Abstract
Transthyretin aggregation-associated proteotoxicity appears to cause several human amyloid diseases. Rate-limiting tetramer dissociation and monomer misfolding of transthyretin (TTR) occur before its aggregation into cross-β-sheet amyloid fibrils. Small molecule binding to and preferential stabilization of the tetrameric state of TTR over the dissociative transition state raises the kinetic barrier for dissociation, imposing kinetic stabilization on TTR and preventing aggregation. This is an effective strategy to halt neurodegeneration associated with polyneuropathy, according to recent placebo-controlled clinical trial results. In three recent papers, we systematically ranked possibilities for the three substructures composing a typical TTR kinetic stabilizer, using fibril inhibition potency and plasma TTR binding selectivity data. Herein, we have successfully employed a substructure combination strategy to use these data to develop potent and selective TTR kinetic stabilizers that rescue cells from the cytotoxic effects of TTR amyloidogenesis. Of the 92 stilbene and dihydrostilbene analogues synthesized, nearly all potently inhibit TTR fibril formation. Seventeen of these exhibit a binding stoichiometry of > 1.5 of a maximum of 2 to plasma TTR, while displaying minimal binding to the thyroid hormone receptor (< 20%). Six analogues were definitively categorized as kinetic stabilizers by evaluating dissociation time-courses. High resolution TTR•(kinetic stabilizer)2 crystal structures (1.31-1.70 Å) confirmed the anticipated binding orientation of the 3,5-dibromo-4-hydroxyphenyl substructure and revealed a strong preference of the isosteric 3,5-dibromo-4-aminophenyl substructure to bind to the inner thyroxine binding pocket.
Introduction
Intrinsic and/or extrinsic challenges to the maintenance of organismal protein homeostasis, in the absence of a biological correction (e.g., induction of a stress-responsive signaling pathway) or a chemical correction (a small molecule that binds to and stabilizes a particular misfolding-prone protein) to rebalance the proteostasis network, can lead to aging-associated proteotoxicity and degenerative diseases.1-5 These include Alzheimer's disease, as well as the transthyretin and gelsolin amyloidoses.3,5-10 These maladies are associated with the accumulation of an insoluble protein(s), including amyloid fibrils after which these diseases have been named, leading to the degeneration of one or more tissues – often those composed of post-mitotic cells, such as neurons or muscle cells.3,6,10 Whether intra- or extracellular aggregates lead to degeneration, which aggregate morphology is responsible, and by what mechanism are key unanswered questions related to the amyloidoses.3-11
Transthyretin (TTR) is a homotetrameric protein composed of 127-amino-acid, β-sheet-rich subunits.12-15 The established physiological functions of TTR are to bind to and transport the thyroid hormone thyroxine (T4) and holo-retinol binding protein in the blood and cerebrospinal fluid (CSF).11,12,16-18 Although TTR serves as the major carrier of thyroxine in the CSF, TTR is a minor carrier in blood because of the presence of two other T4 carrier proteins, thyroid binding globulin and albumin. Thus, more than 99% of TTR's thyroxine binding sites in the blood are unoccupied.11
Transthyretin is one of more than 30 non-homologous human amyloidogenic proteins, whose misfolding and/or misassembly appears to elicit the proteotoxicity and cell degeneration thought to cause the amyloidoses.4,7,11,19 Amyloidogenesis from TTR secreted by the liver appears to require rate-limiting tetramer dissociation, which affords non-amyloidogenic folded monomers that must undergo partial denaturation to misassemble into a variety of aggregate structures, including cross-β-sheet amyloid fibrils.20-25 TTR amyloidogenesis occurs by a thermodynamically favorable or downhill aggregation reaction, and not by a nucleated polymerization that governs many other amyloidogenesis processes.26 Amyloidogenesis could also compete with TTR monomer folding in the endoplasmic reticulum of hepatocytes, although this source of proteotoxicity is still under debate. The demonstrated effectiveness of a kinetic stabilizer in a placebo-controlled clinical trial for polyneuropathy suggests that dissociation of the TTR tetramer is the predominate process that leads to TTR proteotoxicity.4 Accumulation of either wild type (WT) TTR or mutant TTR aggregates outside of cells, and possibly later inside of certain cells, appears to cause the neurodegeneration and/or organ degeneration characteristic of the TTR amyloidoses.
Amyloidogenesis of WT-TTR within the heart leads to the sporadic amyloid disease senile systemic amyloidosis (SSA) – a late onset cardiomyopathy that affects up to 20% of the aged population.27-29 Familial amyloid cardiomyopathy (FAC) appears to be caused by the deposition of one of a few TTR mutants within the heart, the most common variant deposited being V122I-TTR, a mutation found in 3-4% of African Americans that appears to confer complete penetrance of FAC.30,31 Both SSA and FAC result from TTR proteotoxicity in trans, as heart tissue does not synthesize TTR.
Amyloidogenesis of one of over 100 thermodynamically less stable variants of TTR4,32,33 appears to cause familial amyloid polyneuropathy (FAP), a peripheral neuropathy, often exhibiting autonomic nervous system involvement.34 The most common FAP variant is V30M-TTR, affecting largely the Portuguese, Swedish and Japanese populations.35-37 Central nervous system selective amyloidosis (CNSA) appears to be a rarer disease associated with the deposition of the most destabilized TTR variants (e.g., A25T- and D18G-TTR) in the brain, but not in the periphery.4,38,39 The choroid plexus secreting these variants into the CSF appears to be a more permissive secretor of unstable and misfolding-prone TTR variants than the liver, which extensively degrades these highly destabilized variants instead of secreting them into the blood, explaining why the peripheral tissues are not subjected to A25T- and D18G-TTR amyloidogenesis.4,38-40
There are currently no FDA-approved drugs or accepted therapeutic strategies for treating SSA and FAC. The only therapeutic strategy currently being practiced for ameliorating FAP is gene therapy mediated by liver transplantation, wherein the disease-associated TTR variant producing liver is replaced by a WT-TTR producing liver, substantially reducing disease-associated variant TTR levels in the blood.41-43 The drawbacks of utilizing liver transplantation to treat FAP include the shortage of livers, the significant surgical risk, life-long immune suppression requirements, and the expense. In addition, WT-TTR continues to deposit in the heart post-transplantation, leading to cardiomyopathy, suggesting an aging-associated decline in the proteostasis capacity of the heart.1,2,44-46 Because of the limited applicability of liver transplantation to treat the TTR amyloidoses (not applicable to SSA – the disease with the most patients) and especially the risk of death from transplant complications, it is highly desirable to develop a general chemotherapeutic strategy for ameliorating the TTR amyloidoses.11,47,48 Towards this end, we have been developing kinetic stabilizers of TTR, small molecules that differentially stabilize the native tetrameric structure of TTR over the dissociative transition state, making the barrier for TTR tetramer dissociation too high to surmount under physiological conditions – thus preventing TTR amyloidogenesis.5,11,49-56 A phase II/III placebo-controlled clinical trial has recently been completed by FoldRx Pharmaceuticals for one of the kinetic stabilizers synthesized by our laboratory, demonstrating that it halts neurodegeneration (www.foldrx.com). The demonstrated utility of a TTR kinetic stabilizer to ameliorate a TTR amyloid disease is further supported by human genetic evidence – namely, that the incorporation of T119M-TTR trans-suppressor subunits into TTR heterotetramers otherwise composed of FAP-associated subunits kinetically stabilizes the TTR tetramer and ameliorates FAP amyloidogenesis in Portuguese compound heterozygotes.5,36,49
The energetically weaker dimer-dimer interface of tetrameric TTR,54 bisected by the crystallographic C2 axis (Figure 1A), creates the two funnel-shaped T4 binding pockets.11-14,17,57 To date, we have synthesized over one thousand small molecules that exhibit structural complementarity to the T4 binding sites within TTR and bind with a range of affinities and cooperativities.5,11,15,47,48,52,55-72 We have recently conducted a systematic three-part study to optimize the three substructures comprising a typical TTR kinetic stabilizer, the two aromatic rings (aryl-X and aryl-Z) and the linker-Y connecting them (Figure 1B).58-60 In each paper, we rank-ordered the numerous possibilities for one substructure, based on their ability to inhibit TTR fibril formation and to bind selectively to TTR in human plasma, the remainder of the structure being constant. We also evaluated the highly ranked substructures for their ability to bind to the thyroid hormone receptor or cyclooxygenase.58-60 We created a so-called efficacy score, eq. 1 (discussed in detail below) integrating TTR amyloid inhibition efficacy data (dependent on the extent of kinetic stabilization of TTR) and plasma TTR binding stoichiometry data into one score.
Figure 1.
