Abstract
Multinuclear platinum anticancer complexes are a proven option to overcome resistance of established anticancer compounds. Transferring this concept to ruthenium complexes led to the synthesis of dinuclear Ru(II)–arene compounds containing a bis(pyridinone)alkane ligand linker. A pronounced influence of the spacer length on the in vitro anticancer activity was found, which is correlated to the lipophilicity of the complexes. IC50 values in the same dimension as for established platinum drugs were found in human tumor cell lines. No cross-resistance to oxoplatin, a cisplatin prodrug, was observed for the most active complex in three resistant cell lines; in fact, a 10-fold reversal of sensitivity in two of the oxoplatin-resistant lines was found. (Bio)analytical characterization of the representative examples showed that the ruthenium complexes hydrolyze rapidly, forming predominantly diaqua species that exhibit affinity toward transferrin and DNA, indicating that both proteins and nucleobases are potential targets.
Introduction
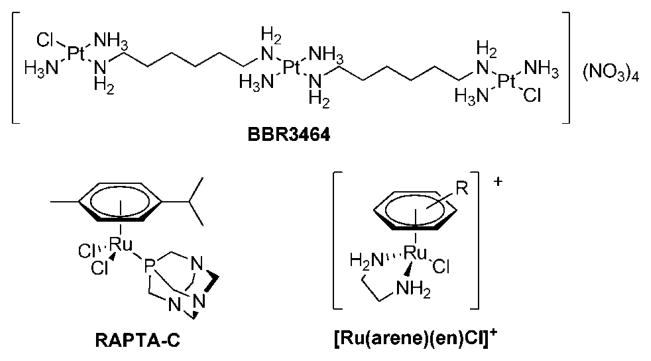
With the objective of developing compounds with a new mode of action in comparison to the established anticancer drugs cisplatin, transplatin, and oxaliplatin for treatment of a broader range of tumors and with fewer side effects, many metal complexes were investigated in recent years for their tumor-inhibiting properties.1 In order to achieve a modified mechanism of activity, several approaches were considered promising; e.g., the exchange of platinum centers by other metals,2,3 the connection of more than one platinum center,1,2 platinum complexes with trans geometry,2 and conjugation of metallo-drugs to proteins or other biomolecules as targeting concepts.2,4–6 In BBR3464 two of these concepts are realized within one compound (Figure 1); i.e., three trans-configured Pt moieties are linked by diaminoalkanes, and the compound shows a much higher degree of DNA interstrand cross-linking than cisplatin.7 Furthermore, a 4-fold positively charged Pt complex was not known before to have antineoplastic activity. The high in vitro activity was also demonstrated for BBR3464 in cisplatin-resistant cell lines; in fact, the activity was found to be 2–3 orders of magnitude higher than that of cisplatin.8 Recently, the compound finished a phase I clinical trial in which preliminary activity against pancreatic, non-small-cell lung, and ovarian cancer as well as melanoma was shown.9–11 However, in subsequent phase II studies no major responses were observed in small cell lung cancer as well as gastric and gastroesphageal adenocarcinoma patients when BBR3464 was applied as a single agent.2,9,11
Figure 1.
Structures of BBR3464 and organometallic Ru(II)–arene complexes of interest as anticancer agents.
Another concept for improving the chemotherapeutic activity was realized by replacing the platinum center with ruthenium. So far only two Ru(III) coordination compounds have reached clinical trials, NAMI-A {H2Im[trans-RuCl4(DMSO)(Him)] (HIm = imidazole)} and KP1019 {H2Ind[trans-RuCl4(HInd)2] (HInd = indazole)}, and both of them are considered promising drug candidates.12,13 Binding to plasma proteins14–21 and the reduction of Ru(III) to Ru(II)22,23 seem to be key steps in their mode of action and the thereby-mediated degree of selectivity is regarded as a reason for their low general toxicity.24,25 More recently organometallic ruthenium(II)–arene complexes were reported to exhibit antitumoral activity.26–28 RAPTA and ethylenediamine compounds (see Figure 1) are the most widely studied representatives of this compound class. RAPTA complexes exhibit selective cytotoxicity in TS/A cancer cells relative to nontumorigenic HBL-100 cells and were found to be, similar to NAMI-A, active in vivo against lung metastases derived from a MCa mammary carcinoma in CBA mice and in Ehrlich ascites carcinoma.27,29 Ethylenediamine (en) complexes with the general formula [Ru(arene)(en)Cl]+ can be optimized in terms of biological activity and minimized side effects by variation of the arene and the nonleaving groups.26 The compounds are equally potent toward cisplatin-sensitive (A2780) and cisplatin-resistant human ovarian cancer cells in culture (A2780cis), and similar results were obtained in vivo.
In contrast to multinuclear platinum complexes, analogous ruthenium compounds have rarely been studied, with just a few examples to be found in literature.30–36 Herein, we report structure–activity relationships for an extended series of di-nuclear pyridinone-derived ruthenium complexes37,38 with different spacer lengths, based on chemical and (bio)analytical characterization, i.e., octanol/water partition, hydrolytic stability, reactivity toward biomolecules, and extraordinary in vitro activity in human tumor cell lines.
Results and Discussion
Synthesis and Characterization of Dinuclear Ru(II)–Arene Complexes
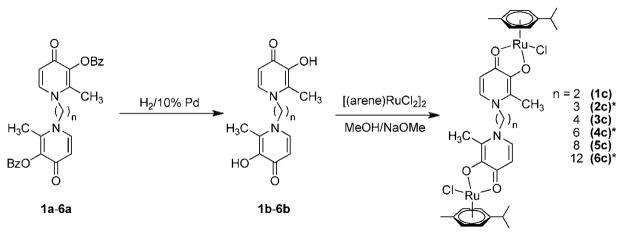
To complete a series of recently developed di-nuclear Ru(II)–arene compounds37 with the purpose of establishing more detailed structure–activity relationships, the Ru complexes 1c, 3c, and 5c with varying aliphatic spacers were prepared in a similar way as reported earlier.37 The complexes were obtained from the reaction between [(η6-p-cymene)RuCl2]2 and sodium methoxide-deprotonated bis(3-hydroxy-2-methyl-4-pyridinon-1-yl)alkanes 1b, 3b, and 5b (alkane = ethane 1b, butane 2b, octane 2c) in good yields (41%, 58%, and 66%, respectively, Figure 2). The complexes were characterized by 1H and 13C NMR spectroscopy, MS, and elemental analysis. As reported earlier for 2c, 4c, and 6c, only one set of signals was observed in the 1H NMR spectra. Electrospray ionization mass spectra contained as the most abundant peaks doubly charged [M–2Cl]2+ ions, while lower intensity peaks were assignable to [M – Cl]+ and [M + Na]+.
Figure 2.
Synthesis of the dinuclear ruthenium(II)–arene complexes 1c–6c. For items marked with an asterisk (*) see ref 37 for synthetic procedure.
Hydrolytic Stability
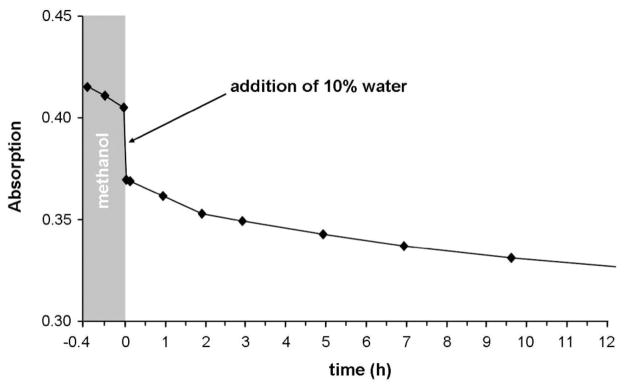
To investigate the hydrolytic stability of this type of complex, the behavior of 4c in water was followed spectroscopically by UV/vis and NMR methods, and the chloride release was determined with a chloride selective electrode. A kinetic UV/vis experiment of 4c in water was performed over 2 d. Two absorption bands at λ 314 and 224 nm were observed that did not change over time (Supporting Information, Figure S1). Time-resolved 1H NMR spectra of 4c in D2O before and after the addition of AgNO3, which guaranteed the full hydrolysis of the Ru complex, also showed no changes over 5 d (Supporting Information, Figure S2). These results suggest that the hydrolysis occurs immediately after dissolution of the sample. To slow the hydrolysis reaction, 4c was dissolved in dry methanol and analyzed again by UV/vis (Figure 3, Supporting Information, Figure S3). After a few measurement cycles, 10% of water was added and the reaction was followed for 10 h, showing a significant change of the absorption bands over time after the addition of water.
Figure 3.
Time course of UV/vis spectra at λ 325 nm for 4c in dry methanol and after addition of 10% water (25 °C).
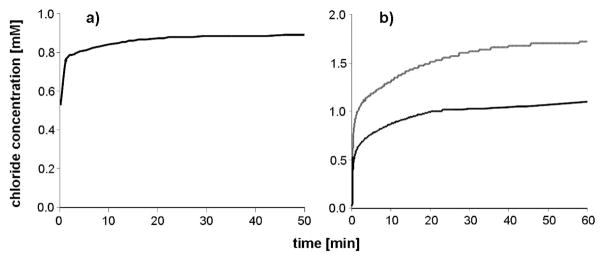
With a chloride selective electrode the release of the chlorido ligands from 4c in water was followed over time. Immediately after preparation of a 0.5 mM solution, the chloride ion concentration was observed to be 0.53 mM and increased rapidly until the complete release of the second chloride ion (Figure 4a). In order to slow this process, 4c was dissolved in 0.5 mL of methanol and added to water (Figure 4b). For 0.5 and 1 mM methanolic solutions, the free chloride concentration was measured immediately after addition to water, and within a few minutes chloride concentrations of 1 and 2 mM, respectively, were detected, indicating full aquation of the complex. These findings are in good agreement with those obtained by 1H NMR and UV/vis. In contrast to the analogous mononuclear complex chlorido(maltolato-κO2)( η6-p-cymene)ruthenium(II) 8, there was no indication of the formation of dimeric hydroxido-bridged species.39
Figure 4.
