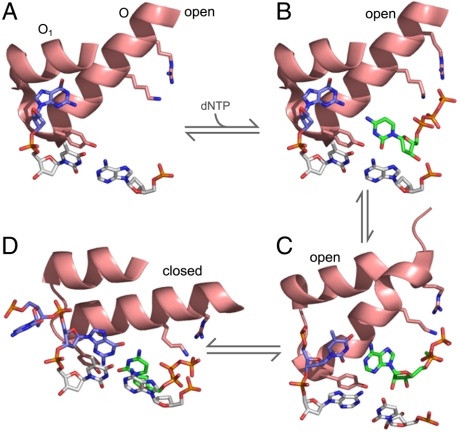
The smFRET approach also enabled us to detect the subtle FRET changes that accompany the binding of ribonucleotides or dNTP mispairs. An intriguing possibility is that the unique molecular species we have observed may provide clues as to the nature of the steps on the reaction pathway that constitute the kinetic checkpoints that discriminate against these inappropriate substrates. With this in mind, we have used known cocrystal structures to derive a plausible sequence of active site structures describing the processes from dNTP binding to fingers-closing (Fig. 5). A and D correspond to the open binary and closed ternary complexes that define the start and finish of the sequence. B shows a possible model for a dNTP entering the Pol-DNA binary complex, derived by combining the structure of a Pol-dNTP complex with the Pol-DNA complex of A. The interactions between the triphosphate moiety of the nucleotide and conserved positively charged side chains on the O-helix may result in a subtle rearrangement of the open conformation, corresponding to the slightly higher FRET of the Pol-dNTP binary complex, compared with the open Pol-DNA binary complex. C, derived from a cocrystal structure of T7 RNA polymerase (30), a structural homologue of DNA polymerase I, is a good candidate for the kinetic checkpoint that discriminates against mismatched dNTPs because it provides a mechanism for testing an incoming nucleotide for complementarity to the templating base within the open conformation. It appears that this structure could be reached with minimal rearrangement of the proposed initial dNTP-bound species (B), consistent with the similar FRET characteristics of Pol-dNTP and Pol-DNA-dNTP(mis) complexes. Specificity would be provided by the failure of a mispair to proceed beyond this stage, whereas a correct template-dNTP pair could be smoothly delivered into the active site by movement of the O-helix into the closed position (D). While the actual intermediates on the reaction pathway may differ significantly from those captured in cocrystal structures, this hypothetical scheme provides a useful starting point for integrating structural and mechanistic aspects of DNA polymerase specificity.
Fig. 5.
Proposed reaction pathway for the prechemistry steps at the polymerase active site, based on cocrystal structures. In (A)–(D), the structures are reduced to the minimal elements that illustrate important features of the reaction. These comprise the O-helix or its structural homologue (which moves during fingers-closing), part of the O1 helix (not mobile), the invariant Tyr at the C terminus of the O-helix, conserved Lys and Arg side chains that interact with the dNTP phosphates, the terminal base pair (Gray) of the primer template, the unpaired base(s) on the template strand (Blue), and the incoming dNTP (Green). (A) The Pol-DNA binary complex of Bst Pol (PDB file 1L3U) represents the start of the reaction. The O-helix is in the open conformation, Tyr is stacked on the template base of the terminal base pair and the next templating base is in a pocket between the O and O1 helices (3). (B) A model for the initial complex when a dNTP associates with the Pol-DNA complex, generated by aligning the structure in (A) with a structure of Klentaq with a bound dNTP in the absence of DNA (PDB file 5KTQ) (20). The positions of the dCTP molecule and the Lys and Arg side chains were based on the Klentaq coordinates. (C) A plausible candidate for the complex in which the incoming nucleotide is tested for complementarity with the templating base, provided by a cocrystal of T7 RNA polymerase (a structural homologue of the A-family DNA polymerases; PDB file 1SOV). This structure is remarkable in that it shows base pairing between the templating base and an incoming nucleotide outside of the active site pocket, while the O-helix, terminal base pair and Tyr side chain maintain positions characteristic of the open complex (30). The nucleotide and templating base have moved only slightly from their positions in (B). (D) The closed Pol-DNA-dNTP complex of Bst Pol (PDB file 1LV5) represents the end of the sequence, with the reactants poised for catalysis (3); this step is the checkpoint for the rejection of rNTPs. Compared to (A), rotation of the O-helix and downwards movement of the Tyr side chain have created a binding pocket for the nascent base pair.