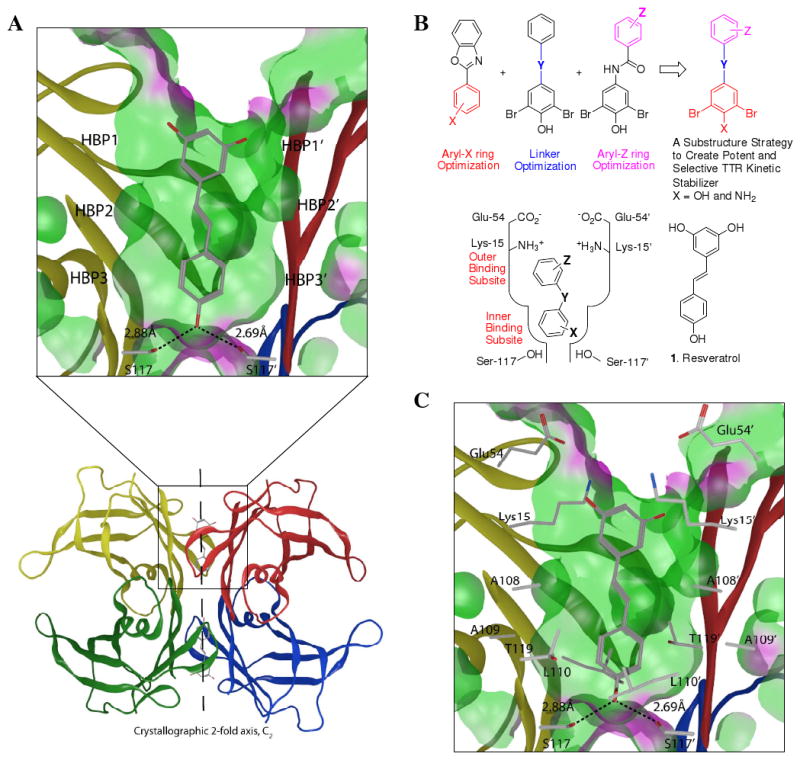
Substructure combination strategy to identify potent and selective TTR kinetic stabilizers. (A) Ribbon diagram depiction of the crystal structure (PDB accession code 1DVS) of resveratrol (1) bound to the two thyroxine (T4) binding pockets within the WT-TTR tetramer, with the monomer subunits individually colored. The top portion represents an expanded view of one T4 binding site with 1 bound with a ‘Connelly’ analytical molecular surface applied to residues within 8Å of ligand in the T4 binding pocket (green = hydrophobic, purple = polar). (B) Schematic depiction of the substructure combination strategy to create TTR kinetic stabilizers (top). The aryl-X and aryl-Z rings as well as the linker-Y can be varied to generate candidate TTR kinetic stabilizers. The most highly ranked aryl-X, aryl-Z and linker-Y substructures from previous studies, based on potency and plasma TTR binding stoichiometry, are combined in this substructure combination strategy to create potent and selective TTR kinetic stabilizers that nearly eliminate amyloidogenesis associated cytotoxicity. The inner and outer T4 binding subsites within one schematically represented TTR T4 binding site are labeled in red font. (C) The innermost halogen binding pockets (HBPs) 3 and 3′ (labeled in A) are composed of the methyl and methylene groups of Ser117/117′, Thr119/119′, and Leu110/110′. HBPs 2 and 2′ (labeled in A) are made up by the side chains of Leu110/110′, Ala109/109′, Lys15/15′ and Leu17/17′. The outermost HBPs 1 and 1′ (labeled in A) are lined by the methyl and methylene groups of Lys15/15′, Ala108/108′ and Thr106/106′. Figure generated using the program MOE (2006.08), Chemical Computing Group, Montreal, Canada.
| (eq. 1) |
While structure-based drug design principles were not employed implicitly in this studies, we were aware that one of the most highly ranked aryl-X substructures strongly prefers to bind in the outer thyroxine binding subsite (Figure 1B, bottom left) for reasons outlined below. In addition, we knew that the trans −CH=CH− and −CH2-CH2− linkers maximized the hydrophobic effect upon binding to TTR. Numerous TTR•(kinetic stabilizer)2 crystal structures reveal the key molecular interactions required for high affinity binding to the two T4 sites.57-60,62-67 Pertinent to this study, aryl-X rings bearing a 4-hydroxyl substituent flanked by bromides strongly prefer to bind in the outer binding subsite, bridging subunits by making salt bridging interactions with the Lys-15/15′ ε-ammonium groups and by maximizing occupancy of halogen binding pockets (HBPs) 1 and 1′.58 Electron withdrawing substituents at the 3 and 5 positions (e.g., Br, Cl) flanking a 4-hyrdoxl group results in phenolate formation (pKa lowering) at neutral pH.58 Benzoxazole, biphenyl, biphenylether, and biphenylamide TTR co-crystal structures all reveal that the 3,5-dibromo-4-hydroxylphenyl substructure comprising them strongly prefers to be placed in the outer binding site to make K15 salt bridges.59,60 Hence, we expect the 3,5-dibromo-4-hydroxyphenyl ring comprising the stilbenes and dihydrostilbenes used in this study to occupy the outer T4 binding pocket. Moreover, we expect the trans double bond linker and the −CH2-CH2− linker used in this exercise to interact with the hydrophobic side chains of Leu-17/17′, Ala-108/108′, Leu-110/110′ and Val-121/121′, maximizing the hydrophobic effect and thus binding affinity, based on crystal structures of WT-TTR•(Resveratrol (1))257 (Figure 1A and C) and the complexes between WT-TTR and other stilbenes having only one of the two aromatic rings bearing substituents.59
Based on the data from the substructure optimization studies,58-60 we tested the hypothesis that we could produce a small molecule library that is rich in potent and highly selective kinetic stabilizers by simply combining the most highly ranked substructures. We utilized the highly ranked 3,5-dibromo-4-hydroxy substituted aryl-X ring and isosteric variants thereof (replacing the hydroxyl group with functional groups like NH2), a variety of highly ranked aryl-Z rings, as well as desirable hydrophobic linkers (trans −CH=CH− and −CH2CH2−) to demonstrate that the substructure combination strategy affords a library with a high proportion of potent and highly selective TTR kinetic stabilizers / amyloidogenesis inhibitors. Structural studies reinforce prior data on the binding preference of the 3,5-dibromo-4-hydroxyphenyl substructure to the outer thyroxine binding subsite58-60 and, importantly, reveal the strong preference of the 3,5-dibromo-4-aminophenyl substructure to bind to the inner thyroxine binding subsite. The ability of all the TTR kinetic stabilizers tested in this study to nearly eliminate TTR-induced cytotoxicity in the recently developed cell culture assay suggests that they merit further study as potential drug candidates to ameliorate the human TTR amyloidoses.
Results
Having established previously that a stilbene comprising one 3,5-dibromo-4-hydroxyphenyl ring and one unsubstituted aryl ring exhibits a binding stoichiometry to TTR of 1.5 out of a maxima of 2 in human plasma and completely inhibits TTR amyloidogenesis (7.2 μM59; see below), herein we first set out to optimize the structure of stilbene-based TTR kinetic stabilizers using previously determined aryl-Z ring efficacy scores to guide this substructure combination strategy. Secondly, we used the substructure combination strategy to evaluate the highly ranked −CH2-CH2− linker, readily synthesized by hydrogenation of the double bond of the stilbenes. Thirdly, we used the substructure combination strategy to demonstrate that by further optimizing the aryl-X ring we could remove an off-target binding activity. The chemical purity of all compounds used in this study was ≥ 95 %, Figures S1-S3.
Inhibition of Acid-mediated WT-TTR Amyloid Formation by Candidate Kinetic Stabilizers
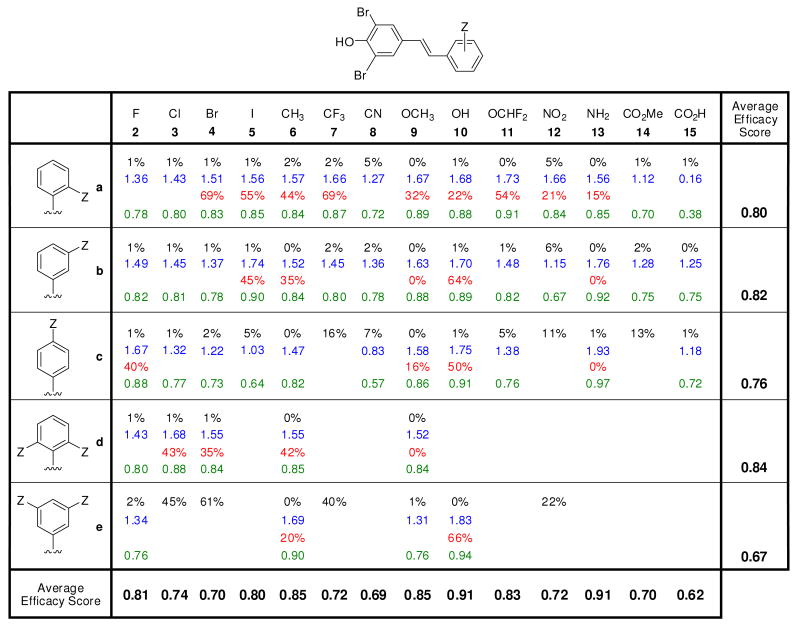
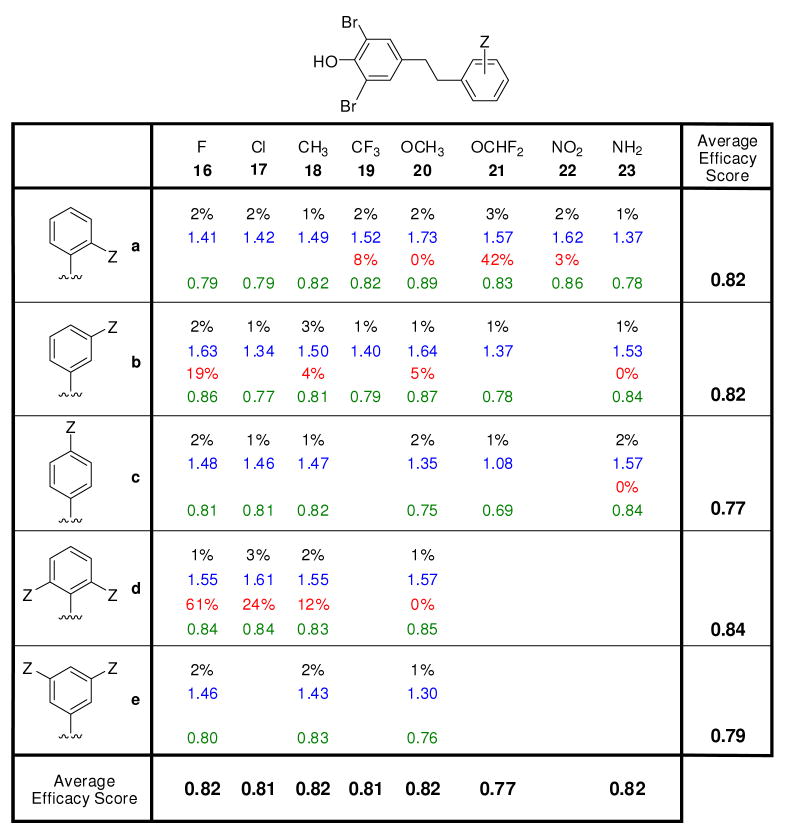
The efficacy with which stilbene and dihydrostilbene library members inhibit WT-TTR amyloidogenesis was evaluated using the previously established acid-mediated fibril formation assay.20,58-60 Briefly, each compound (7.2 μM, the minimum concentration required to occupy both T4 binding sites) was preincubated with a physiologically relevant concentration of WT-TTR (3.6 μM) for 30 min and TTR amyloidogenesis was initiated by adjusting the pH to 4.4. The extent of TTR fibril formation was quantified by measuring turbidity, previously shown to be equivalent to thioflavin T quantification. At this pH, a 90% yield of TTR fibril formation is observed after a 72 h incubation period in the absence of a TTR kinetic stabilizer. TTR amyloidogenesis in the presence of a candidate kinetic stabilizer was expressed as a percentage relative to that exhibited by WT-TTR in the absence of inhibitor (assigned to be 100 %). Potent TTR kinetic stabilizers allow < 10% of WT-TTR fibril formation at a concentration of 7.2 μM and < 40% of WT-TTR fibril formation at a concentration equal to that of WT-TTR (3.6 μM) (0% fibril formation = 100% inhibition).5,15,55,56,58-60,62,64-67,69-72 Of the 88 3,5-dibromo-4-hydroxyphenyl–based stilbene- and dihydrostilbene kinetic stabilizers synthesized and evaluated here, 77 are excellent amyloidogenesis inhibitors, allowing < 10% of WT-TTR fibril formation at a concentration of 7.2 μM (> 90% inhibition, Figures 2-4, black font = % fibril formation). Compounds exhibiting more than 95% inhibition were reevaluated at a concentration equal to the TTR tetramer (3.6 μM). At this concentration, the library of compounds allows between 15–30% of WT-TTR fibril formation (Figures S4-S6), demonstrating that the substructure combination strategy is capable of producing potent TTR kinetic stabilizers.
