Abstract
The NF-κB transcription factor is a critical regulator of the immune system, and is responsive to a large number of stimuli. Different stimuli engage signaling pathways to activate NF-κB, and effect distinct cellular responses. Mathematical modeling of the NF-κB network has been useful in studying the dynamic and cross-talk regulation of NF-κB. In this review, we discuss the regulation of NF-κB activity in response to different types of stimuli, including inflammatory signals, developmental cues, metabolic stress, and DNA damage. The distinct molecular mechanisms engaged in each pathway for activating and terminating NF-κB activity are discussed. In addition, we summarize the evidence for cross-talk mechanisms that allow for different stimuli to be integrated within the NF-κB signaling module to produce synergistic or qualitatively different signaling outcomes.
INTRODUCTION
NF-κB refers to a family of transcription factors involved in the regulation of inflammation, innate and adaptive immune responses and development, cell survival, and proliferation. They are the effectors of a signaling system that is responsive to a large number of stimuli, mediated by most members of the tumor necrosis factor receptor (TNFR) and toll-like receptor (TLR) superfamilies, as well metabolic or genotoxic stress inducers. As a critical regulator of immunity, NF-κB signaling components can be found in almost all multicellular organisms, including mammals, insects, urchins, and mollusks but not Caenorhabditis elegans.1,2 Mice mutated in NF-κB signaling components show a diverse range of phenotypes in immune development, apoptosis, and immune response regulation, and in humans, misregulated NF-κB is a hallmark of a variety of chronic inflammatory diseases and cancers.3,4 Further, mutations in signaling components directly involved in the NF-κB pathway have been implicated in several inherited genetic diseases and immune disorders.5
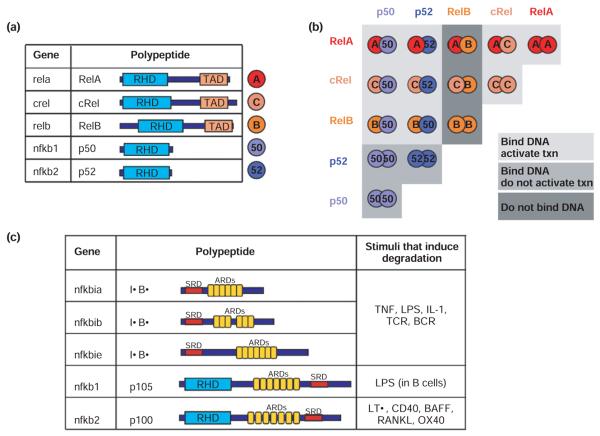
In mammals, the genes rela, relb, crel, nfkb1, and nfkb2 encode the five NF-κB protein family members RelA (p65), RelB, c-Rel, p50, and p52, respectively, which form homo- and heterodimeric DNA-binding complexes (Figure 1(a)). All five family members have a characteristic Rel homology domain (RHD) responsible for DNA binding and dimerization. Whereas RelA, c-Rel, and RelB each possess a transcriptional activation domain (TAD), p50 and p52 do not. Thus, of the 15 theoretically possible NF-κB dimers, some function as transcriptional activators (notably the ubiquitous RelA:p50 heterodimer), but others (notably the p50:p50 homodimer) do not unless they recruit specific coactivator proteins, and some dimers are not known to bind DNA at all6 (Figure 1(b)).
FIGURE 1.
The components of the NF-κB signaling system a) The NF-κB family members. The gene name is indicated for each polypeptide. Each NF-κB family member has a Rel Homology Domain (RHD) for dimerization and DNA binding. RelA, cRel, and RelB have Transcriptional Activation Domains (TAD). b) The 5 NF-κB monomers can combine to form 15 potential dimers. Of these, 9 can bind DNA and activate gene transcription (light grey), 3 (the p50 or p52 only containing dimers) bind DNA but do not activate transcription (medium grey), and 3 do not bind DNA (dark grey). c) The IκB protein family members and signals that induce the degradation of each. The ARDs on p105 and p100 (which are proteolytically processed to p50 and p52 NF-κB monomers, respectively) can act to self-inhibit p50 and p52. p100 can form a multimeric complex in which it can inhibit other latent NF-κB dimers. BCR = B cell receptor; TCR = T cell receptor RHD = Rel Homology Domain; ARD = ankyrin repeat domain; TAD = transcriptional activation domain, SRD = signal response domain.