Chloride ion concentration over time of (a) 0.5 mM 4c solution in water, (b) 0.5 and 1 mM 4c solutions prepared in 0.5 mL of methanol and added to 4.4 mL of water (containing final 5 M NaNO3) at 25 °C.
pKa Determination
The pKa values of 2c, 4c, and 6c were determined by NMR titration and are exemplary for the whole series of compounds. The titration curves were plotted as a function of the chemical shift of the aromatic protons of the p-cymene arene group versus the measured pD values (Supporting Information, Figure S4). The pKa values were obtained from the inflection points of the curves and corrected to consider the difference between H2O and D2O. The pKa values for 2c, 4c, and 6c indicate that the complexes are present as aquated species under biological conditions (pH 7.4), with the linker length having only a very small influence on the pKa (see Table 1). The pKa values obtained range from 9.60 to 9.83 and are close to the one of the mononuclear analogue chlorido(maltolato-κO2)(η6-p-cymene)ruthenium(II), i.e., 9.23.39 The coordination of strong electron donating ligands such as maltol-derived pyridinone to a ruthenium(II) center facilitates the formation of aqua over hydroxido complexes. Note that the pKa values can be easily modified by replacing the O,O chelating ligands by ethylenediamine, which leads to a decrease of the pKa values by about 1 unit (pKa ≈ 8).39 Replacing the ruthenium center by osmium results in a pKa being 1.6 units lower than that of the Ru(II) species.39
Table 1.
pKa Values Obtained by the pD Titration
| complex | pKa values |
|---|---|
| 2c | 9.70 ± 0.03 |
| 4c | 9.60 ± 0.02 |
| 6c | 9.83 ± 0.06 |
| [RuCl(cym)(mal)] 8a | 9.23 |
From ref 39.
Determination of Lipophilicity: log Poct/water Values
The lipophilicity of the complexes 1c–6c was measured as the n-octanol/water partition coefficient, log P, by applying the shake flask method40 and quantification of the ruthenium contents in the aqueous and octanol phases with ICP-MS and UV/vis, respectively. Because of the fast hydrolysis of the compounds in aqueous solution, the log Poct/water values that were determined were actually those of the aqua complexes, which were in the range from −1.42 to −0.56 (see Table 2). These values show a marked correlation to the spacer length linking the pyridinone moieties.
Table 2.
The log Poct/water Values, Determined by Using the Shake Flask Method and Analysis by ICP-MS and UV/Visible Spectroscopy, and Cytotoxicity of Complexes 1c–6c Compared to the Mononuclear Complexes 7 and [Ru(cym)(mal)Cl] 8 in SW480 and A2780 Human Cancer Cells, Determined by the MTT Assay
| log Poct/water |
IC50, μM |
||||
|---|---|---|---|---|---|
| compd | n | ICP-MS | UV/vis | SW480 | A2780 |
| 1c | 2 | −1.42 | −1.42 | 76 ± 4 | 84 ± 3 |
| 2ca | 3 | −1.39 | −1.40 | 62 ± 14a | 25 ± 2a |
| 3c | 4 | −1.33 | −1.38 | 28 ± 1 | 41 ± 2 |
| 4ca | 6 | −1.36 | −1.34 | 26 ± 8a | 30 ± 6a |
| 5c | 8 | −1.28 | −1.23 | 2.5 ± 0.2 | 5.7 ± 0.5 |
| 6ca | 12 | −0.56 | −0.58 | 0.29 ± 0.05a | 1.5 ± 0.3a |
| 7 | 42 ± 1 | 88 ± 12 | |||
| [RuCl(cym)(mal)] 8 | >100 | >100b | |||
Cytotoxicity in Cancer Cell Lines
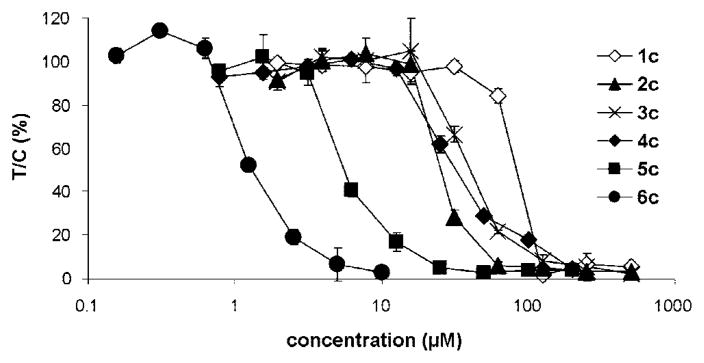
The antiproliferative activity of the dinuclear complexes was determined in the two human cancer cell lines SW480 (colon) and A2780 (ovary) by means of the colorimetric MTT assay and in the other cell lines with the crystal violet assay. The IC50 values are listed in Tables 2 and 3, and representative concentration–effect curves in A2780 cells are shown in Figure 5. Chain length–activity relationships in the human cell lines are similar, indicating that cytotoxicity increases with the chain length of the alkane spacer from IC50 values in the high 10−5 M range in the case of 1c, decreasing to very low micromolar or even submicromolar concentrations in the case of 6c (Table 2). Remarkably, the former compound is roughly equieffective in the ovarian and colon cancer cell lines, while the latter reveals large differences in their sensitivity. In other words, the cytotoxic effects of 6c are to a greater extent dependent on the cell type whereas 1c exerts rather unspecific effects. This same trend is apparent in seven of the eight cell lines shown in Table 3; i.e., 6c is more active than 4c and 2c. As with the smaller panel of cells, the eight cell line panel shows a large difference in sensitivity to 6c, with a more than 10-fold difference in IC50 values between MCF-7 and KYSE70 cells.
Table 3.
Cytotoxicity of the Complexes 2c, 4c, and 6c Compared to Cisplatin and Oxoplatin in Nine Human Cancer Cell Lines and Three Oxoplatin Resistant Cancer Cell Lines, Determined by the Crystal Violet Method
| IC50, μMa |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| compd | LCLC-103H | A-427 | RT-4 | MCF-7 | DAN-G | 5637 | 5637-oxo | SISO | SISO-oxo | KYSE70 | KYSE70-oxo |
| 2c | >20 | 5.2 ± 2.4 μM | >20 | >20 | >20 | >20 | >20 | 21.2b | 32.2b | >20 | >20 |
| 4c | >20 | 13.0 ± 9.5 μM | >20 | 15.2b | >20 | >20 | >20 | 9.5b | 15.7b | >20 | >20 |
| 6c | 2.5 ± 1.1 | 10.4 ± 4.0 | 6.0 ± 3.2 | 2.1b | 4.4 ± 3.0 | 20.0b | 1.1 ± 0.8 (0.06)c | 2.5 ± 2.3 | 2.5 ± 1.9 (1.0)c | 22.5 ± 8.3 | 1.8 ± 1.2 (0.08)c |
| cisplatin | 0.90 ± 0.19d | 1.96 ± 0.54d | 1.61 ± 0.16d | 1.38 ± 0.29d | 0.73 ± 0.34d | 0.37 ± 0.01 | 1.1 ± 0.4 (2.9)c | 0.28 ± 0.04 | 0.58 ± 0.08 (2.1)c | 0.72 ± 0.11 | 1.7 ± 0.4 (2.4)c |
| oxoplatin | 17.2 ± 5.1 | 13.0 ± 2.7 | 26 ± 8 | 5.5 ± 1.1 | 11.8 ± 3.2 | 3.1 ± 1.5 | 10.6 ± 1.3 (3.4)c | 1.7 ± 0.4 | 5.2 ± 0.9 (3.1)c | 4.2 ± 2.8 | 8.8 ± 5.2 (2.1)c |
Mean values ± SD of three independent determinations unless otherwise noted.
Average of two independent determinations.
Resistance factor.
From ref 54.
Figure 5.
Concentration–effect curves of the dinuclear Ru(II)–arene complexes 1c–6c in A2780 cells, as obtained by the MTT assay (96 h exposure).
Chain length–activity relationships correlate to some extent with the hydrophobicity of the complexes as reflected by their log P values (Table 2), although no quantitative relationship could be established. As highlighted above, the complexes attain a more hydrophobic character with increasing chain length between the two pyridinone moieties. Hydrophobicity may contribute to an increased uptake of the complex into the cells, thereby enhancing antiproliferative activity. The fact that no direct correlation to lipophilicity was observed supports the idea that other effects are also important for the mode of action.
Comparison of the in vitro activity with that of cisplatin, oxoplatin [a Pt(IV) cisplatin precursor with two hydroxido ligands in the axial positions] and oxaliplatin shows that the most cytotoxic compound 6c is as active as oxaliplatin in the colon carcinoma cell line SW480 and more active than cisplatin and the anticancer drug candidate KP1019 in A2780 cells.37 In five of the eight cell lines LCLC-103, A-427, RT-4, MCF-7, DAN-G, 5637, SISO, and KYSE70, 6c is less active than cisplatin but more active than oxoplatin (Table 3).