Figure 2.
Evaluation of the potency and selectivity of stilbene-based TTR kinetic stabilizers. Percent (%) fibril formation (F.F.) values are in black font representing the extent of in vitro WT-TTR (3.6 μM) fibril formation in the presence of inhibitor (7.2 μM) relative to aggregation in the absence of inhibitor (assigned to be 100%). The TTR tetramer binding stoichiometry of potent aggregation inhibitors (defined as those exhibiting < 10% F.F.) bound to human plasma TTR ex vivo are shown in blue font (10.8 μM kinetic stabilizer concentration, maximum binding stoichiometry is 2 due to the two thyroxine binding sites per TTR tetramer). Extent of competitive binding of potent (based on % F.F. as defined above) and highly selective (defined as those exhibiting a human plasma TTR binding stoichiometry > 1.5) TTR kinetic stabilizers to the thyroid hormone receptor is shown in red font. Individual efficacy scores (as defined by eq. 2) of TTR kinetic stabilizers are shown in green font, whereas average efficacy scores (defined by eq. 1) are shown at the bottom of the columns (reflecting the average value in a column) and at the right side of the rows, reflecting the average value of the compounds in that row.
Figure 4.
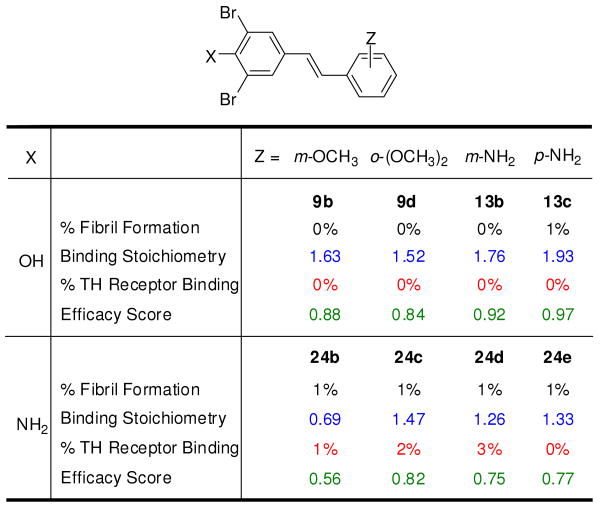
Effect of replacement of a hydroxyl group by an amino group in selected TTR kinetic stabilizers. Percent fibril formation (% F.F.) values (black font), plasma TTR binding stoichiometry (blue font), T3 displacement from thyroid hormone receptor (red font) as well as individual efficacy scores (green font) are shown, precisely as defined in Figure 2, with the exception that average efficacy scores are not shown.
Binding Stoichiometry of Potent Kinetic Stabilizers to TTR in Human Blood Plasma
The 77 stilbene and dihydrostilbene kinetic stabilizers that allowed < 10% of WT-TTR fibril formation in vitro were further evaluated for their ability to bind selectively to TTR in human blood plasma (3.6 – 5.4 μM) using the established ex vivo TTR plasma binding selectivity assay.73 Briefly, the candidate kinetic stabilizer (10.8 μM) was incubated with human blood plasma at 37 °C for 24 h. Any unbound inhibitors, endogenous small molecules, or macromolecules that could bind to the resin used in the next step were removed by addition of quenched sepharose resin.73 The TTR and TTR•(kinetic stabilizer)n complexes in the plasma were immunocaptured using a sepharose-resin-conjugated anti-TTR antibody. After extensive washing, TTR and any TTR-small molecule complexes bound to the resin were dissociated by high pH treatment. The ratio of TTR monomer to bound candidate kinetic stabilizer was calculated from HPLC peak areas using standard curves.73 The numbers in blue font in Figures 2-4 represent the average candidate kinetic stabilizer binding stoichiometry to TTR in human blood plasma, the maximum being 2, due to two thyroxine binding sites per TTR tetramer. Of the 77 potent (< 10% fibril formation) TTR aggregation inhibitors evaluated, only three kinetic stabilizers display modest binding stoichiometry (< 1 equivalent bound) whereas 74 inhibitors exhibit average binding stoichiometries exceeding 1 equivalent bound per tetramer. Notably, 37 of these exhibit exceptional binding selectivity to plasma TTR, exhibiting > 1.5 equivalents bound per TTR tetramer. It is reassuring that the substructure combination strategy yields stilbenes and dihydrostilbenes that generally exhibit very high individual efficacy scores (eq. 2), reflecting desirable amyloid inhibition efficacy and plasma TTR binding selectivity (Figures 2-4, numbers in green font, 1 being a perfect kinetic stabilizer).
| (eq. 2) |
Average efficacy scores (eq. 1) are also depicted in bold black font in Figures 2 and 3 in the bottom row and the rightmost column. In this calculation, we average % fibril formation (% F.F.ave) and plasma TTR binding stoichiometry (P.S.ave) values in each column where the substituent(s) is constant but the positioning on the aryl ring varies and in each row where the substituent(s) vary but the positioning on the aryl ring is constant.
Figure 3.
Evaluation of the potency and selectivity of dihydrostilbene-based WT-TTR kinetic stabilizers. This figure is organized and defined strictly analogous to the descriptions in the legend to Figure 2.
Evaluating TTR Kinetic Stabilizers for Thyroid Hormone Receptor Binding
Because the majority of the members of our stilbene and dihydrostilbene library are composed of the 3,5-dibromo-4-hydroxyphenyl thyroxine-like substructure, we were concerned about their binding selectivity to TTR over the thyroid hormone receptor. Thus, TTR kinetic stabilizers were evaluated for their ability to bind to the thyroid hormone receptor [analyses carried out by Cerep laboratories in Redmond, WA (see experimental section for details)]. The results are expressed as percent displacement of the control radioligand (125I-labeled triiodothyronine, [125I]−T3). Greater than 20% displacement of [125I]−T3 by candidate kinetic stabilizers indicates undesirable library members. Of the 41 TTR kinetic stabilizers evaluated, nearly half (11 stilbene analogues and 10 dihydrostilbene analogues) exhibit insignificant thyroid hormone receptor binding (< 20%) (Figures 2-4, % values in red font). These compounds are promising lead TTR kinetic stabilizers. Given the rarity of thyroid hormone receptor binding in the previous study that identified the 3,5-dibromo-4-hydroxy substructure as optimal,58 it was disappointing, but perhaps not shocking, that half of the stilbenes and dihydrostilbenes tested bound to thyroid hormone receptor.
Replacing the Phenolic Group in 3,5-dibromo-4-hydroxyphenyl-based Stilbenes
In a modest effort towards further aryl-X substructure optimization, we asked whether we could substitute the hydroxyl group in the context of a simple stilbene composed of one unsubstituted aryl ring (aryl-Z) and one ring bearing 3,5-dibromo-substituents and a variable 4-position substituent without losing potency and selectivity. We considered a variety of functional groups at the 4-position (Figure S7). The amino group was equivalent to the hydroxyl group in terms of potency: an encouraging result as anilines are functional groups in FDA-approved drugs. However, some anilines are carcinogenic in animals, and possibly in humans. Motivated by this result, four stilbenes with aniline-based aryl-X rings and substituted aryl-Z substituents were synthesized (Figure 4, compounds 24b-e) to assess both potency and selectivity. Given the limited number of compounds that were synthesized and the diversity of Z-substituents, the potency of these compounds (Figure 4, % F.F. numbers in black font) was impressive, demonstrating the merit of further aryl-X optimization. Moreover, three of these compounds exhibit excellent TTR binding selectivity in plasma (Figure 4, numbers in blue font). Notably, none of these compounds bind to the thyroid hormone receptor (Figure 4, percentage in red font), suggesting that further aryl-X optimization may be an effective strategy to reduce TTR kinetic stabilizer thyroid hormone receptor binding. This limited series exhibits impressive individual efficacy scores (Figure 4, numbers in green font). Moreover, as discussed in detail below, the distinct inner binding subsite preference of the 3,5-dibromo-4-aminophenyl substructure nicely complements the outer binding site preference of the 3,5-dibromo-4-hydroxyphenyl substructure discussed above, providing another substructure to include in future substructure combination strategies.