In the absence of stimuli, NF-κB dimers are retained in the cytosol through association with an inhibitor of κB activity, termed IκB. All IκB proteins have an ankyrin repeat domain (ARD), which forms a large interaction surface around the NF-κB dimer (Figure 1(c)). The three classical or so-called canonical IκB proteins, IκBα, IκBβ, and IκBε, are encoded by the nfκbia, nfκbib, and nfκbie genes, respectively, and a fourth IκB activity was recently characterized.7 This noncanonical IκB activity, which we term IκBδ, results from the multimeric association of the nfκb2-encoded p52 precursor protein p100. Together, these four IκBs prevent DNA binding by NF-κB dimers by shifting their cellular localization to the cytoplasm. Each IκB contains a signal responsive domain (SRD) that contains phosphorylation and ubiquitination sites for signal responsive degradation. Such signals therefore liberate NF-κB from the canonical and noncanonical IκB proteins, allowing binding of κB site sequences (defined by a loose consensus of GGRNNN(N)YCC) in the promoters and enhancers of a myriad of genes.8
Other IκB-like proteins with different functions have been identified. The p50 and p52 proteins are produced via proteolytic processing of the precursor proteins p105 and p100, respectively, each of which contains a C-terminal ARD. Therefore, dimerization via the RHD by p100 or p105 with other NF-κB proteins results in self-inhibited dimers. In addition, the IκB protein family members Bcl3, IκBζ/MAIL, and IκBNS bind subsets of NF-κB dimers, but do not inhibit DNA binding—instead these proteins may function as coactivators, for example, for the TAD-deficient NF-κB dimers p50:p50 or p52:p52.9-13
NF-κB is generally thought to be an antiapoptotic, proproliferative, and proinflammatory regulator. Indeed, it is often upregulated in human cancers, where it has been shown to contribute to proliferation, tumor growth, metastasis, and chemoresistance. Deregulated NF-κB activity is also found in chronic inflammatory diseases, such as rheumatoid arthritis, Crohn's disease, and ulcerative colitis. As such, the NF-κB activation mechanism has been studied in detail in the hope of developing specific pharmacological strategies to control deregulated NF-κB activity.
INFLAMMATORY SIGNALING
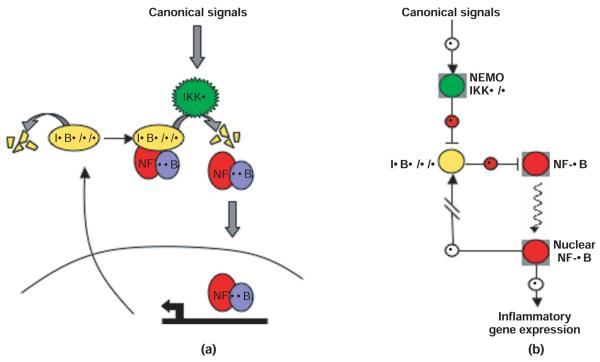
Endogenous inflammatory stimuli (e.g., cytokines TNFα, IL–1β) or pathogen-derived substances (e.g., lipopolysaccharide (LPS) or CpG) activate the ubiquitous RelA:p50 dimer through the ‘canonical’ or ‘classical’ NF-κB pathway. Engagement of the TNF receptor (TNFR), interleukin–1β receptor (IL–1βR), or TLRs causes phosphorylation-dependent activation of the IκB kinase (IKK) complex, composed of the two catalytic subunits, IKKα and IKKβ, and the scaffolding protein, IKKγ/NF-κB essential modulator (NEMO). Once activated, the canonical IKK complex phosphorylates IκBα, −β, and −ε at two specific serine residues. IκBα is the predominant IκB in most cells, bound to RelA:p50. Phosphorylation of IκBβ at serines 32 and 36 signals as a docking site for the E3 ubiquitin ligase β-TrCp, which catalyzes K48-linked ubiquitination at lysines 21 and 22 of IκBα leading to its subsequent degradation by the 26 S proteasome.14 Degradation of IκBα releases RelA:p50, allowing it to localize to the nucleus to bind DNA and activate gene expression (Figure 2). Interestingly, several IκB proteins are among the large number of NF-κB response genes, thus potentially functioning as negative feedback regulators. Indeed, NF-κB activity induced by inflammatory stimuli shows complex and diverse temporal or dynamic profiles.
FIGURE 2.