Comparison of the dinuclear complexes with [RuCl(cym)-(mal)] 8 and chlorido[2-methyl-3-(oxo-κO)-1-propyl-4(1H)-pyridinonato-κO4](η6-p-cymene)ruthenium(II) 7 (see supporting information for structures of 7 and 8), a mononuclear analogue of 4c, reveals that a change from the maltolato to a pyridinonato ligand clearly increases the antiproliferative activity. Most notably, the linking of two metal centers results in a more than 2-fold enhancement of activity in both cell lines (Table 2).
In three oxoplatin-resistant cell lines, 5637-oxo, SISO-oxo, and KYSE70-oxo, which are also completely cross-resistant to cisplatin, no cross-resistance to 6c was observed (Table 3). In fact, the cell lines 5637-oxo and KYSE70-oxo are more than 10-fold more sensitive to 6c than the parent cell lines 5637 and KYSE70, respectively. A dramatic reversal of resistance was not apparent for 2c and 4c, however. These data provide evidence that 6c acts by a mechanism that is amplified when cells become resistant to oxoplatin/cisplatin.
Reactivity toward Proteins and Nucleotides
Protein Affinity
Proteins are known to be primary targets for anticancer metallodrugs.20 With the goal of gaining deeper insight into the mode of action of the complexes, mass spectrometric experiments were conducted to elucidate the affinity toward transferrin (Tf), ubiquitin (Ub), and cytochrome c (Cyt-c). The stoichiometry of the binding to transferrin, which is the most important iron transport protein in the body41 and may thus act as a selective vehicle for ruthenium and other antitumor metal compounds to the iron-demanding tumor cells, as well as to Ub42 and Cyt-c,43 which were chosen as model cellular proteins, was evaluated. Complex 2c was investigated as a representative complex because it has good aqueous solubility while still having a considerable spacer length between the two Ru moieties. Samples containing varying drug-to-protein ratios from 1:1 to 8:1 were prepared and analyzed by electro-spray ionization mass spectrometry (ESI-MS). In the cases of Ub and Cyt-c, no adduct formation could be observed even at an 8-fold excess of the drug (data not shown). This observation also argues against unspecific gas phase reactions that could feign adduct formation.
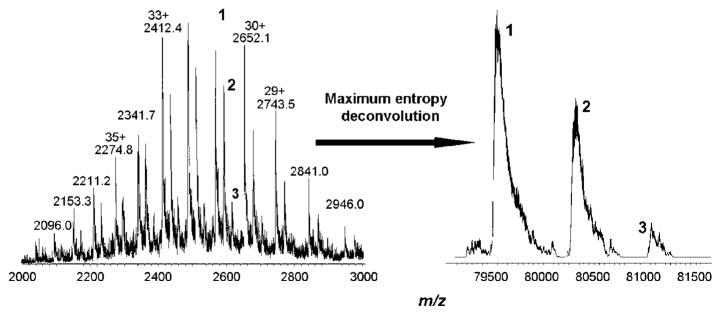
By contrast, adduct formation of 2c with Tf was already observed at a 1:1 ratio. A molecular weight of 79 565 ± 16 Da was determined for the protein, which is in good accordance with the literature.44 A mass increase of approximately 760 Da was assigned to a conjugate formed with the dechlorinated complex, whereas a mass increase of approximately 1520 Da was assigned to the addition of a second dechlorinated species (Figure 6). Interestingly, no adducts corresponding to the binding of more than two complex molecules (mass increase of approximately 1520 Da, exchange of chlorido ligands) were detected, even at 8-fold excess of the complex. One could speculate that specific binding of the Ru moieties to the iron-binding pockets of the serum protein results in very stable conjugates, which might also be enhanced by electrostatic attraction: the calculated pI of the protein is 6.81,45 resulting in an overall negative charge under physiological conditions (pH 7.4), whereas the complex is doubly positively charged because of hydrolysis. An exact elucidation of the structure was not possible because of the inaccuracy of the mass spectrometer equipped with an ion trap analyzer; thus, a higher resolution instrument is required.
Figure 6.
Recorded (left) and maximum entropy deconvoluted (right) mass spectra of transferrin with 2-fold excess of 2c, recorded after 30 min of incubation in 20 mM ammonium carbonate buffer at 37 °C and pH 7.4. Peak 1 corresponds to the protein itself, peak 2 to the monoadduct (mass increase of ~760 Da), peak 3 to the bisadduct (mass increase of ~1520 Da).
Affinity toward DNA and DNA Model Compounds
DNA is regarded as one of the most important targets for Pt anticancer compounds. Binding to their preferred target, i.e., guanine residues, leads to a modification of the DNA secondary structure and inhibition of transcription and replication.1
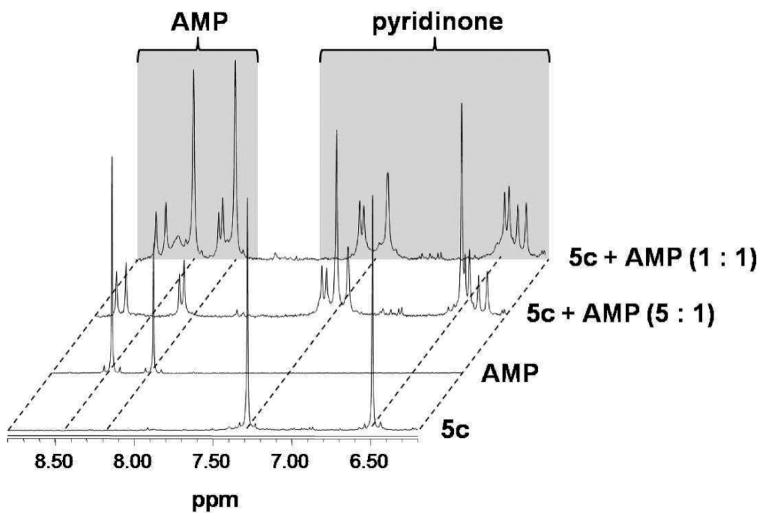
The dinuclear complexes 4c and 5c were reacted with the nucleotides guanosine 5′-monophosphate (GMP), adenosine 5′-monophosphate (AMP), cytidine 5′-monophosphate (CMP), uridine 5′-monophosphate (UMP), and thymidine 5′-monophosphate (TMP) at molar ratios of 20:1, 10:1, 5:1, 3.3:1, 2:1, 1.6:1, and 1:1, and the reaction was followed by 1H NMR and 31P NMR spectroscopy. For both complexes similar behavior was observed: The formation of GMP and AMP adducts proceeded rapidly and quantitatively up to a ratio of 2:1 (complex/nucleotide), as evidenced by a shift of the H8 proton of GMP from approximately 8.1 to 7.8 ppm and of the H8 and H2 protons of AMP from 8.4 to 8.7 and from 8.2 to 8.3, respectively.46 Coordination of AMP resulted in a doublet formation of the signals assigned to the purine protons, and for both AMP and GMP the arene and pyridone regions (6–7 and 5–6 ppm, respectively) of the 1H NMR spectra contained a high number of signals that change over time (Figure 7 for the reaction of AMP with 5c). For CMP, TMP, and UMP no reaction was observed, as also reported for other metal–arene compounds.39 In the 31P NMR spectra no indication for coordination of the phosphate moiety to the metal center was observed.
Figure 7.
1H NMR spectra for the reaction of 5c with AMP at different ratios of metal complex/nucleotide in NaClO4/D2O.
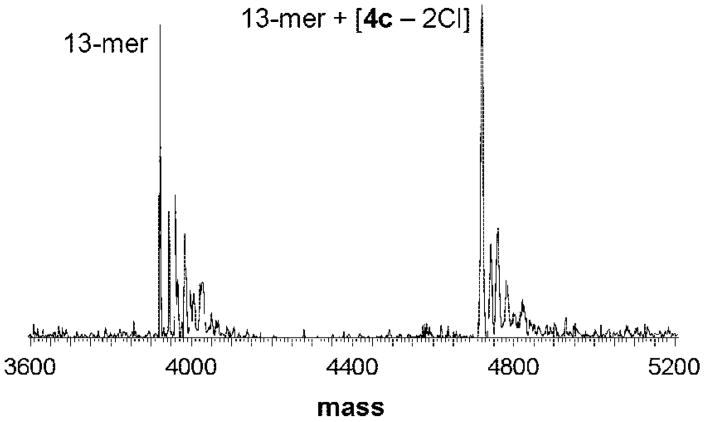
Furthermore, the affinity of 4c to a DNA 13-mer (3921 Da) was studied by means of ESI-MS, and the reaction was followed over 24 h (for experimental details, see Materials and Methods). Different DNA/complex ratios were prepared, and the ruthenation increased with the rB. For example, at rB = 0.15 the 13-mer and an adduct signal, assignable to 13-mer + [4c – 2Cl] (4721 Da), were observed at a relative intensity of approximately 1:1 (Figure 8). When rB > 0.1, the formation of a bis-adduct was attained (13-mer + [2 4c – 4Cl], 5521 Da). At rB = 0.77, the ratio between these two adducts was found to be 1:1, with no unruthenated 13-mer being detectable. It was not possible to further increase the concentration of 4c, since this induced the precipitation of the 13-mer. Furthermore, precipitation was observed in all samples with rB > 0.1 after incubation for more than 24 h, and in the remaining mother liquor neither adduct nor unreacted 13-mer was detectable. As observed for the nucleotides AMP and GMP, the reaction proceeded very rapidly and no changes over time were observed. It was also considered that the dinuclear complex could cross-link two 13-mer molecules; however, such species were not observed in the mass spectra but cannot be excluded to be present in the precipitate.