Materials and Methods
The following procedures are described in SI Materials and Methods: preparation of proteins and DNA, labeling of Pol I(KF) derivatives, single-molecule confocal experiments, and in-gel FCS experiments.
Supplementary Material
Acknowledgments.
We thank Anna M. Pyle for the use of her stopped-flow instrument, Christina Grindley for making the K550C mutation, and Aleš Benda for help with Monte-Carlo simulations. The work was supported by a UK Engineering and Physical Sciences Research Council Grant EP/D058775 (to A.N.K.) , theEuropean Union’s 7th Framework Programme Grant HEALTH-F4-2008-201418, entitled READNA, (to.A.N.K.), an E.P. Abraham Cephalosporin Scholarship (Linacre College, University of Oxford; to Y.S.), and National Institutes of Health Grant GM-28550 (to C.M.J., O.P., and N.D.F G). N.D.F.G. thanks Brasenose College, University of Oxford, for a visiting fellowship for the 2007-8 academic year.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910909107/DCSupplemental.
References
- 1.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of Thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doublié S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure. 1999;7:R31–R35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 5.Rothwell PJ, Mitaksov V, Waksman G. Motions of the fingers subdomain of Klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol Cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 6.Joyce CM, et al. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008;47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 7.Tsai Y-C, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purohit V, Grindley NDF, Joyce CM. Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment) Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 9.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 10.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 11.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 12.Hanson JA, et al. Illuminating the mechanistic roles of enzyme conformational dynamics. Proc Natl Acad Sci USA. 2007;104:18055–18060. doi: 10.1073/pnas.0708600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mekler V, et al. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 14.Kapanidis AN, et al. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee NK, et al. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys J. 2005;88:2939–2953. doi: 10.1529/biophysj.104.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nir E, et al. Shot-noise limited single-molecule FRET histograms: Comparison between theory and experiments. J PhysChem B. 2006;110:22103–22124. doi: 10.1021/jp063483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung HS, Louis JM, Eaton WA. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc Natl Acad Sci USA. 2009;106:11837–11844. doi: 10.1073/pnas.0901178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santoso Y, Kapanidis AN. Probing biomolecular structures and dynamics of single molecules using in-gel alternating-laser excitation. Anal Chem. 2009;81:9561–9570. doi: 10.1021/ac901423e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres T, Levitus M. Measuring conformational dynamics: A new FCS-FRET approach. J PhysChem B. 2007;111:7392–7400. doi: 10.1021/jp070659s. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Kong Y, Korolev S, Waksman G. Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates. Protein Sci. 1998;7:1116–1123. doi: 10.1002/pro.5560070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beese LS, Friedman JM, Steitz TA. Crystal structures of the Klenow fragment of DNA polymerase I complexed with deoxynucleoside triphosphate and pyrophosphate. Biochemistry. 1993;32:14095–14101. doi: 10.1021/bi00214a004. [DOI] [PubMed] [Google Scholar]
- 22.DeLucia AM, Grindley NDF, Joyce CM. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res. 2003;31:4129–4137. doi: 10.1093/nar/gkg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astatke M, Ng K, Grindley NDF, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc Natl Acad Sci USA. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 25.Kiefer JR, et al. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 Å resolution. Structure. 5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 26.Korolev S, Nayal M, Barnes WM, Di Cera E, Waksman G. Crystal structure of the large fragment of Thermus aquaticus DNA polymerase I at 2.5-Å resolution: Structural basis for thermostability. Proc Natl Acad Sci USA. 1995;92:9264–9268. doi: 10.1073/pnas.92.20.9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase β: Evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 28.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 29.Dahlberg ME, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow fragment): Identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 30.Temiakov D, et al. Structural basis for substrate selection by T7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 31.Kapanidis AN, et al. Retention of transcription initiation factor σ70 in transcription elongation: Single-molecule analysis. Mol Cell. 2005;20:347–356. doi: 10.1016/j.molcel.2005.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.