Demonstrating that Potent Amyloidogenesis Inhibitors are Kinetic Stabilizers of WT-TTR
TTR fibril formation is rate limited by tetramer dissociation.5,20-26 Thus, the imposition of kinetic stabilization on TTR by small molecule binding to the TTR tetramer can be measured by the rate of tetramer dissociation, which is assessed by linking the slow tetramer dissociation step to rapid monomer unfolding, easily quantified using far-UV circular dichroism (CD) or fluorescence spectroscopy.5,11,54 To demonstrate kinetic stabilization of TTR, we preincubated WT-TTR (1.8 μM) with candidate kinetic stabilizers (1.8 or 3.6 μM) for 30 min. Dissociation was then accelerated and made measurable on a convenient laboratory timescale by adding urea to a final concentration of 6 M. The rate and extent of dissociation was monitored by the thermodynamically linked monomer unfolding of WT-TTR at 25 °C, monitored by far-UV CD over 144 h. All candidate kinetic stabilizers at a concentration of 3.6 μM dramatically slowed dissociation, allowing less than 15% of the TTR to dissociate (Figure 5A). Tetramer dissociation was also clearly slowed at a concentration of 1.8 μM, equal to that of the TTR tetramer, allowing 20 to 33% tetramer dissociation, indicating that the binding of one small molecule to one of the two T4 binding sites is sufficient to stabilize the ground state over the dissociative transition state – imposing kinetic stabilization on the entire TTR tetramer (Figure 5B).
Figure 5.
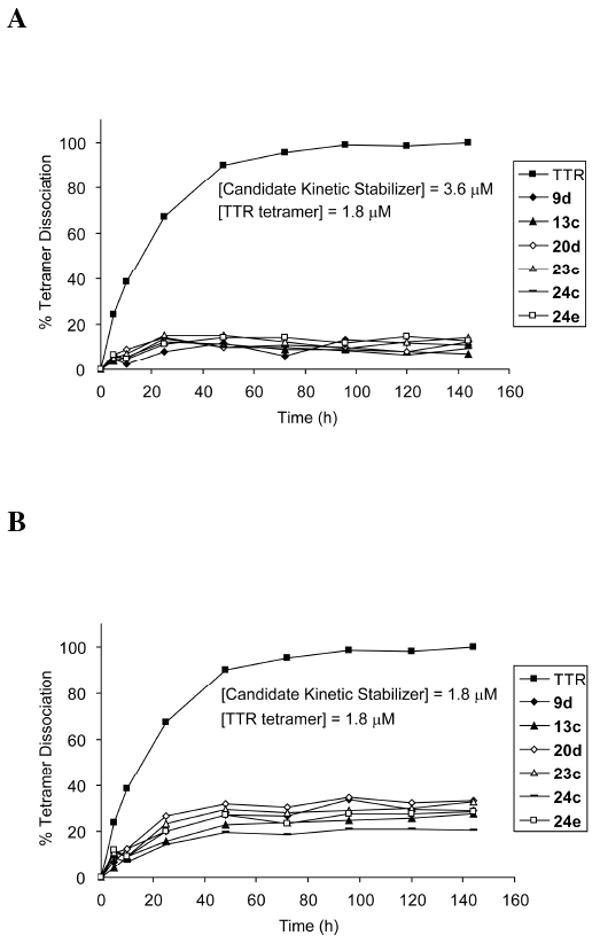
The influence of kinetic stabilizer binding on the rate of TTR tetramer dissociation. WT-TTR (1.8 μM) tetramer dissociation time courses in 6 M urea without (■) and in the presence of putative kinetic stabilizers 9d, 13c, 20d, 23c, 24c, and 24e at a concentration of 3.6 μM (A) and 1.8 μM (B), evaluated by linking the slow tetramer dissociation process to rapid and irreversible monomer denaturation in 6 M urea, as measured by far-UV circular dichroism at 215-218 nm over a time course of 144 h.
Inhibition of WT- and V30M-TTR-induced Cytotoxicity by Potent Kinetic Stabilizers
Previous studies demonstrate that treatment of cell lines derived from tissues that are targets of TTR deposition with recombinant WT-TTR or V30M-TTR homotetramers results in proteotoxicity.19,74 Moreover, these studies establish that TTR cytotoxicity can be inhibited by small molecules that kinetically stabilize TTR against dissociation.19,74,75 Previous experiments conclude that partially folded monomers and/or oligomers resulting from TTR dissociation and misassembly are the major cytotoxic species in cell culture.19 Resveratrol (1) is known to bind to and kinetically stabilize WT- and V30M-TTR and reduce their cytotoxicity, and therefore was used as the positive control. WT-TTR and V30M-TTR were preincubated without and with equimolar amounts of candidate kinetic stabilizers or 1 at 4 °C for 18 h. The human neuroblastoma IMR-32 cells were treated with these solutions (8 μM final concentration of TTR and compounds) for 24 h at 37° C after which cell viability was assessed by the resazurin reduction assay.76 Viable cells reduce the substrate resazurin to resorufin, a fluorescent compound, which was monitored by fluorescence emission at 590 nm. The results are expressed as the percentage of fluorescence generated by viable cells treated with vehicle only (100% viable). Cells treated with cytotoxic TTR in the absence of kinetic stabilizers exhibit 45-65% viability. All the kinetic stabilizers tested nearly completely inhibit WT-TTR and V30M-TTR-induced cytotoxicity at a final concentration equimolar to that of TTR (Figure 6A). Importantly, none of the compounds evaluated were cytotoxic to IMR-32 cells in the absence of TTR (> 80% cell viability, (Figure 6B)). Thus, the substructure combination strategy affords compounds that protect against TTR proteotoxicity.
Figure 6.
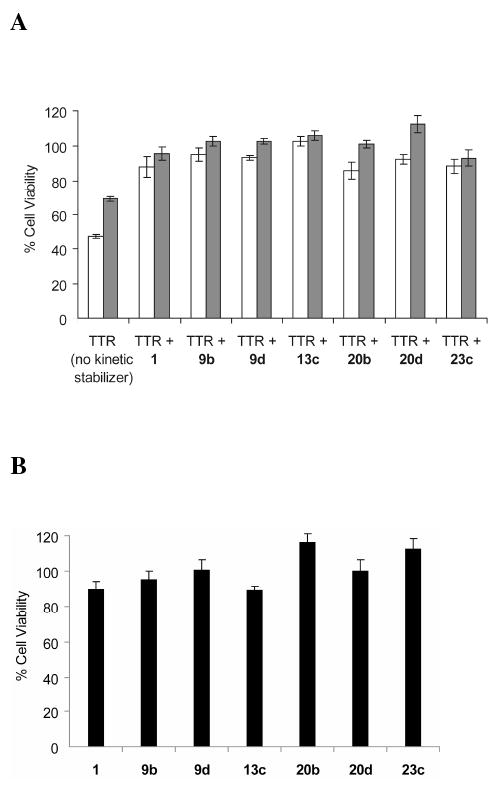
(A) Demonstration that kinetic stabilizers of WT-TTR and V30M-TTR can prevent cytotoxicity associated with the process of TTR amyloidogenesis. IMR-32 human neuroblastoma cells were treated with WT-TTR (8 μM, white bar) or V30M-TTR (8 μM, grey bar) exhibiting cytotoxicity that is prevented by preincubating WT-TTR (white bars) or V30M-TTR (grey bars) with the stilbene and dihydrostilbene-based kinetic stabilizers or resveratrol (1) (a kinetic stabilizer previously shown to be effective) included as a positive control (8 μM each). Cell viability was measured after 24 h by the resazurin reduction assay and all the experimental conditions were compared to cells treated with vehicle only (100% cell viability). (B) Cytotoxicity of selected compounds (8 μM) to the human neuroblastoma cell line IMR-32. Columns represent the average values of 2 independently performed experiments (6 experimental replicates). The error bars represent standard error.
Crystallographic Analysis of WT-TTR•(kinetic stabilizer)2 Complexes
Crystal structures of WT-TTR in complex with 3d, 13c, 17d, and 24c-e were determined to 1.70, 1.40, 1.40, 1.32, 1.40 and 1.48 Å resolutions, respectively (see Table 1 for data collection and refinement statistics). In all 6 structures, the electron density was clear and allowed unambiguous placement of the ligand (unbiased 2Fo-Fc electron density maps contoured at 3σ shown in Figure S8). As anticipated, the 3,5-dibromo-4-hydroxyphenyl rings of stilbene 3d and dihydrostilbene 17d contribute to binding by their placement in the complementary thyroxine outer binding subsite,58,59 positioning the bromides in HBPs 1 and 1′. The phenolate anion of 3d and 17d appears to engage in a salt bridge interaction with the ε-ammonium group of Lys-15/15′ (Figure 7A and C). The 2,6-dichlorophenyl ring in 3d and 17d occupies the inner binding subsite, directing the chloride atoms into HBPs 3 and 3′, stacking the face of the aryl ring between the two symmetry-related Leu110 residues (Figure 7A and C). In the case of stilbene 13c, composed of a 3,5-dibromo-4-hydrophenyl aryl-X ring and a p-amino aryl-Z ring, a mixture of two binding orientations is observed in the electron density. The 3,5-dibromo-4-hydroxyphenyl ring occupies the thyroxine outer binding subsite ∼90% of the time for reasons described above, orientating the p-amino group from the aryl-Z ring to make bridging hydrogen bonds with Ser-117/117′ side chains (Figure 7B). In the minor binding orientation, the 3,5-dibromo-4-hydroxyphenyl ring of 13c occupies the inner binding subsite and the phenol/phenolate makes hydrogen bonds with Ser-117/117′ side chains while the bromides occupy HBPs 3 and 3′ (Figure 7B).
Table 1.