(a) Intuitive depiction of canonical NF-κB activation. Canonical (inflammatory) signals activate IKKβ-containing complexes (also contain NEMO and possibly IKKα), which target NF-κB-bound IκBα, β, and ε for degradation. IκBα, β, and ε that is not bound to NF-κB is constitutively degraded through an IKK- and ubiquitin-independent mechanism. Liberated NF-κB (primarily RelA:p50 dimers) translocates to the nucleus and activates gene expression, including the IκBα and IκBε genes. (b) Navigational map of canonical NF-κB activation. Canonical signals activate the IKK complex, which inhibits the IκBs, thus removing the inhibitory effect of IκB on NF-κB. NF-κB is then free to translocate to the nucleus and activate gene expression. The color coding corresponds to the intuitive depiction in (a).
A mathematical model of the IκB–NF-κB signaling module recapitulates the signaling events triggered by TNFα stimulation observed experimentally in mouse embryonic fibroblasts (MEFs). Combined experimental and computational studies showed that the three canonical IκB proteins IκBα, −β and −ε each have distinct roles in the dynamic control of NF-κB activation and termination.15 In response to TNFα, IκBα is rapidly degraded and then rapidly resynthesized in an NF-κB-dependent manner. Continued TNFα stimulation propagates a cycle of synthesis and degradation of IκBα, which can result in oscillations of nuclear NF-κB activity. IκBε expression is also strongly induced by NF-κB activity, but with a distinct 45-min delay.16 As IκBε protein accumulates at later time points, this antiphase negative feedback loop acts to dampen the IκBα-driven oscillations.
The ability of the IκB–NF-κB signaling module to mediate complex temporal control over NF-κB activity has led to research into the functionality of NF-κB dynamics. Different inflammatory stimuli elicit different IKK activation profiles, which induce distinct temporal profiles of NF-κB activity. For example, in MEFs, transient TNFα stimulation provides for 1 h of strong NF-κB activation, but equivalent stimulation with LPS, however, induces lower and longer lasting IKK activity profiles.17 Whereas TNFα signaling is limited by the negative regulator A20, LPS activates gene expression of cytokines that provide for positive autocrine feedback, amplifying late IKK activity. Further experimental investigation and expansion of the current NF-κB mathematical model to include other cell types, for example, macrophages, B and T cells, dendritic cells, and neurons, will be necessary to decipher how NF-κB dynamics controls mammalian physiology.
STRESS RESPONSE SIGNALING
Recent work has uncovered homeostatic mechanisms of regulation of NF-κB in resting cells (i.e., in the absence of a stimulus) that determines the responsiveness of the NF-κB system to inducers, including stress stimuli. Two IκB degradation pathways largely control steady-state regulation of NF-κB. Free IκB is intrinsically unstable with a 5–10-min half life, being degraded in an IKK- and ubiquitin-independent manner.18,19 Accumulating evidence indicates that the 20S proteasome can mediate degradation of free IκBα, whereby weakly folded regions in C-terminal ankyrin repeats and other C-terminal sequences mediate its instability.20,21 NF-κB association triggers complete IκB protein folding and removes IκB from the 20S proteasome pathway. Because NF-κB-bound IκB is remarkably stable in resting cells, NF-κB-bound IκB degradation is controlled by the level of IKK activity. As such, NF-κB sensitizes IκB to regulation by the IKK-mediated ubiquitin–proteasome pathway.