Figure 8.
Deconvoluted ESI mass spectrum for a mixture of 4c and a DNA 13-mer (molar ratio 1:1; rB = 0.15) after 10 min of incubation time.
The affinity of dinuclear Ru(II) complexes toward DNA was assayed utilizing plasmid DNA, which showed that the addition of 2c and 4c induced slight unwinding of supercoiled plasmid DNA. The reaction of 2c and 4c with calf thymus DNA showed a high degree of ruthenation (nearly 50% of the applied amount). Similar observations were made in a more detailed study on the DNA binding, most notably with a high degree of DNA–protein and DNA interduplex cross-linking, which is again dependent on the spacer length.47
The linearization of plasmid DNA by 2c and 6c was assayed by gel electrophoresis. For this purpose, the two restriction enzymes BamH1 and HindIII, which are known to cleave DNA sequence specifically at GG and AA sites, respectively, were added to 48 h preincubated complex–pUC18 plasmid mixtures. In contrast to traditional platinum anticancer agents,48 both 2c and 6c do not inhibit the linearization of the plasmid DNA.
Conclusions
Ruthenium anticancer drug candidates tend to be significantly less toxic both in vitro and in vivo than the established platinum complexes. Therefore, there exists the potential to increase cytotoxicity without damaging healthy tissue. One proven option for increasing the cytotoxicity of platinum compounds is to link several platinum atoms by diaminoalkane ligands, resulting in compounds such as BBR3464 that are even active in cisplatin-resistant cell lines.
Dinuclear Ru(II)–arene compounds with the Ru centers acting cooperatively in human tumor cell lines have been developed and (bio)analytically characterized with respect to structure–activity relationships. As usual for many organometallic ruthenium compounds, rapid hydrolysis was observed, with the aqua complexes exhibiting pKa values of about 9.7. Similar to other Ru complexes, affinity for transferrin was observed, but surprisingly no interaction with the smaller cellular proteins ubiquitin and cytochrome c was detected. The compounds were found to react rapidly with DNA and model nucleotides, as observed by 1H NMR spectroscopy, DNA precipitation, mass spectrometry, and gel electrophoresis. Their in vitro anticancer activity in several human tumor cell lines correlates with spacer length and lipophilicity. Interestingly, two cell lines resistant to a cisplatin analogue, oxoplatin, are more sensitive to the dinuclear Ru(II)–arene 6c than the native cell lines.
Materials and Methods
All reactions were carried out in dry solvents and under argon atmosphere. Bis[dichlorido(η6-p-cymene)ruthenium(II)]49 and 3-benzyloxy-2-methyl-4-pyrone50 as well as 2c, 4c, 6c,37 chlorido[2-methyl-3-(oxo-κO)-1-propyl-4(1H)-pyridinonato-κO4](η6-p-cymene)ruthenium(II) 7,51 and chlorido(maltolato-κO2)(η6-p-cymene)ruthenium(II) 839 were prepared according to literature procedures. Purchased chemicals were used without further purifications. Melting points were determined with a Büchi B-540 apparatus and are uncorrected. Elemental analyses were carried out with a Perkin-Elmer 2400 CHN elemental analyzer at the Microanalytical Laboratory of the University of Vienna. NMR spectra were recorded at 25 °C on a Bruker Avance DPX400 spectrometer (Ultrashield magnet) at 400.13 MHz (1H) and 100.63 MHz (13C) in MeOH-d4, DMSO-d6, or D2O/CF3COOH (9: 1). Electrospray ionization mass spectrometry (ESI-MS) was performed on a Bruker Esquire3000 instrument (Bruker Daltonics, Bremen, Germany), and theoretical and experimental isotope distributions were compared. Inductively coupled plasma mass spectrometric (ICP-MS) analyses were made on an Agilent 7500ce quadrupole mass spectrometer (Agilent, Waldbronn, Germany) and ultraviolet–visible spectroscopic (UV/vis) measurements on a Perkin-Elmer Lambda 650 instrument (from 500 to 200 nm). Apotransferrin (97%, lot 074K1370), ubiquitin (from bovine red blood cells, min 90%, lot 075K7405), cytochrome c (from horse heart, 99%, lot 065K7001), and ammonium bicarbonate (purum, pa) were products of Sigma-Aldrich (Vienna, Austria). High-purity water used for the MS experiments was obtained from a Millipore Synergy 185 UV Ultrapure Water system (Molsheim, France).
Synthesis
General Procedure for 1a, 3a, and 5a
Sodium hydroxide (1.40 g, 35.0 mmol) was added to a solution of 3-benzyloxy-2-methyl-4-pyrone (10.0 g, 46.2 mmol) and 1,n-diamine (15.4 mmol) in a methanol–water mixture (2:1, 210 mL). The reaction mixture was refluxed for 48 h and then allowed to cool to room temperature. The product was extracted with dichloromethane (3 × 50 mL), and afterward the solvent was removed in vacuum. The pure product was isolated by column chromatography on silica gel.
1,2-Bis[3-benzyloxy-2-methyl-4(1H)-pyridinon-1-yl]ethane (1a)
From NaOH (1.40 g, 35.0 mmol), 3-benzyloxy-2-methyl-4-pyrone (10.0 g, 46.2 mmol), and 1,2-ethylenediamine (0.92 g, 15.4 mmol) 1a was obtained. Yield 2.03 g (29%), mp 198–199 °C. Anal. (C28H28N2O4 · ⅔H2O) C, H, N. MS (ESI+) m/z 457 [M + H]+. 1H NMR in MeOH-d4: δ = 2.08 [s, 6H, CH3], 4.29 [s, 4H, CH2–N], 5.12 [s, 4H, CH2–Ph], 6.36 [d, 2H, CH–C–O, 3J = 7.3 Hz], 7.17 [d, 2H, CH, 3J = 7.6 Hz], 7.35–7.42 [m, 10H, CHarom]. 13 C NMR in MeOH-d4: δ = 11.7 [CH3], 53.2 [CH2–N], 73.4 [CH2–Ph], 116.8 [CH–C–O], 128.5 [CH], 128.5 [CH], 129.2 [CH], 137.4 [Carom], 140.0 [CH], 143.5 [C–CH3], 146.2 [C–O], 174.2 [C–O].
1,4-Bis[3-benzyloxy-2-methyl-4(1H)-pyridinon-1-yl]butane (3a)
From NaOH (1.40 g, 35.0 mmol), 3-benzyloxy-2-methyl-4-pyrone (10.0 g, 46.2 mmol), and 1,4-diaminobutane (1.36 g, 15.4 mmol) 3a was obtained. Yield: 3.91 g (52%), mp 202–204 °C. Anal. (C30H32N2O4 · ⅓H2O) C, H, N. MS (ESI+) m/z 485 [M + H]+. 1H NMR in MeOH-d4: δ = 1.65 [brs, 4H, CH2], 2.17 [s, 6H, CH3], 3.96 [brs, 4H, CH2–N], 5.11 [s, 4H, CH2–Ph], 6.48 [d, 2H, CH–C–O, 3J = 7.0 Hz], 7.32–7.34 [m, 10H, CHarom], 7.67 [d, 2H, CH, 3J = 7.5 Hz]. 13C NMR in MeOH-d4: δ = 11.8 [CH3], 27.2 [CH2CH2–N], 53.5 [CH2–N], 73.4 [CH2–Ph], 116.4 [CH–C–O], 128.4 [CH], 128.4 [CH], 129.3 [CH], 137.4 [Carom], 140.0 [CH], 143.9 [C–CH3], 146.0 [C–O], 173.8 [C–O].
1,8-Bis[3-benzyloxy-2-methyl-4(1H)-pyridinon-1-yl]octane (5a)
From NaOH (1.40 g, 35.0 mmol), 3-benzyloxy-2-methyl-4-pyrone (10.0 g, 46.2 mmol), and 1,8-diaminooctane (2.22 g, 15.4 mmol) 5a was obtained. Yield: 4.04 g (49%), mp 96–98 °C. Anal. (C34H40N2O4 · 2H2O) C, H, N. MS (ESI+) m/z 541 [M + H]+. 1H NMR in MeOH-d4: δ = 1.33 [brs, 8H, CH2], 1.64–1.67 [m, 4H, CH2CH2–N], 2.17 [s, 6H, CH3], 3.94–3.98 [t, 4H, CH2–N, 3J = 7.5 Hz], 5.10 [s, 4H, CH2–Ph], 6.48 [d, 2H, CH–C–O, 3J = 7.6 Hz], 7.33–7.41 [m, 10H, CHarom], 7.69 [d, 2H, CH, 3J = 7.5 Hz]. 13C NMR in MeOH-d4: δ = 11.8 [CH3], 26.1 [CH2(CH2)3–N], 29.1 [CH2(CH2)2–N], 30.5 [CH2CH2–N], 54.3 [CH2–N], 73.4 [CH2–Ph], 116.3 [CH–C–O], 128.3 [CH], 128.4 [CH], 129.4 [CH], 137.4 [Carom], 140.2 [CH], 144.0 [C-CH3], 145.9 [C–O], 173.7 [C–O].