Crystallographic Data and Refinement Statistics
| WT-TTR•3d | WT-TTR•13c | WT-TTR•17d | WT-TTR•24c | WT-TTR•24d | WT-TTR•24e | |
|---|---|---|---|---|---|---|
| Data Collection | ||||||
| Beamline | SSRL 9-2 | APS GM/CA-CAT 23-IDB | SSRL 11-1 | SSRL 11-1 | SSRL 11-1 | SSRL 11-1 |
| Wavelength (Å) | 0.9194 | 1.0333 | 0.9797 | 0.9797 | 0.9797 | 0.9797 |
| Resolution (Å) | 1.70 (1.70-1.76)a | 1.40 (1.40-1.45) | 1.40 (1.40-1.45) | 1.31 (1.31-1.36) | 1.40 (1.40-1.45) | 1.48 (1.48-1.53) |
| Space group | P21212 | P21212 | P21212 | P21212 | P21212 | P21212 |
| a, b, c (Å) | 42.92,85.0,64.50 | 42.26, 84.79, 63.24 | 42.50,85.11,63.32 | 42.8, 85.38, 64.39 | 42.7, 85.29, 64.2 | 42.78, 85.16, 64.16 |
| No. of molecules in the a.u. | 2 | 2 | 2 | 2 | 2 | 2 |
| No. of observations | 376,642 (30,797)a | 322,489 (31,176) | 165,495 (16,232) | 268,463 (20,385) | 220,973 (18,236) | 192,765 (18,569) |
| No. of unique reflections | 26,903 (2,588) | 44,988 (4,391) | 45,971 (4,509) | 56,323 (4,972) | 46,374 (4,342) | 39,462 (3,951) |
| Redundancy | 14.0 (11.9)a | 7.2 (7.1) | 3.6 (3.6) | 4.8 (4.1) | 4.8 (4.2) | 4.9 (4.7) |
| Completeness (%) | 99.5 (97.0)a | 98.8 (98.0) | 99.7 (99.6) | 97.6 (87.3) | 98.2 (93.6) | 97.7 (99.4) |
| Rsym (%)b | 5.7 (69.5)b | 5 .0 (57.4) | 4.0 (45.2) | 3.1 (46.1) | 2.9 (48.3) | 3.4 (52.3) |
| Average I/sigma | 34.6 (7.3)a | 28.3 (3.6) | 26.1 (3.1) | 37.5 (2.8) | 45.5 (2.2) | 36.5 (3.3) |
| Refinement statistics | ||||||
| Resolution (Å) | 1.70-64.55 | 1.40-84.82 | 1.40-63.37 | 1.31-85.44 | 1.40-85.13 | 1.47-85.13 |
| No. of reflections (working set) | 25,229 (1,781)a | 42,702 (2,843) | 43,555 (3,170) | 53,423 (3,720) | 44,010 (2,940) | 37,452 (2,478) |
| No. of reflections (test set) | 1,333 (93)a | 2,257 (154) | 2,332 (177) | 2,869 (203) | 2,340 (142) | 1,981 (121) |
| Rcryst (%)c | 16.9 (17.9)a, c | 16.3 (21.4) | 16.5 (19.8) | 16.7 (28.8) | 16.8 (25.3) | 16.7 (23.4) |
| Rfree (%)d | 21.6 (26.4)a, d | 19.0 (27.0) | 19.7 (25.8) | 19.7 (30.7) | 19.0 (32.7) | 20.0 (26.7 |
| Average B-values | ||||||
| TTR | 22.3 | 17.1 | 14.7 | 18.6 | 20.5 | 19.6 |
| Ligand | 45.3 | 20.4R, 27.2F | 15.9 | 18.4 | 28.6 | 26.4 |
| Wilson B-value | 23.6 | 19.0 | 17.1 | 18.2 | 22.0 | 21.7 |
| Ramachandran plot | ||||||
| Most favored (%) | 92.1 | 93.0 | 91.1 | 93.0 | 93.5 | 93.5 |
| Additionally allowed (%) | 7.9 | 7.0 | 8.9 | 7.0 | 6.5 | 6.5 |
| Generously allowed (%) | 0 | 0 | 0 | 0 | 0 | 0 |
| Disallowed (%) | 0 | 0 | 0 | 0 | 0 | 0 |
| R.M.S deviations | ||||||
| Bond lengths (Å) | 0.019 | 0.018 | 0.018 | 0.016 | 0.018 | 0.018 |
| Angles (°,) | 1.81 | 1.71 | 1.74 | 1.78 | 1.64 | 1.61 |
Numbers in parentheses are for highest resolution shell of data.
Rsym = Σhkl |I-<I>| Σhkl I
Rcryst = Σhkl |Fo-Fc| Σhkl Fo
Rfree is the same as Rcryst, but for 5% of data excluded from the refinement.
F/R = Forward or Reverse conformations of the ligand.
Figure 7.
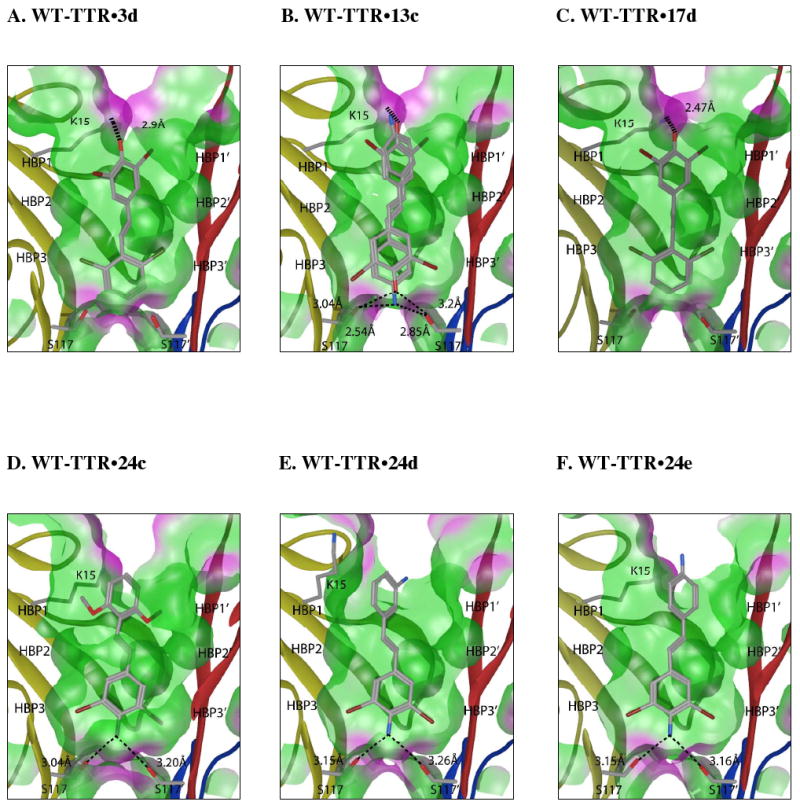
Crystal structures of homotetrameric WT-TTR in complex with inhibitors 3d, 13c, 17d, 24c, 24d, and 24e. Ribbon diagram depiction of a close-up view of one of the two identical T4 binding sites (see Figure 1A). A ‘Connelly’ analytical molecular surface was applied to residues within 8Å of ligand in the T4 binding pocket (green = hydrophobic, purple = polar). Polar residues K15 and S117/117′ are shown with bonds depicted where interactions are observed. In the case of stilbene 13c, a mixture of two binding orientations is observed. The 3,5-dibromo-4-hydroxyphenyl ring occupies the thyroxine outer binding subsite ∼90% of the time (see Results section for a more complete description and explanation). Figure generated using the program MOE (2006.08), Chemical Computing Group, Montreal, Canada.
In stilbenes 24c-e, the 3,5-dibromo-4-aminophenyl substructure was placed in the inner binding subsite in all cases. This binding orientation appears to be stabilized by bridging hydrogen bonds between the amino substituent and the Ser-117/117′ hydroxyl side chains and by simultaneous placement of the bromides into HBPs 3 and 3′ (Figure 7D, E and F). The p- or m-amino substituent on the other ring in 24d and 24e does not appear to interact with TTR, and thus the aryl-Z ring can be further optimized by the substructure combination strategy. The dimethoxyphenyl groups on 24c were able to interact with HBPs 1 and 1′ (Figure 7D).
In all 6 structures, the trans −CH=CH− or −CH2CH2− linker occupies the hydrophobic pocket created by residues Leu-17/17′, Ala-108/108′, Leu-110/110′ and Val-121/121′. These linkers afford the aryl rings some degree of rotational freedom enabling optimal placement of the two substituted rings into HBPs 1, 1′ and 3, 3′.
Discussion
A substructure combination strategy was employed herein, based on previous aryl-X, linker-Y, and aryl-Z substructure rankings,58-60 to guide the synthesis of a library of stilbenes and dihydrostilbenes. This effort afforded a high fraction of potent and highly selective TTR kinetic stabilizers that rescue cells from the cytotoxic effects of TTR amyloidogenesis thought to cause the human amyloidoses, without exhibiting cytotoxicity themselves. The stilbene libraries depicted in Figures 2 and 3 were made up of combinations of substructural components exhibiting high average efficacy scores in the aforementioned 3-part study.58-60 Even though more of the 3,5-dibromo-4-hydroxyphenyl-based stilbenes and dihydrostilbenes afforded thyroid hormone binding capacity than the previous substructure optimization studies would have predicted58-60, a further aryl-X ring and/or aryl-Z substructure optimization and combination strategy may produce potent and selective compounds lacking this property.
We calculated individual efficacy scores according to equation 2 for each compound synthesized and evaluated in this study, Figures 2-4. The individual efficacy scores displayed in Figure 2 reveal that the substructure combination strategy was very effective, in that 96% of the stilbenes examined exhibit an efficacy score > 0.57 out of a maximum of 1. The efficacy score of 0.57 was chosen as a benchmark, because this is the efficacy score of the compound that ameliorated familial amyloid polyneuropathy in a placebo-controlled clinical trial (www.foldrx.com). Compounds exhibiting > 10% fibril formation at a concentration twice that of TTR, for which plasma binding stoichiometry was not assessed, were assumed to have an efficacy score < 0.57.