Homeostatic regulation of IκB synthesis and degradation renders the NF-κB system surprisingly insensitive to a variety of perturbations. First, the differential degradation of free and bound IκB allows for compensation between IκBs, evident by studies in IκB knockout cells.18 Second, the very short half life of free IκB necessitates a high rate of constitutive IκB synthesis to maintain a small excess of free IκB in the cell (estimated at 15% of total IκB amounts22) that is critical for keeping basal NF-κB activity levels low. One consequence of the very high constitutive IκB synthesis and degradation flux is that the NF-κB system is remarkably resistant to transient alterations in translation rates that are a hallmark of metabolic stress agents. Indeed, ultraviolet radiation (UV), unfolded protein response (UPR), or other ribotoxic stress that cause partial inhibition of IκB synthesis rates were found to activate NF-κB only modestly.23
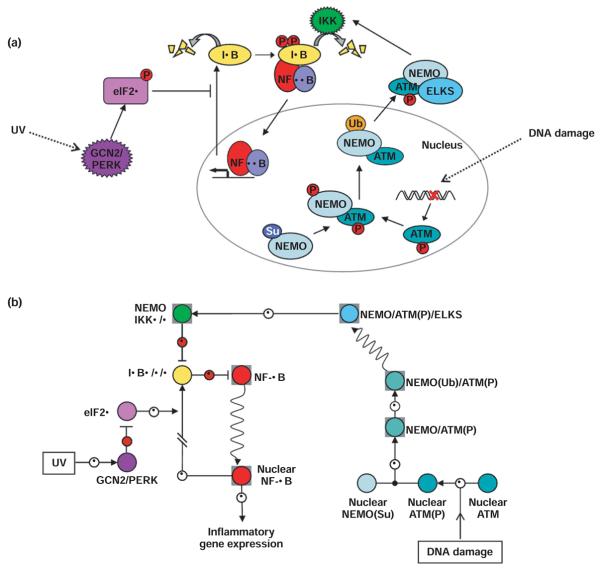
Understanding homeostatic control of the IκB–NF-κB system also resolved some seemingly conflicting observations with regard to NF-κB activation by UV. Although UV does not enhance IKK activity, IKK was found to be required.23,24 The systems model revealed how basal IKK activity is critical for slowly degrading NF-κB-bound IκBα, allowing for accumulation of nuclear NF-κB in response to translational inhibition. Further, mutations that stabilize free IκB (but allow for IKK-dependent degradation of NF-κB-bound IκBα) abolish activation of NF-κB by UV, as the free IκB pool must also be depleted in response to translation inhibition (Figure 3).23 UV-induced CK2 activity, which can cause phosphorylation of the C-terminus of IκBα, may further accelerate IκBα degradation, but the molecular mechanisms of this pathway remain unclear.25
FIGURE 3.
(a) Intuitive depiction of NF-κB activation by metabolic stress (UV) and by DNA damage. UV irradiation activates the stress response kinases GCN2 or PERK, which phosphorylate the initiation factor elF2α. Phosphorylated elF2α prevents translation initiation, blocking IκB synthesis. Free IκB is rapidly depleted, preventing the replenishment of NF-κB-bound IκB that is slowly degraded through constitutive IKK activity, and NF-κB is slowly liberated and passes to the nucleus. Certain agents cause double stranded breaks in DNA. This triggers the phosphorylation of ATM, which associates with sumoylated NEMO in the nucleus, causing subsequent phosphorylation and ubiquitination of NEMO, and export to the cytoplasm where the complex of NEMO and phosphorylated ATM associate with the ELKS protein and stimulate activity of IKKβ containing complexes, causing degradation of IκBα and subsequent NF-κB translocation to the nucleus. (b) Navigational map of NF-κB activation by metabolic stress (UV) and DNA damage. UV irradiation induced activation of the stress kinases GCN2 and PERK results in the inhibition of the translation inhibition factor elF2α. The resulting inhibition of IκB synthesis allows for NF-κB translocation to the nucleus. DNA damage activates ATM, which associates with nuclear sumoylated NEMO, causing ubiquitination of NEMO and subsequent export to the cytoplasm. In the cytoplasm, the NEMO/ATM complex associates with the ELKs protein and activates IKK to induce NF-κB translocation to the nucleus. The color coding corresponds to the intuitive depiction in (a).