General Procedure for 1b, 3b, and 5b
Hydrogen was passed through a mixture of 1,n-bis[3-benzyloxy-2-methyl-4(1H)-pyridinon-1-yl]alkane and palladium on activated carbon (10% Pd) in 100% acetic acid. The conversion was monitored by means of TLC, and the reaction was terminated when the spot of the starting compound disappeared. The catalyst was filtered off, the solvent was removed, and the product was dried in vacuum.
1,2-Bis[3-hydroxy-2-methyl-4(1H)-pyridinon-1-yl]ethane (1b)
From 1a (0.50 g, 1.10 mmol) and palladium on activated carbon (75 mg, 10% Pd) in acetic acid (100%, 40 mL) 1b was obtained. Yield: 0.26 g (86%), mp 300–302 °C (dec). Anal. (C14H16N2O4 · 7/6H2O) C, H, N. MS (ESI+) m/z 277 [M + H]+. 1H NMR in D2O/CF3COOH: δ = 0.79 [s, 6H, CH3], 3.06 [s, 4H, CH2], 5.37 [d, 2H, CH–C–O, 3J = 7.0 Hz], 5.89 [d, 2H, CH, 3J = 7.1 Hz]. 13C NMR in D2O/CF3COOH: δ = 10.1 [CH3], 53.2 [CH2–N], 110.1 [CH–C–O], 136.3 [CH], 140.1 [C–CH3], 142.7 [C–O], 158.0 [C–O].
1,4-Bis[3-hydroxy-2-methyl-4(1H)-pyridinon-1-yl]butane (3b)
From 3a (0.80 g, 1.65 mmol) and palladium on activated carbon (80 mg, 10% Pd) in acetic acid (100%, 40 mL) 3b was obtained. Yield: 0.20 g (40%), mp 296–298 °C (dec). Anal. (C16H20N2O4) C, H, N. MS (ESI+) m/z 305 [M + H]+. 1H NMR in D2O/CF3COOH: δ = 0.70 [brs, 4H, CH2], 1.34 [s, 6H, CH3], 3.10 [brs, 4H, CH2–N], 5.92 [d, 2H, CH–C–O, 3J = 7.0 Hz], 6.64 [d, 2H, CH, 3J = 7.1 Hz]. 13C NMR in D2O/CF3COOH: δ = 10.5 [CH3], 25.5 [CH2CH2–N], 54.8 [CH2–N], 110.3 [CH–C–O], 136.6 [CH], 140.8 [C–CH3], 142.6 [C–O], 157.5 [C–O].
1,8-Bis[3-hydroxy-2-methyl-4(1H)-pyridinon-1-yl]octane (5b)
From 5a (1.0 g, 1.85 mmol) and palladium on activated carbon (100 mg, 10% Pd) in acetic acid (100%, 20 mL) 5b was obtained. Yield: 0.45 g (68%), mp 261–263 °C (dec). Anal. (C20H28N2O4 · ⅔H2O) C, H, N. MS (ESI+) m/z 361 [M + H]+. 1H NMR in D2O/CF3COOH: δ = 1.07 [brs, 8H, CH2], 1.57 [m, 4H, CH2CH2–N], 2.35 [s, 6H, CH3], 4.08 [t, 4H, CH2–N, 3J = 7.5 Hz], 6.89 [d, 2H, CH–C–O, 3J = 6.5 Hz], 7.80 [d, 2H, CH, 3J = 7.0 Hz]. 13C NMR in D2O/CF3COOH: δ = 12.3 [CH3], 25.4 [CH2(CH2)3–N], 28.2 [CH2(CH2)2–N], 29.6 [CH2CH2–N], 57.1 [CH2–N], 111.0 [CH–C–O], 138.6 [CH], 142.8 [C–CH3], 142.8 [C–O] 158.1 [C–O].
General Procedure for 1c, 3c, and 5c
A solution of bis[dichlorido(η 6-p-isopropyltoluene)ruthenium(II)] in methanol was added to a suspension of 1,n-bis[3-hydroxy-2-methyl-4(1H)-pyridinon-1-yl]alkane and sodium methoxide in methanol. The reaction mixture was stirred at room temperature under argon atmosphere for 2 days. The unconverted ligand was removed by filtration, and the solvent was evaporated under vacuum. The pure complex was obtained by extraction with a dichloromethane–diethyl ether (2:1) mixture. The solvents were removed, and the orange-red compound was dried in vacuum.
1,2-Bis{chlorido[3-(oxo-κO)-2-methyl-4(1H)-pyridinonato-κO4](η 6-p-cymene)ruthenium(II)}ethane (1c)
From bis[dichlorido(η 6-p-cymene)ruthenium(II)] (155 mg, 0.25 mmol), 1b (100 mg, 0.36 mmol), and sodium methoxide (43 mg, 0.80 mmol), 1c was obtained. Yield: 85 mg (41%), mp 160–164 °C (dec). Anal. (C34H42N2O4Ru2Cl2) C, H, N. MS (ESI+) m/z 781 [M–Cl]+ and 373 [M – 2Cl]2+. 1H NMR in MeOH-d4: δ = 1.33 [d, 12H, (CH3)2CH, 3J = 6.8 Hz], 2.28 [s, 6H, CH3], 2.31 [s, 6H, CH3], 2.80–2.88 [m, 2H, CH(CH3)2], 4.44 [s, 4H, CH2–N], 5.43 [d, 4H, CHarom, 3J = 5.8 Hz], 5.65 [d, 4H, CHarom, 3J = 5.8 Hz], 6.31 [d, 2H, CH–C–O, 3J = 6.8 Hz], 6.87 [d, 2H, CH, 3J = 6.8 Hz]. 13C NMR in MeOH-d4: δ = 10.6 [CH3], 17.5 [CH3], 21.6 [(CH3)2CH], 31.4 [CH(CH3)2], 53.9 [CH2–N], 77.9 [CH], 79.7 [CH], 96.2 [Carom], 97.1 [Carom], 109.7 [CH–C–O], 133.0 [CH], 133.9 [C–CH3], 159.9 [C–O], 175.1 [C–O].
1,4-Bis{chlorido[3-(oxo-κO)-2-methyl-4(1H)-pyridinonato-κO4](η 6-p-cymene)ruthenium(II)}butane (3c)
From bis[dichlorido(η 6-p-cymene)ruthenium(II)] (109 mg, 0.18 mmol), 3b (60 mg, 0.20 mmol), and sodium methoxide (23 mg, 0.43 mmol), 3c was obtained. Yield: 85 mg (57%), mp 280–282 °C (dec). Anal. (C36H46N2O4Ru2Cl2) C, H, N. MS (ESI+) m/z 387 [M – 2Cl]2+. 1H NMR in MeOH-d4: δ = 1.33 [d, 12H, (CH3)2CH, 3J = 6.5 Hz], 1.72 [brs, 4H, CH2CH2–N], 2.27 [s, 6H, CH3], 2.45 [s, 6H, CH3], 2.82–2.89 [m, 2H, CH(CH3)2], 4.07 [brs, 4H, CH2–N], 5.43 [d, 4H, CHarom, 3J = 6.0 Hz], 5.66 [d, 4H, CHarom, 3J = 6.0 Hz], 6.48 [d, 2H, CH–C–O, 3J = 6.5 Hz], 7.38 [d, 2H, CH, 3J = 7.0 Hz]. 13C NMR in MeOH-d4: δ = 10.8 [CH3], 17.5 [CH3], 21.6 [(CH3)2CH], 27.4 [CH2CH2–N], 31.4 [CH(CH3)2], 54.2 [CH2–N], 77.9 [CH], 79.6 [CH], 96.0 [Carom], 98.9 [Carom], 109.4 [CH–C–O], 133.6 [CH], 134.2 [C–CH3], 156.4 [C–O], 174.2 [C–O].
1,8-Bis{chlorido[3-(oxo-κO)-2-methyl-4(1H)-pyridinonato-κO4](η 6-p-cymene)ruthenium}octane (5c)
From bis[dichlorido(η 6-p-cymene)ruthenium(II)] (76 mg, 0.12 mmol), 5b (50 mg, 0.14 mmol), and sodium methoxide (17 mg, 0.31 mmol), 5c was obtained. Yield: 73 mg (65%), mp 288–290 °C (dec). Anal. (C40H54N2O4Ru2Cl2) C, H, N. MS (ESI+) m/z 865 [M–Cl]+ and 415 [M – 2Cl]2+. 1H NMR in MeOH-d4: δ = 1.33–1.35 [m, 20H, CH2(CH2)3–N, CH2(CH2)2–N,(CH3)2CH], 1.71 [m, 4H, CH2CH2–N], 2.28 [s, 6H, CH3], 2.49 [s, 6H, CH3], 2.82–2.89 [m, 2H, CH(CH3)2], 4.06 [t, 4H, CH2–N, 3J = 7.5 Hz], 5.45 [d, 4H, CHarom, 3J = 6.0 Hz], 5.67 [d, 4H, CHarom, 3J = 6.0 Hz], 6.51 [d, 2H, CH–C–O, 3J = 6.5 Hz], 7.42 [d, 2H, CH, 3J = 7.0 Hz]. 13C NMR in MeOH-d4: δ = 10.8 [CH3], 17.5 [CH3], 21.6 [(CH3)2CH], 26.1 [CH2(CH2)3–N], 29.0 [CH2(CH2)2–N], 30.6 [CH2CH2–N], 31.4 [CH(CH3)2], 54.9 [CH2–N], 78.0 [CH], 79.6 [CH], 96.0 [Carom], 99.0 [Carom], 109.3 [CH–C–O], 133.6 [CH], 134.1 [C–CH3], 159.6 [C–O], 174.0 [C–O].