The individual efficacy scores displayed in Figure 3 reveal that the linker substructure combination strategy was also very effective. All of the dihydrostilbenes examined exhibit an efficacy score > 0.57 out of a maximum of 1. We did not prepare quite as many dihydrostilbenes as we did stilbenes and it could be argued that we hydrogenated the stilbenes exhibiting high efficacy scores, hence it is probably not appropriate to conclude that dihydrostilbenes are superior to stilbenes. Nonetheless, we sampled quite a range of substituents and substitution patterns in both libraries, indicating that the substructure combination strategy was quite effective in the context of both stilbenes and dihydrostilbenes.
Surprisingly, only 30% of the promising stilbene compounds evaluated displaced < 20% of T3 from the thyroid hormone receptor. This was unexpected because the compounds resulting from the individual substructure optimization studies displayed much less T3 displacement.58-60 However, the problem of T3 displacement seems to be easily remedied by a linker replaccement affected by simply hydrogenating the thyroid receptor binding stilbenes. More than 76% of the promising dihydrostilbenes evaluated displaced < 20% of T3 from the thyroid hormone receptor. Another means of correcting the T3 displacement problem could be to perform additional aryl-X substructure optimization and combination strategies.
The structural data presented herein provides further evidence for the strong preference of the 3,5-dibromo-4-hydroxyphenyl ring to occupy the outer binding pocket of TTR in the context of stilbenes and dihydrostilbenes (Figure 7A-C). In stark contrast, the structural data reveal that the 3,5-dibromo-4-aminophenyl ring strongly prefers to occupy the inner binding subsite in the context of stilbenes (Figure 7D-F). Based on this result, we predict that stilbenes and dihydrostilbenes composed of one 3,5-dibromo-4-hydroxyphenyl ring, which prefers to bind in the outer thyroxine binding subsite, and one 3,5-dibromo-4-aminophenyl ring, which prefers to bind to the inner thyroxine binding subsite, will exhibit high individual efficacy scores. The substructure combination strategy used herein to generate potent and selective stilbene and dihydrostilbene TTR kinetic stabilizers could likely be successfully applied to generate potent and selective 2-aryl benzoxazole-based TTR kinetic stabilizers as well. Not only is the hetero-aromatic ring of a benzoxazole a highly ranked linker,59 but previous studies that were much more limited in scope demonstrate that benzoxazoles are excellent TTR kinetic stabilizers62.
The kinetic studies carried out herein demonstrate that selective binding of small molecules to the native tetrameric state of TTR drastically slows down the rate of tetramer dissociation, by increasing the energy barrier for dissociation. We also showed that the kinetic stabilization of TTR imposed by the binding of these compounds precludes TTR-mediated cytotoxicity using a recently introduced cytotoxicity assay; apparently by preventing tetramer dissociation that leads to monomers and oligomers previously shown to be the cytotoxic species. Since amyloidogenesis-linked neurotoxicity and cardiotoxicity is thought to cause FAP and SSA, respectively, we envision that this assay, not utilized in the prior substructure optimization studies, will be useful for predicting compounds that will show efficacy in human patients.
Conclusions
We have successfully utilized the data generated in three previous substructure optimization papers to identify potent and selective stilbene and dihydrostilbene kinetic stabilizers using a substructure combination strategy. We also showed that this strategy is useful for eliminating off-target activities, such as thyroid hormone receptor binding. Notably, the majority of the kinetic stabilizers emerging from the substructure combination strategy exhibit high individual efficacy scores and prevent TTR amyloidogenesis-associated cytotoxicity in a cell culture model that we believe is predictive for kinetic stabilizer efficacy in the human TTR amyloidoses. There is reason to be optimistic that a subset of the potent and selective stilbene- and dihydrostilbene-based TTR kinetic stabilizers produced in this study will exhibit the appropriate pharmacokinetic and pharmacodynamic properties to motivate their further development.
Experimental Section
Wild Type Transthyretin Fibril Formation Assay
WT-TTR was expressed and purified from an E.coli expression system as described previously.77 To assess fibril formation, a test compound (5 μL of a 1.44 mM solution in DMSO) was added to 495 μL of a solution of TTR (0.4 mg/mL) in 10 mM phosphate, 100 mM KCl and 1 mM EDTA (pH 7.0) in a disposable cuvette. The mixture was vortexed, and incubated at room temperature for 30 min. An acidic buffer solution (500 μL of 100 mM acetate, 100 mM KCl, 1 mM EDTA, pH 4.2) was then added to decrease the pH of assay solution to 4.4. The cuvettes were incubated at 37 °C for 72 h without agitation. After the solution was vortexed to evenly distribute any precipitate, the turbidity of the solution at 400 nm was measured using a Hewlett Packard model 8453 UV-VIS spectrophotometer.
Evaluating the Binding Stoichiometry of Candidate Kinetic Stabilizers to TTR in Human Blood Plasma
The plasma TTR binding selectivity assay that evaluates the binding stoichiometry of a test compound to TTR in human blood plasma has been previously described,73 and is used with minor modifications. Briefly, to a 1 mL sample of human blood plasma in a 2 mL Eppendorf tube was added a test compound (7.5 μL of a 1.44 mM solution in DMSO) and then the plasma solution was incubated at 37 °C for 24 h on a rocker plate (30 rpm). A 1:1 (v/v) slurry of unfunctionalized sepharose resin in TSA (10 mM Tris, 140 mM NaCl, pH 8.0) buffer was added and the solution was incubated for 1 h at 4 °C on a rocker plate (18 rpm). The solution was then centrifuged and the supernatant was divided into two 400 μL aliquots, which were added to 200 μL of 1:1 (v:v) slurry of anti-TTR antibody conjugated sepharose resin in TSA. The solution was gently rocked (18 rpm) at 4 °C for 20 min, then centrifuged and the supernatant removed. The resin was washed three times by shaking for 1 min with 1 mL of TSA containing 0.05% saponin and then twice more with 1 mL of TSA. After centrifugation to remove the supernatant, 155 μL of triethylamine (100 mM, pH 11.5) was added to the sepharose resin to dissociate the TTR and bound test compound from the resin and the suspension was vortexed for 1 min. After centrifugation, 144 μL of the supernatant containing the test compound and TTR was neutralized by addition of 0.84 μL of glacial acetic acid before HPLC analysis. The supernatant was analyzed by reverse phase HPLC, as described previously,73 on a Water 600 E multi-solvent delivery system, using a Waters 486 tunable absorbance detector, a 717 autosampler, and a ThermoHypersil Keystone Betabasic-18 column (150 Å pore size, 3 μm particle size). The “A” mobile phase comprises 0.1% TFA in 94.9% H2O + 5% CH3CN and the “B” mobile phase is made up of 0.1% TFA in 94.9% CH3CN + 5% H2O. Linear gradients were run from 100:0 A:B to 0:100 A:B for 9 min.
Binding of Potent TTR Amyloidogenesis Inhibitors to the Thyroid Hormone Receptor
The potential binding of TTR kinetic stabilizers to the thyroid hormone receptor was evaluated by Cerep laboratories in Redmond, WA using the following experimental protocol. Membrane homogenates of liver (100 μg of total protein) are incubated for 18 h at 4 °C with 0.1 nM [125I]T3 in the absence or presence of a TTR kinetic stabilizer in buffer (20 mM Tris-HCl, pH 7.6, 50 mM NaCl, 2 mM EDTA, 10% glycerol, and 5 mM β-mercaptoethanol). Nonspecific binding is corrected for in the presence of 1 μM T3. Following the incubation period, the samples are filtered rapidly under vacuum through glass fiber filters (GF/B, Packard) and rinsed several times with an ice-cold buffer containing 50 mM Tris-HCl and 150 mM NaCl (pH 7.4), employing a 96-sample cell harvester (Unifilter, Packard). The filters are dried, then counted for radioactivity in a scintillation counter (Topcount, Packard) using a scintillation cocktail (Microscint 0, Packard).
Urea-induced Dissociation Kinetics Study
Slow TTR tetramer dissociation is not detectable by far-UV circular dichroism (CD) spectroscopy, however, dissociation is linked to rapid (∼500,000 × faster) monomer unfolding under denaturing conditions, which is easily detectable by far-UV CD spectroscopy. Test compounds (5 and 10 mM in DMSO) were diluted 10 fold in EtOH to give 0.5 and 1 mM stock solutions. Such stocks (7.2 μL) were added to 200 μL of TTR (18 μM in 10 mM sodium phosphate buffer, 100 mM KCl, 1 mM EDTA (pH 7.0)) in 2 mL Eppendorf tubes. These mixtures were briefly vortexed and incubated for 30 min at 25 °C. TTR/test compounds (100 μL) were added to 900 μL of a 6.67 M urea solution in 10 mM sodium phosphate buffer, 100 mM KCl and 1 mM EDTA (pH 7.0) to give a final concentration of 1.8 μM for TTR and final concentrations of test compound of 1.8 μM (1×) and 3.6 μM (2×), respectively. The mixtures were vortexed and incubated in the dark at 25 °C without agitation. CD spectra at a final urea concentration of 6 M were measured at 215-218 nm (0.5 nm steps and 5 times scan) after 0, 5, 10, 25, 48, 72, 96, 120, 144 h of incubation.
IMR-32 Cell-based Assay
Cells
The IMR-32 human neuroblastoma cell line was maintained in Opti-MEM (cell culture media, Invitrogen), supplemented with 5% fetal bovine serum, 1 mM Hepes buffer, 2 mM L-glutamine, 100 units/mL penicillin, 100 μg/mL streptomycin and 0.05 mg/ml CaCl2 (complete cell medium). Two- to 3-day-old cultures (70% confluent) were used for the cytotoxicity experiments.
Recombinant WT-TTR and V30M-TTR purified at 4 °C and capable of amyloidogenesis were used as cytotoxic insults to IMR-32 cells. The proteins were buffer exchanged in Opti-MEM at 10 °C using a Centriprep device (10 kDa MWCO, Millipore).
Stocks of the TTR kinetic stabilizers were prepared at 1 mM in 0.5% DMSO/Opti-MEM and stored at -20 °C. For the experiments, test compounds (32 μM in Opti-MEM) were mixed 1:1 with filtered-sterilized TTR (32 μM in Opti-MEM) or with Opti-MEM only, vortexed and incubated for 18 h at 4 °C. TTR (16 μM) alone and Opti-MEM alone containing the same amount of DMSO used above were prepared in parallel and incubated under the same conditions.