Determination of log P Values
The shake flask method was used to determine the n-octanol/water partition coefficient of the dinuclear ruthenium(II)–arene compounds.40 UV/vis spectroscopy and ICP-MS were used to determine the concentration of the ruthenium complexes in the organic and aqueous phase, respectively. For UV/vis quantification, the wavelength of the absorption maximum was determined for each complex (λ 327, 325, and 325 nm for 1c, 3c, and 5c, respectively), and an external calibration was performed in the range from 1 to 100 μM with standards prepared in n-octanol. The ruthenium concentration in the aqueous phase was determined by ICP-MS using external calibration from 1 to 30 ppb. Both 101Ru and 102Ru isotopes were employed for quantification and gave essentially the same results. All solutions were prepared in 5% HNO3 (HNO3, pa from Fluka, further purified with a quartz sub-boiling system from Milestone-MLS GmbH, Leutkirch, Germany; 18.2 MΩ H2O was obtained from a Synergy 185, Millipore, Bedford, MA), and 1 ppb of 185Re was added for internal standardization to the samples and standard solutions.
Determination of pKa Values
The pH values of the NMR samples of the aqua complexes (prepared in D2O and stirred overnight in the presence of 2 equiv of AgNO3) were measured with an Eco Scan pH meter equipped with a combination pH glass electrode–microelectrode (Orion 9826BN) and calibrated with standard buffer solutions of pH 4, 7, and 10. The pH titration was performed by addition of sodium deuteroxide (40% in D2O) or D-nitric acid (65% in D2O), and the curves were fitted to the Henderson–Hasselbalch equation by KALEIDAGRAPH (version 4.03) software, assuming that the observed chemical shifts are weighted averages according to the populations of the protonated and deprotonated species. The pKa values determined experimentally were corrected by using eq 1,52 which converts the pKa value determined in D2O (pKa*) into the corresponding pKa in aqueous solutions.
| (1) |
Determination of the Chloride Ion Concentration
The chloride ion concentration after dissolution of 4c in water or in water/methanol was recorded with a CyberScan 2100 pH/ion meter over time at 25 °C. The instrument was equipped with an Eutech chloride ion combination epoxy-body electrode calibrated from 10 to 1000 ppm. The ionic strength of the samples was maintained constant by addition of 0.1 mL of 5 M NaNO3 solution to 4.9 mL of sample solution (0.5 or 1 mM).
DNA and Nucleotide Binding Studies
Gel Electrophoresis
The plasmid pUC18 (2686 bp) was transformed in XL1 blue E. coli bacteria, isolated, purified according to standard procedures,53 and dissolved in TE buffer (10 mM Tris-Cl, pH 7.4, 1 mM Na2EDTA).
In order to investigate the type of interaction between plasmid DNA and the dinuclear complexes, an amount of 100 ng of pUC18 was incubated with 2c and 6c, applying a concentration gradient from 0 to 50 μM in 20 mM phosphate buffer at pH 7.4 and 37 °C. After 48 h of incubation in the dark, the split sample was incubated with the restriction enzymes BamH1 and HindIII for 2 h at 37 °C. Afterward, both the digested and the nondigested samples were loaded on a 1% agarose gel and separated at 25 °C and 100 V in TAE buffer (0.04 M Tris-acetate, 1 mM EDTA, pH 7.0). The gel was stained with ethidium bromide (0.2 μg/mL) in TAE buffer for 30 min and visualized by UV.
Mass Spectrometry
For the MS studies, the non-self-complementary single-stranded DNA 13-mer 5′-ATC TGT TTG TCT T-3′ (3921 Da; Midland Certified Reagents, Midland TX) was incubated with 4c in H2O at DNA (40 μM), with complex ratios ranging from 5:1 (rB = 0.031; rb is the ruthenium: nucleotide ratio) to 1:5 (rB = 0.77). Before use, the purified 13-mer was further desalted by using a custom built dialysis chamber equipped with a hollow microdialysis fiber with a 13 kDa molecular weight cut-off (MWCO) from Spectrum Laboratories (Rancho Dominguez, CA) in 25 mM ammonium acetate solution. The desalted oligonucleotide was lyophilized to dryness and reconstituted in diionized water (18 MΩ).
The sample mixtures were analyzed immediately after mixing and after 0.5 and 24 h of incubation with a Waters/Micromass Qtof-2 (Manchester, U.K.) instrument equipped with a custom built nanospray source operated in negative ion mode over a mass range of 500–2000 m/z. Samples were introduced into the inlet at 1.0 μL/min with a capillary voltage of −1.9 kV and a cone voltage of 36 V. The source temperature was maintained constant at 110 °C throughout the experiments. A 1:1 mixture of methanol/water (with 25 mM ammonium acetate) was used as the spray solvent. Data were collected and processed by using the Mass Lynx 4.0 software, and the deconvolution to molecular mass scale was performed with the maximum entropy (Max Ent) software supplied with the instrument.
NMR Spectroscopy
For studying the reaction of the dinuclear complex 5c (1 mM) with nucleotides, the metal complex was reacted with guanosine 5′-monophosphate (GMP), adenosine 5′-monophosphate (AMP), cytidine 5′-monophosphate (CMP), uridine 5′-monophosphate (UMP), and thymidine 5′-monophosphate (TMP) at molar ratios of 20:1, 10:1, 5:1, 3.3:1, 2:1, 1.6:1, and 1:1 in 10 mM NaClO4 in D2O, and the reaction was followed by 1H NMR and 31P NMR spectroscopy up to 7 days.
Protein Binding Studies
An Esquire3000 ion trap mass spectrometer (Bruker Daltonics, Bremen, Germany), equipped with an orthogonal ESI ion source, was used for mass spectrometric determination of the complex–protein interactions. The instrument was operated in positive ion mode for characterizing proteins and protein–metal adducts. To ensure the best performance and to simulate physiological pH, samples containing varying drug to protein ratios (from 1:1 to 8:1, protein concentrations: transferrin, 50 μM; ubiquitin, 25 μM; cytochrome c, 25 μM) were incubated at 37 °C in 20 mM ammonium bicarbonate buffer adjusted to pH 7.4 by titration with 0.1 M formic acid. Prior to measurement, 30% acetonitrile (cytochrome c, ubiquitin) or isopropanol (transferrin) and 1% formic acid (10% in water) were added to the samples to ensure good spraying conditions; the samples were measured twice after incubation periods of 30 min and 24 h. The solutions were introduced via flow injection at a rate of 4 μL/min by using a Cole-Parmer 74900 single-syringe infusion pump (Vernon Hills, IL). The ESI-MS instrument was controlled by means of the Esquire Control software (version 5.2), and all data were processed using Data Analysis software (version 3.2) (both Bruker Daltonics).
Cytotoxicity in Cancer Cell Lines
Cell Lines and Culture Conditions
Human SW480 (colon carcinoma) and A2780 (ovarian carcinoma) cells were kindly provided by Brigitte Marian (Institute of Cancer Research, Medical University of Vienna, Austria) and Evelyn Dittrich (General Hospital, University of Vienna, Austria). Cells were grown in 75 cm2 culture flasks (Iwaki/Asahi Technoglass, Gyouda, Japan) as adherent monolayer cultures in complete culture medium, i.e., Minimal Essential Medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum, 1 mM sodium pyruvate, 4 mM L-glutamine, and 1% nonessential amino acids (100×) (all purchased from Gibco/Invitrogen, Paisley, U.K.). The cell lines 5637 and RT-4 (bladder cancer), LCLC-103H and A-427 (lung cancer), DAN-G (pancreatic cancer), MCF-7 (breast cancer), KYSE70 (esophagus cancer), and SISO (cervical cancer) were obtained from the German Collection of Microorganisms and Cell Culture (DSMZ, Braunschweig, FRG). The three oxoplatin-resistant cell lines were established in our laboratories (Greifswald) through weekly exposure of cells to increasing concentrations of oxoplatin over a period of several months. Cells were grown in culture flasks (Sarstedt, Germany) as adherent monolayer cultures in 90% RPMI medium supplemented with 10% heat-inactivated fetal bovine serum and the antibiotics benzyl-penicillin and streptomycin. To the medium for the MCF-7 cells, 1 mM sodium pyruvate and 1% nonessential amino acids were added. Cultures were maintained at 37 °C in a humidified atmosphere containing 5% CO2.
MTT Assay Conditions
Cytotoxicity was determined by means of a colorimetric microculture assay (MTT assay, MTT = 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide). For this purpose, SW480 and A2780 cells were harvested from culture flasks by trypsinization and seeded into 96-well microculture plates (Iwaki/Asahi Technoglass, Gyouda, Japan) in densities of 2.5 × 103 and 5.0 × 103 cells/well, respectively, in order to ensure exponential growth throughout drug exposure. After a 24 h preincubation, cells were exposed to serial dilutions of the test compounds in 200 μL/well complete culture medium for 96 h. At the end of exposure, drug solutions were replaced by 100 μL/well RPMI 1640 culture medium (supplemented with 10% heat-inactivated fetal bovine serum and 2 mM L-glutamine) and 20 μL/well MTT solution in phosphate-buffered saline (5 mg/mL). After incubation for 4 h, the medium/MTT mixtures were removed and the formazan crystals formed by vital cells were dissolved in 150 μL of DMSO per well. Optical densities at λ = 550 nm were measured with a microplate reader (Tecan Spectra Classic) by using a reference wavelength of λ = 690 nm to correct for unspecific absorption. Quantities of vital cells were expressed as T/C values by comparison to untreated control microcultures, and 50% inhibitory concentrations (IC50) were calculated from concentration–effect curves by interpolation. Evaluation is based on mean values from at least three independent experiments, each comprising at least six replicates per concentration level.
Crystal Violet Assay Conditions
This assay has been described in detail elsewhere.54 Culture conditions were the same as used in the MTT assay. Briefly, cells were seeded into 96-well microculture plates (Sarstedt, Germany) in cell densities of 1.0 × 103 cells/well except for LCLC-103H, which was seeded at 250 cells/well. After a 24 h preincubation, cells were treated with test substance for 96 h. Stock solutions of test substance were prepared to 20 mM in DMF and diluted 1000-fold in RPMI 1640 culture medium containing 10% fetal calf serum. Substances that showed a ≥50% growth inhibition at 20 μM were tested at five serial dilutions in 4 wells/concentration to determine the IC50 values as described.54 The staining of the cells was done for 30 min with a 0.02% crystal violet solution in water followed by a washing out of the excess dye. Cell bound dye was redissolved in 70% ethanol/water solution, and the optical densities at λ = 570 nm were measured with a microplate reader (Anthos 2010).
Supplementary Material
Acknowledgments
We thank the University of Vienna, the Hochschuljubiläumsstiftung Vienna, the Theodor-Körner-Fonds, the Austrian Council for Research and Technology Development, the FFG—Austrian Research Promotion Agency (Project FA 526003), the FWF—Austrian Science Fund (Schrödinger Fellowship J2613-N19 [C.G.H.]), the EPFL, and COST D39 for financial support. This research was supported by a Marie Curie Intra European Fellowship within the 7th European Community Framework Programme Projects 220890-SuRuCo (A.A.N.). We gratefully acknowledge Dr. Markus Galanski for recording the NMR spectra.
Footnotes
Supporting Information Available: Results from elemental analysis, hydrolysis of 4c, and pKa determination. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Galanski M, Jakupec MA, Keppler BK. Update of the preclinical situation of anticancer platinum complexes: novel design strategies and innovative analytical approaches. Curr Med Chem. 2005;12:2075–2094. doi: 10.2174/0929867054637626. [DOI] [PubMed] [Google Scholar]
- 2.Zhang CX, Lippard SJ. New metal complexes as potential therapeutics. Curr Opin Chem Biol. 2003;7:481–489. doi: 10.1016/s1367-5931(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 3.Hartinger CG, Dyson PJ. Bioorganometallic chemistry – from teaching paradigms to medicinal applications. Chem Soc Rev. 2009 doi: 10.1039/B707077M. [DOI] [PubMed] [Google Scholar]
- 4.van Zutphen S, Reedijk J. Targeting platinum anti-tumour drugs: overview of strategies employed to reduce systemic toxicity. Coord Chem Rev. 2005;249:2845–2853. [Google Scholar]
- 5.Haag R, Kratz F. Polymer therapeutics: concepts and applications. Angew Chem, Int Ed. 2006;45:1198–1215. doi: 10.1002/anie.200502113. [DOI] [PubMed] [Google Scholar]
- 6.Hartinger CG, Nazarov AA, Ashraf SM, Dyson PJ, Keppler BK. Carbohydrate–metal complexes and their potential as anticancer agents. Curr Med Chem. 2008;15:2574–2591. doi: 10.2174/092986708785908978. [DOI] [PubMed] [Google Scholar]
- 7.Kasparkova J, Nováková O, Vrana O, Farrell N, Brabec V. Effect of geometric isomerism in dinuclear platinum antitumor complexes on DNA interstrand cross-linking. Biochemistry. 1999;38:10997–11005. doi: 10.1021/bi990245s. [DOI] [PubMed] [Google Scholar]
- 8.Manzotti C, Pratesi G, Menta E, Domenico RD, Cavalletti E, Fiebig HH, Kelland LR, Farrell N, Polizzi D, Supino R, Pezzoni G, Zunino F. BBR 3464: a novel triplatinum complex, exhibiting a preclinical profile of antitumor efficacy different from cisplatin. Clin Cancer Res. 2000;6:2626–2634. [PubMed] [Google Scholar]
- 9.Jodrell DI, Evans TRJ, Steward W, Cameron D, Prendiville J, Aschele C, Noberasco C, Lind M, Carmichael J, Dobbs N, Camboni G, Gatti B, De Braud F. Phase II studies of BBR3464, a novel tri-nuclear platinum complex, in patients with gastric or gastro-oesophageal adenocarcinoma. Eur J Cancer. 2004;40:1872–1877. doi: 10.1016/j.ejca.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 10.Kostova I. Platinum complexes as anticancer agents. Recent Pat Anti-Cancer Drug Discovery. 2006;1:1–22. doi: 10.2174/157489206775246458. [DOI] [PubMed] [Google Scholar]
- 11.Hensing TA, Hanna NH, Gillenwater HH, Camboni MG, Allievi C, Socinski MA. Phase II study of BBR 3464 as treatment in patients with sensitive or refractory small cell lung cancer. Anti-Cancer Drugs. 2006;17:697–704. doi: 10.1097/01.cad.0000215054.62942.7f. [DOI] [PubMed] [Google Scholar]
- 12.Rademaker-Lakhai JM, Van Den Bongard D, Pluim D, Beijnen JH, Schellens JHM. A phase I and pharmacological study with imidazolium-trans-DMSO-imidazole-tetrachlororuthenate, a novel ruthenium anticancer agent. Clin Cancer Res. 2004;10:3717–3727. doi: 10.1158/1078-0432.CCR-03-0746. [DOI] [PubMed] [Google Scholar]
- 13.Hartinger CG, Zorbas-Seifried S, Jakupec MA, Kynast B, Zorbas H, Keppler BK. From bench to bedside—preclinical and early clinical development of the anticancer agent indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019 or FFC14A) J In-org Biochem. 2006;100:891–904. doi: 10.1016/j.jinorgbio.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Piccioli F, Sabatini S, Messori L, Orioli P, Hartinger CG, Keppler BK. A comparative study of adduct formation between the anticancer ruthenium(III) compound HInd trans-[RuCl4(Ind)2] and serum proteins. J Inorg Biochem. 2004;98:1135–1142. doi: 10.1016/j.jinorgbio.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Timerbaev AR, Aleksenko KS, Polec-Pawlak K, Ruzik R, Semenova O, Hartinger CG, Oszwaldowski S, Galanski M, Jarosz M, Keppler BK. Platinum metallodrug–protein binding studies by capillary electrophoresis–inductively coupled plasma-mass spectrometry: characterization of interactions between Pt(II) complexes and human serum albumin. Electrophoresis. 2004;25:1988–1995. doi: 10.1002/elps.200305984. [DOI] [PubMed] [Google Scholar]
- 16.Hartinger CG, Hann S, Koellensperger G, Sulyok M, Grössl, Timerbaev AR, Rudnev AV, Stingeder G, Keppler BK. Interactions of a novel ruthenium-based anticancer drug (KP1019 or FFC14a) with serum proteins—significance for the patient. Int J Clin Pharmacol Ther. 2005;43:583–585. doi: 10.5414/cpp43583. [DOI] [PubMed] [Google Scholar]
- 17.Sulyok M, Hann S, Hartinger CG, Keppler BK, Stingeder G, Koellensperger G. Two dimensional separation schemes for investigation of the interaction of an anticancer ruthenium(III) compound with plasma proteins. J Anal At Spectrom. 2005;20:856–863. [Google Scholar]
- 18.Timerbaev AR, Rudnev AV, Semenova O, Hartinger CG, Keppler BK. Comparative binding of antitumor indazolium [trans-tetrachlorobis(1H-indazole)ruthenate(III)] to serum transport proteins assayed by capillary zone electrophoresis. Anal Biochem. 2005;341:326–333. doi: 10.1016/j.ab.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 19.Polec-Pawlak K, Abramski JK, Semenova O, Hartinger CG, Timerbaev AR, Keppler BK, Jarosz M. Platinum group metallodrug–protein binding studies by capillary electrophoresis–inductively coupled plasma-mass spectrometry: A further insight into the reactivity of a novel antitumor ruthenium(III) complex toward human serum proteins. Electrophoresis. 2006;27:1128–1135. doi: 10.1002/elps.200500694. [DOI] [PubMed] [Google Scholar]
- 20.Timerbaev AR, Hartinger CG, Aleksenko SS, Keppler BK. Interactions of antitumor metallodrugs with serum proteins: advances in characterization using modern analytical methodology. Chem Rev. 2006;106:2224–2248. doi: 10.1021/cr040704h. [DOI] [PubMed] [Google Scholar]
- 21.Groessl M, Reisner E, Hartinger CG, Eichinger R, Semenova O, Timerbaev AR, Jakupec MA, Arion VB, Keppler BK. Structure–activity relationships for NAMI-A-type complexes (HL)[trans-RuCl4L(S-dmso)ruthenate(III)] (L = imidazole, indazole, 1,2,4-triazole, 4-amino-1,2,4-triazole, and 1-methyl-1,2,4-triazole): aquation, redox properties, protein binding, and antipro-liferative activity. J Med Chem. 2007;50:2185–2193. doi: 10.1021/jm061081y. [DOI] [PubMed] [Google Scholar]