IMR-32 cells were seeded in black wall, clear bottom, 96-well tissue culture plates (Costar) at a density of 6,000 cells/well in complete cell culture medium and incubated overnight at 37 °C. The incubated TTR/test compound samples, TTR and Opti-MEM were diluted 1:1 with freshly prepared Opti-MEM supplemented with 0.8 mg/mL bovine serum albumin, 2 mM Hepes buffer, 4 mM L-glutamine, 200 units/mL penicillin, 200 μg/mL streptomycin and 0.1 mg/mL CaCl2. The medium from the cells was then removed and replaced immediately by the TTR/test compound mixtures, TTR or Opti-MEM. The 96-well plates were spun at 2000 × g for 30 min at 4 °C to allow the IMR-32 cells to re-attach to the bottom of the wells. The cells were then incubated 24 h at 37 °C after which cell viability was measured. The final concentration of both TTR and test compound was 8 μM.
Cell viability assay
The viability of the cells treated with TTR or TTR/test compound mixtures were evaluated by resazurin reduction assay. Briefly, 10 μL/well of resazurin (500 μM in PBS) was added to each well and incubated for 2 h at 37 °C. Viable cells reduce resazurin to the highly fluorescent resorufin dye, which is quantitated in a multiwell plate reader (Exc/Em 530/590nm, Tecan Safire2, Austria). Cell viability was calculated as percentage of fluorescence relative to cells treated with vehicle only (100% viability) after subtraction of blank fluorescence (wells without cells). All the experimental conditions were performed at least in triplicate. Averages and standard error corresponding to 2 independently performed experiments were calculated using GraphPrism (San Diego, CA, US).
Crystallization and Structure Determination of the WT-TTR/ligand Complexes
The TTR protein was concentrated to 4 mg/mL in 10 mM sodium phosphate buffer and 100 mM KCl (pH 7.6) and co-crystallized at room temperature with a 5 molar excess of each ligand using the vapor-diffusion sitting drop method. All crystals were grown from 1.395 M sodium citrate, 3.5% v/v glycerol at pH 5.5. The crystals were cryo-protected with 10% v/v glycerol. Data for 13c were collected at beamline GM/CA-CAT 23-IDB at the Advanced Photon Source (APS) at a wavelength of 1.0333 Å. Data for 3d were collected at beam line 9-2 and data for 17d and 24c-e at 11-1 at the Stanford Synchrotron Radiation Lightsource (SSRL) at wavelengths of 0.9194 Å and 0.9795 Å, respectively. All diffraction data were indexed, integrated and scaled using HKL200078 in space group P21212 with two subunits observed per asymmetric unit. The structure was determined by molecular replacement using the model coordinates of 2FBR72 in the program Phaser.79 Further model building and refinement were completed using Refmac.80 Hydrogens were added during refinement and anisotropic B values calculated. Final models were validated using the JCSG quality control server incorporating Molprobity,81 ADIT (http://rcsb-deposit.rutgers.edu/validate) WHATIF,82 Resolve,83 and Procheck.84 Data collection and refinement statistics are displayed in Table 1.
Protein Data Bank Accession Code
Atomic coordinates and structure factors have been deposited in the Protein Data Bank and are available under accession codes 3IMR (WT-TTR in complex with 3d), 3IMT (WT-TTR in complex with 13c), 3IMS (WT-TTR in complex with 17d), 3IMW (WT-TTR in complex with 24c), 3IMU (WT-TTR in complex with 24d), and 3IMV (WT-TTR in complex with 24e).
Supplementary Material
Acknowledgments
We thank the NIH (DK46335 to J.W.K and CA58896 and AI42266 to I.A.W.), the American Heart Association (0865061F to N.R.), as well as the Skaggs Institute for Chemical Biology and the Lita Annenberg Hazen Foundation for financial support. Technical support from Mike Saure and Gina Dendle is also greatly appreciated. We thank the General Clinical Research Center of the Scripps Research Institute for providing human blood. X-ray Diffraction data for 13c were collected at GM/CA-CAT 23-IDB beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. X-ray diffraction data for 3d, 17d, and 24c-e carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. The authors would also like to acknowledge Drs Stanfield, Dai, Yoon, Xu and Ekiert for assisting with x-ray data collection.
Footnotes
Supporting Information Available: Detailed synthesis, characterization and purities of stilbenes and dihydrostilbenes, efficacy of stilbenes and dihydrostilbenes at a concentration of 3.6 μM, evaluation of a variety of related functional groups to replace the hydroxyl group, unbiased 2Fo-Fc electron density maps for the ligands in the WT-TTR•(kinetic stabilizer)2 complexes, and cytotoxicity of selected kinetic stabilizers to the IMR-32 cell line. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Balch WE, Morimoto RI, Dillin A, Kelly JW. Science. 2008;319:916–9. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Annu Rev Biochem. 2009;78:959–91. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 3.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Science. 2006;313:1604–10. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 4.Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 5.Hammarstrom P, Wiseman RL, Powers ET, Kelly JW. Science. 2003;299:713–6. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 6.Page LJ, Suk JY, Bazhenova L, Fleming SM, Wood M, Jiang Y, Guo LT, Mizisin AP, Kisilevsky R, Shelton GD, Balch WE, Kelly JW. Proc Natl Acad Sci USA. 2009;106:11125–30. doi: 10.1073/pnas.0811753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coelho T. Curr Opin Neurol. 1996;9:355–9. [PubMed] [Google Scholar]
- 8.Dobson CM. Nature. 2003;426:884–90. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 9.Cohen FE, Kelly JW. Nature. 2003;426:905–9. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 10.Selkoe DJ. Nature. 2003;426:900–4. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 11.Johnson SM, Wiseman RL, Sekijima Y, Green NS, Adamski-Werner SL, Kelly JW. Acc Chem Res. 2005;38:911–21. doi: 10.1021/ar020073i. [DOI] [PubMed] [Google Scholar]
- 12.Monaco HL, Rizzi M, Coda A. Science. 1995;268:1039–41. doi: 10.1126/science.7754382. [DOI] [PubMed] [Google Scholar]
- 13.Blake CC, Geisow MJ, Oatley SJ, Rerat B, Rerat C. J Mol Biol. 1978;121:339–56. doi: 10.1016/0022-2836(78)90368-6. [DOI] [PubMed] [Google Scholar]
- 14.Hornberg A, Eneqvist T, Olofsson A, Lundgren E, Sauer-Eriksson AE. J Mol Biol. 2000;302:649–69. doi: 10.1006/jmbi.2000.4078. [DOI] [PubMed] [Google Scholar]
- 15.Sacchettini JC, Kelly JW. Nat Rev Drug Discov. 2002;1:267–75. doi: 10.1038/nrd769. [DOI] [PubMed] [Google Scholar]
- 16.White JT, Kelly JW. Proc Natl Acad Sci USA. 2001;98:13019–24. doi: 10.1073/pnas.241406698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wojtczak A, Luft J, Cody V. J Biol Chem. 1992;267:353–7. [PubMed] [Google Scholar]
- 18.Soprano DR, Herbert J, Soprano KJ, Schon EA, Goodman DS. J Biol Chem. 1985;260:1793–8. [PubMed] [Google Scholar]
- 19.Reixach N, Deechongkit S, Jiang X, Kelly JW, Buxbaum JN. Proc Natl Acad Sci USA. 2004;101:2817–22. doi: 10.1073/pnas.0400062101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colon W, Kelly JW. Biochemistry. 1992;31:8654–60. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 21.Lai Z, Colon W, Kelly JW. Biochemistry. 1996;35:6470–82. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 22.Lashuel HA, Lai Z, Kelly JW. Biochemistry. 1998;37:17851–64. doi: 10.1021/bi981876+. [DOI] [PubMed] [Google Scholar]
- 23.Liu K, Cho HS, Lashuel HA, Kelly JW, Wemmer DE. Nat Struct Biol. 2000;7:754–7. doi: 10.1038/78980. [DOI] [PubMed] [Google Scholar]
- 24.Jiang X, Smith CS, Petrassi HM, Hammarstrom P, White JT, Sacchettini JC, Kelly JW. Biochemistry. 2001;40:11442–52. doi: 10.1021/bi011194d. [DOI] [PubMed] [Google Scholar]
- 25.Palaninathan SK, Mohamedmohaideen NN, Snee WC, Kelly JW, Sacchettini JC. J Mol Biol. 2008;382:1157–67. doi: 10.1016/j.jmb.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 26.Hurshman AR, White JT, Powers ET, Kelly JW. Biochemistry. 2004;43:7365–81. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- 27.Westermark P, Bergstrom J, Solomon A, Murphy C, Sletten K. Amyloid. 2003;10 1:48–54. [PubMed] [Google Scholar]