- 22.Jakupec MA, Reisner E, Eichinger A, Pongratz M, Arion VB, Galanski M, Hartinger CG, Keppler BK. Redox-active antineoplastic ruthenium complexes with indazole: correlation of in vitro potency and reduction potential. J Med Chem. 2005;48:2831–2837. doi: 10.1021/jm0490742. [DOI] [PubMed] [Google Scholar]
- 23.Schluga P, Hartinger CG, Egger A, Reisner E, Galanski M, Jakupec MA, Keppler BK. Redox behavior of tumor inhibiting ruhenium(III) complexes and effects of physiological reductants on their binding to GMP. Dalton Trans. 2006:1796–1802. doi: 10.1039/b511792e. [DOI] [PubMed] [Google Scholar]
- 24.Clarke MJ, Zhu F, Frasca DR. Non-platinum chemotherapeutic metallopharmaceuticals. Chem Rev. 1999;99:2511–2533. doi: 10.1021/cr9804238. [DOI] [PubMed] [Google Scholar]
- 25.Clarke MJ. Ruthenium metallopharmaceuticals. Coord Chem Rev. 2003;236:209–233. [Google Scholar]
- 26.Yan YK, Melchart M, Habtemariam A, Sadler PJ. Organometallic chemistry, biology and medicine: ruthenium arene anticancer complexes. Chem Commun. 2005;4764:4776. doi: 10.1039/b508531b. [DOI] [PubMed] [Google Scholar]
- 27.Ang WH, Dyson PJ. Classical and non-classical ruthenium-based anticancer drugs: towards targeted chemotherapy. Eur J Inorg Chem. 2006:4003–4018. [Google Scholar]
- 28.Allardyce CS, Dyson PJ. Medicinal properties of organometallic compounds. Top Organomet Chem. 2006;17:177–210. [Google Scholar]
- 29.Chatterjee S, Kundu S, Bhattacharyya A, Hartinger CG, Dyson PJ. The ruthenium(II)-arene compound RAPTA-C induces apoptosis in EAC cells through mitochondrial and p53-JNK pathways. J Biol Inorg Chem. 2008;13:1149–1155. doi: 10.1007/s00775-008-0400-9. [DOI] [PubMed] [Google Scholar]
- 30.Huxham LA, Cheu ELS, Patrick BO, James BR. The synthesis, structural characterization, and in vitro anti-cancer activity of chloro(p-cymene) complexes of ruthenium(II) containing a disulfoxide ligand. Inorg Chim Acta. 2003;352:238–246. [Google Scholar]
- 31.Bergamo A, Stocco G, Gava B, Cocchietto M, Alessio E, Serli B, Iengo E, Sava G. Distinct effects of dinuclear ruthenium(III) complexes on cell proliferation and on cell cycle regulation in human and murine tumor cell lines. J Pharmacol Exp Ther. 2003;305:725–732. doi: 10.1124/jpet.102.047803. [DOI] [PubMed] [Google Scholar]
- 32.Hotze ACG, Kariuki BM, Hannon MJ. Dinuclear double-stranded metallosupramolecular ruthenium complexes: potential anticancer drugs. Angew Chem, Int Ed. 2006;45:4839–4842. doi: 10.1002/anie.200601351. [DOI] [PubMed] [Google Scholar]
- 33.Therrien B, Ang WH, Cherioux F, Vieille-Petit L, Juillerat-Jeanneret L, Suess-Fink G, Dyson PJ. Remarkable anticancer activity of triruthenium-arene clusters compared to tetraruthenium-arene clusters. J Cluster Sci. 2007;18:741–752. [Google Scholar]
- 34.Schmitt F, Govindaswamy P, Suess-Fink G, Ang WH, Dyson PJ, Juillerat-Jeanneret L, Therrien B. Ruthenium porphyrin compounds for photodynamic therapy of cancer. J Med Chem. 2008;51:1811–1816. doi: 10.1021/jm701382p. [DOI] [PubMed] [Google Scholar]
- 35.Auzias M, Therrien B, Suess-Fink G, Stepnicka P, Ang WH, Dyson PJ. Ferrocenoyl pyridine arene ruthenium complexes with anticancer properties: synthesis, structure, electrochemistry, and cytotoxicity. Inorg Chem. 2008;47:578–583. doi: 10.1021/ic7018742. [DOI] [PubMed] [Google Scholar]
- 36.Therrien B, Suess-Fink G, Govindaswamy P, Renfrew AK, Dyson PJ. The “complex-in-a-complex” cations [(acac)2MRu6(p-iPrC6H4Me)6(tpt)2(dhbq)3]6+: a Trojan horse for cancer cells. Angew Chem, Int Ed. 2008;47:3773–3776. doi: 10.1002/anie.200800186. [DOI] [PubMed] [Google Scholar]
- 37.Mendoza-Ferri MG, Hartinger CG, Eichinger RE, Stolyarova N, Severin K, Jakupec MA, Nazarov AA, Keppler BK. Influence of the spacer length on the in vitro anticancer activity of dinuclear ruthenium-arene compounds. Organometallics. 2008;27:2405–2407. [Google Scholar]
- 38.Mendoza-Ferri MG, Hartinger CG, Nazarov AA, Kandioller W, Severin K, Keppler BK. Modifying the structure of dinuclear ruthenium complexes with antitumor activity. Appl Organomet Chem. 2008;22:326–332. [Google Scholar]
- 39.Peacock AFA, Melchart M, Deeth RJ, Habtemariam A, Parsons S, Sadler PJ. Osmium(II) and ruthenium(II) arene maltolato complexes rapid hydrolysis and nucleobase binding. Chem Eur J. 2007;13:2601–2613. doi: 10.1002/chem.200601152. [DOI] [PubMed] [Google Scholar]
- 40.OECD Guidelines for Testing of Chemicals. 107 Organisation for Economic Co-operation and Development; Paris: 1995. [Google Scholar]
- 41.Gomme PT, McCann KB, Bertolini J. Transferrin: structure, function and potential therapeutic actions. Drug Discovery Today. 2005;10:267–273. doi: 10.1016/S1359-6446(04)03333-1. [DOI] [PubMed] [Google Scholar]
- 42.Hartinger CG, Ang WH, Casini A, Messori L, Keppler BK, Dyson PJ. Mass spectrometric analysis of ubiquitin–platinum interactions of leading anticancer drugs: MALDI versus ESI. J Anal At Spectrom. 2007;22:960–967. [Google Scholar]
- 43.Casini A, Gabbiani C, Mastrobuoni G, Messori L, Moneti G, Pieraccini G. Exploring metallodrug–protein interactions by ESI mass spectrometry: the reaction of anticancer platinum drugs with horse heart cytochrome c. Chem Med Chem. 2006;1:413–417. doi: 10.1002/cmdc.200500079. [DOI] [PubMed] [Google Scholar]
- 44.Feng R, Konishi Y, Bell AW. High accuracy molecular weight determination and variation characterization of proteins up to 80 ku by ionspray mass spectrometry. J Am Soc Mass Spectrom. 1991;2:387–401. doi: 10.1016/1044-0305(91)85005-Q. [DOI] [PubMed] [Google Scholar]
- 45.Swiss Institute of Bioinformatics ExPASy Proteomics Server Basel. Switzerland: 2003. http://www.expasy.org/ [Google Scholar]
- 46.Dorcier A, Hartinger CG, Scopelliti R, Fish RH, Keppler BK, Dyson PJ. Studies on the reactivity of organometallic Ru–, Rh–and Os–pta complexes towards DNA model compounds. J Inorg Biochem. 2008;102:1066–1076. doi: 10.1016/j.jinorgbio.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 47.Nováková O, Nazarov AA, Hartinger CG, Keppler BK, Brabec V. DNA interactions of dinuclear RuII arene antitumor complexes in cell-free media. Biochem Pharmacol. 2009 doi: 10.1016/j.bcp.2008.1010.1021. [DOI] [PubMed] [Google Scholar]
- 48.Huq F, Daghriri H, Yu JQ, Tayyem H, Beale P, Zhang M. Synthesis, characterisation, activities, cell uptake and DNA binding of [{trans-PtCl(NH3) 2}{μ-(H2N(CH26 )NH2)}{trans-PdCl-(NH3) 2}](NO3)Cl. Eur J Med Chem. 2004;39:947–958. doi: 10.1016/j.ejmech.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 49.Bennett MA, Huang TN, Matheson TW, Smith AK. (η6-Hexamethylbenzene)ruthenium complexes. Inorg Synth. 1982;21:74–78. [Google Scholar]
- 50.Harris RLN. Potential wool growth inhibitors. Synthesis of DL-α-amino-β-(5-hydroxy-4-oxo-3,4-dihydropyrimidin-2-yl)propionic acid, a pyrimidine analog of mimosine. Aust J Chem. 1976:1335–1339. [Google Scholar]
- 51.Lang R, Polborn K, Severin T, Severin K. Halfsandwich complexes of ruthenium(II), rhodium(III) and iridium(III) with N-substituted 3-hydroxy-2-methyl-4-pyridone ligands. Inorg Chim Acta. 1999;294:62–67. [Google Scholar]
- 52.Krezel A, Bal W. A formula for correlating pKa values determined in D2O and H2O. Biochemistry. 2004;98:161–166. doi: 10.1016/j.jinorgbio.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bracht K, Boubakari, Grünert R, Bednarski PJ. Correlations between the activities of 19 anti-tumor agents and the intracellular glutathione concentrations in a panel of 14 human cancer cell lines: comparisons with the National Cancer Institute data. Anti-Cancer Drugs. 2006;17:41–51. doi: 10.1097/01.cad.0000190280.60005.05. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.