- 28.Westermark P, Sletten K, Johansson B, Cornwell GG. Proc Natl Acad Sci USA. 1990;87:2843–5. doi: 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buxbaum JN, Tagoe CE. Annu Rev Med. 2000;51:543–69. doi: 10.1146/annurev.med.51.1.543. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X, Buxbaum JN, Kelly JW. Proc Natl Acad Sci USA. 2001;98:14943–8. doi: 10.1073/pnas.261419998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobson DR, Pastore RD, Yaghoubian R, Kane I, Gallo G, Buck FS, Buxbaum JN. N Engl J Med. 1997;336:466–73. doi: 10.1056/NEJM199702133360703. [DOI] [PubMed] [Google Scholar]
- 32.Hammarstrom P, Jiang X, Hurshman AR, Powers ET, Kelly JW. Proc Natl Acad Sci USA. 2002;99 4:16427–32. doi: 10.1073/pnas.202495199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurshman Babbes AR, Powers ET, Kelly JW. Biochemistry. 2008;47:6969–84. doi: 10.1021/bi800636q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrade C. Brain. 1952;75:408–27. doi: 10.1093/brain/75.3.408. [DOI] [PubMed] [Google Scholar]
- 35.Plante-Bordeneuve V, Said G. Curr Opin Neurol. 2000;13:569–73. doi: 10.1097/00019052-200010000-00011. [DOI] [PubMed] [Google Scholar]
- 36.Coelho T, Chorao R, Sausa A, Alves I, Torres MF, Saraiva MJ. Neuromusc Disord. 1996;6:27. [Google Scholar]
- 37.Saraiva MJ. Amyloid. 2003;10 1:13–6. [PubMed] [Google Scholar]
- 38.Sekijima Y, Hammarstrom P, Matsumura M, Shimizu Y, Iwata M, Tokuda T, Ikeda S, Kelly JW. Lab Invest. 2003;83:409–17. doi: 10.1097/01.lab.0000059937.11023.1f. [DOI] [PubMed] [Google Scholar]
- 39.Hammarstrom P, Sekijima Y, White JT, Wiseman RL, Lim A, Costello CE, Altland K, Garzuly F, Budka H, Kelly JW. Biochemistry. 2003;42:6656–63. doi: 10.1021/bi027319b. [DOI] [PubMed] [Google Scholar]
- 40.Gambetti P, Russo C. Nephrol Dial Transplant. 1998;13 7:33–40. doi: 10.1093/ndt/13.suppl_7.33. [DOI] [PubMed] [Google Scholar]
- 41.Holmgren G, Ericzon BG, Groth CG, Steen L, Suhr O, Andersen O, Wallin BG, Seymour A, Richardson S, Hawkins PN. Lancet. 1993;341:1113–6. doi: 10.1016/0140-6736(93)93127-m. [DOI] [PubMed] [Google Scholar]
- 42.Tan SY, Pepys MB, Hawkins PN. Am J Kidney Dis. 1995;26:267–85. doi: 10.1016/0272-6386(95)90647-9. [DOI] [PubMed] [Google Scholar]
- 43.Suhr OB, Herlenius G, Friman S, Ericzon BG. Liver Transplant. 2000;6:263–76. doi: 10.1053/lv.2000.6145. [DOI] [PubMed] [Google Scholar]
- 44.Olofsson BO, Backman C, Karp K, Suhr OB. Transplantation. 2002;73:745–51. doi: 10.1097/00007890-200203150-00015. [DOI] [PubMed] [Google Scholar]
- 45.Dubrey SW, Davidoff R, Skinner M, Bergethon P, Lewis D, Falk RH. Transplantation. 1997;64:74–80. doi: 10.1097/00007890-199707150-00014. [DOI] [PubMed] [Google Scholar]
- 46.Yazaki M, Tokuda T, Nakamura A, Higashikata T, Koyama J, Higuchi K, Harihara Y, Baba S, Kametani F, Ikeda S. Biochem Biophys Res Commun. 2000;274:702–6. doi: 10.1006/bbrc.2000.3203. [DOI] [PubMed] [Google Scholar]
- 47.Tojo K, Sekijima Y, Kelly JW, Ikeda SI. Neurosci Res. 2006;56:441–9. doi: 10.1016/j.neures.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 48.Sekijima Y, Dendle MA, Kelly JW. Amyloid. 2006;13:236–49. doi: 10.1080/13506120600960882. [DOI] [PubMed] [Google Scholar]
- 49.Hammarstrom P, Schneider F, Kelly JW. Science. 2001;293:2459–62. doi: 10.1126/science.1062245. [DOI] [PubMed] [Google Scholar]
- 50.Lai Z, McCulloch J, Lashuel HA, Kelly JW. Biochemistry. 1997;36:10230–9. doi: 10.1021/bi963195p. [DOI] [PubMed] [Google Scholar]
- 51.Foss TR, Kelker MS, Wiseman RL, Wilson IA, Kelly JW. J Mol Biol. 2005;347:841–54. doi: 10.1016/j.jmb.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 52.Wiseman RL, Johnson SM, Kelker MS, Foss T, Wilson IA, Kelly JW. J Am Chem Soc. 2005;127:5540–51. doi: 10.1021/ja042929f. [DOI] [PubMed] [Google Scholar]
- 53.Wiseman RL, Green NS, Kelly JW. Biochemistry. 2005;44:9265–74. doi: 10.1021/bi050352o. [DOI] [PubMed] [Google Scholar]
- 54.Foss TR, Wiseman RL, Kelly JW. Biochemistry. 2005;44:15525–33. doi: 10.1021/bi051608t. [DOI] [PubMed] [Google Scholar]
- 55.Miroy GJ, Lai Z, Lashuel HA, Peterson SA, Strang C, Kelly JW. Proc Natl Acad Sci USA. 1996;93:15051–6. doi: 10.1073/pnas.93.26.15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peterson SA, Klabunde T, Lashuel HA, Purkey H, Sacchettini JC, Kelly JW. Proc Natl Acad Sci USA. 1998;95:12956–60. doi: 10.1073/pnas.95.22.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klabunde T, Petrassi HM, Oza VB, Raman P, Kelly JW, Sacchettini JC. Nat Struct Biol. 2000;7:312–21. doi: 10.1038/74082. [DOI] [PubMed] [Google Scholar]
- 58.Johnson SM, Connelly S, Wilson IA, Kelly JW. J Med Chem. 2008;51:260–70. doi: 10.1021/jm0708735. [DOI] [PubMed] [Google Scholar]
- 59.Johnson SM, Connelly S, Wilson IA, Kelly JW. J Med Chem. 2008;51:6348–58. doi: 10.1021/jm800435s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson SM, Connelly S, Wilson IA, Kelly JW. J Med Chem. 2009;52:1115–25. doi: 10.1021/jm801347s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petrassi HM, Johnson SM, Purkey H, Chiang KP, Walkup T, Jiang X, Powers ET, Kelly JW. J Am Chem Soc. 2005;127:6662–71. doi: 10.1021/ja044351f. [DOI] [PubMed] [Google Scholar]
- 62.Razavi H, Palaninathan SK, Powers ET, Wiseman RL, Purkey HE, Mohamedmohaideen NN, Deechongkit S, Chiang KP, Dendle MT, Sacchettini JC, Kelly JW. Angew Chem Int Ed Engl. 2003;42:2758–61. doi: 10.1002/anie.200351179. [DOI] [PubMed] [Google Scholar]
- 63.Purkey HE, Palaninathan SK, Kent KC, Smith C, Safe SH, Sacchettini JC, Kelly JW. Chem Biol. 2004;11:1719–28. doi: 10.1016/j.chembiol.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 64.Oza VB, Smith C, Raman P, Koepf EK, Lashuel HA, Petrassi HM, Chiang KP, Powers ET, Sachettinni J, Kelly JW. J Med Chem. 2002;45:321–32. doi: 10.1021/jm010257n. [DOI] [PubMed] [Google Scholar]
- 65.Adamski-Werner SL, Palaninathan SK, Sacchettini JC, Kelly JW. J Med Chem. 2004;47:355–74. doi: 10.1021/jm030347n. [DOI] [PubMed] [Google Scholar]
- 66.Petrassi HM, Klabunde T, Sacchettini JC, Kelly JW. J Am Chem Soc. 2000;122:2178–92. [Google Scholar]
- 67.Johnson SM, Petrassi HM, Palaninathan SK, Mohamedmohaideen NN, Purkey H, Nichols C, Chiang KP, Walkup T, Sacchettini JC, Sharpless KB, Kelly JW. J Med Chem. 2005;48:1576–87. doi: 10.1021/jm049274d. [DOI] [PubMed] [Google Scholar]
- 68.Wojtczak A, Cody V, Luft JR, Pangborn W. Acta Crystallogr D Biol Crystallogr. 1996;52:758–65. doi: 10.1107/S0907444996003046. [DOI] [PubMed] [Google Scholar]
- 69.Baures PW, Oza VB, Peterson SA, Kelly JW. Bioorg Med Chem. 1999;7:1339–47. doi: 10.1016/s0968-0896(99)00066-8. [DOI] [PubMed] [Google Scholar]
- 70.Baures PW, Peterson SA, Kelly JW. Bioorg Med Chem. 1998;6:1389–401. doi: 10.1016/s0968-0896(98)00130-8. [DOI] [PubMed] [Google Scholar]
- 71.McCammon MG, Scott DJ, Keetch CA, Greene LH, Purkey HE, Petrassi HM, Kelly JW, Robinson CV. Structure. 2002;10:851–63. doi: 10.1016/s0969-2126(02)00771-2. [DOI] [PubMed] [Google Scholar]
- 72.Green NS, Palaninathan SK, Sacchettini JC, Kelly JW. J Am Chem Soc. 2003;125:13404–14. doi: 10.1021/ja030294z. [DOI] [PubMed] [Google Scholar]
- 73.Purkey HE, Dorrell MI, Kelly JW. Proc Natl Acad Sci USA. 2001;98:5566–71. doi: 10.1073/pnas.091431798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reixach N, Adamski-Werner SL, Kelly JW, Koziol J, Buxbaum JN. Biochem Biophys Res Commun. 2006;348:889–97. doi: 10.1016/j.bbrc.2006.07.109. [DOI] [PubMed] [Google Scholar]
- 75.Sörgjerd K, Klingstedt T, Lindgren M, Kågedal K, Hammarström P. Biochem Biophys Res Commun. 2008;377:1072–8. doi: 10.1016/j.bbrc.2008.10.121. [DOI] [PubMed] [Google Scholar]
- 76.O'Brien J, Wilson I, Orton T, Pognan F. Eur J Biochem. 2002;267:5421–6. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 77.Lashuel HA, Wurth C, Woo L, Kelly JW. Biochemistry. 1999;38:13560–73. doi: 10.1021/bi991021c. [DOI] [PubMed] [Google Scholar]
- 78.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 79.Storoni LC, McCoy AJ, Read RJ. Acta Crystallogr D Biol Crystallogr. 2004;60:432–8. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 80.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 81.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 82.Vriend G. J Mol Graph. 1990;8:52–6. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 83.Terwilliger TC. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laskowski R, MacArthur M, Moss D, Thornton J. J Appl Crystallogr. 1993;26:283–91. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.