DNA damage, caused by irradiation or chemotherapeutic drugs for example, does induce IKK activation. Until recently, it was unclear how a nuclear signal could relay back to the inhibited NF-κB in the cytoplasm to trigger its activation. It was found that DNA damage not only initiates the activation of the nuclear kinase ataxia telangiectasia mutated (ATM), the primary regulator of the tumor suppressor and transcription factor p53, but also initiates the sumoylation of NEMO by the sumo ligase PIASy, promoting the nuclear localization of NEMO.26 It was known that activated ATM was required for NF-κB activation by DNA damage, and recent work has uncovered the connection between ATM activity and IKK activation. Wu et al. showed that nuclear sumoylated NEMO associates with and is phosphorylated by the activated ATM, promoting monoubiquitination of NEMO, which triggers its export to the cytoplasm. The cytoplasmic ATM–NEMO complex associates with the protein ELKS, facilitating ATM-dependent activation of the canonical IKK complex, leading to IκBα degradation and NF-κB activation (Figure 3).27
DEVELOPMENTAL SIGNALING
A group of noninflammatory signals have been shown to activate NF-κB through the (noncanonical) NF-κB signaling pathway. These developmental signals of the TNF-receptor superfamily, such as B-cell activation factor (BAFF) critical for B-cell survival, lymphotoxin β (LTβ) involved in lymphnode development, and receptor activator of NF-κB ligand (RANKL) essential for osteoclast differentiation, have been described to activate NF-κB at a low level for hours or days. The noncanonical pathway is not transduced by a NEMO/IKKβ-containing kinase complex, but rather by an IKKα-containing kinase complex, whose activation requires NF-κB-inducing kinase (NIK). In addition to the noncanonical IKKα-dependent NF-κB degradation, these signals may also, in certain cellular conditions and contexts, activate the canonical IKKβ-dependent NF-κB activation pathway.28
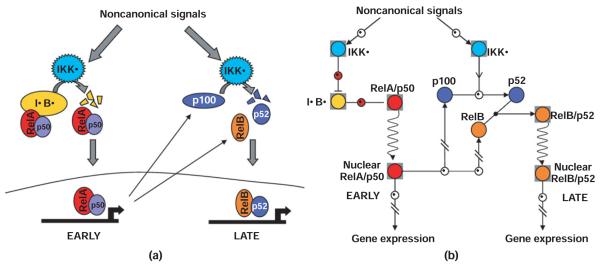
Initial studies of NF-κB activation by these developmental signals focused on the generation of the RelB:p52 dimer by cotranslational proteolytic processing of de novo synthesized p100 to p52. The p100 protein contains C-terminal serines whose phosphorylation by IKKα is critical for stimulus-responsive processing. More recently, it was shown that the same developmental signals can also activate the canonical RelA:p50 NF-κB complexes not through IKKβ-dependent degradation of IκBα, but rather via the degradation of a fourth IκB activity, termed IκBδ (Figure 4). IκBδ activity is the result of the dimerization of two p100 molecules (through their RHD-interacting domains), whereby the ARD of one p100 molecule folds back onto the RHD–RHD dimer interface to effect self-inhibition (‘in cis’), leaving the ARD of the second p100 molecule capable of binding latent NF-κB dimers (primarily RelA:p50, but also RelB:p50) ‘in trans’.29 As trans-inhibition of preformed NF-κB dimers is a hallmark of IκB activities, this latter activity has been termed ‘IκBδ’
FIGURE 4.
(a) Intuitive depiction of NF-κB activation by non-canonical signals. Non-canonical signals (developmental cues) activate IKKα containing complexes, which phosphorylate the C-terminal region of a p100 molecule within a multimeric complex (IκBδ), causing partial degradation of the p100 molecule (processing) and releasing associated RelA containing complexes. Hours later, the ratio of RelB associated with p100 is increased, and IKKα-dependent processing of de novo synthesized p100 leads to more RelB and p52 containing dimers. This requires RelA driven constitutive synthesis of RelB and p100. (b) Navigational map of NF-κB activation by non-canonical signals. Early activation of IKKα-containing complexes initiates the processing of a p100 molecule within the IκBδ complex. This removes the inhibitory action of IκBδ activity on RelA-containing NF-κB dimers, allowing for translocation to the nucleus. Hours later, the ratio of RelB associated with p100 is increased, and IKKα-dependent processing of de novo synthesized p100 leads to more RelB and p52 containing dimers, which translocate to the nucleus to effect gene expression. The color coding corresponds to the intuitive depiction in (a).
Noncanonical signals first induce the degradation of IκBδ, releasing associated RelA:p50 and RelB:p50 to the nucleus. Beginning at around 3 h, the effect of stimulus-induced cotranslational processing of newly synthesized p100 to p52 becomes apparent, effecting a change in the predominant dimer composition from RelA:p50 (and RelB:p50) to RelB : p52 (and RelA:p52) (Figure 4). Whether these dimers have specific target gene functions or primarily exhibit dimer exchange on the same promoters to allow for long-lasting NF-κB driven transcription remains to be investigated in further detail.
NF-κB AS A SIGNALING CROSS-TALK MEDIATOR
Inflammatory signaling determines responsiveness to stresses via control of homeostatic IκB turnover
Computational simulations first suggested that the level of constitutive IKK activity predetermines the responsiveness of NF-κB to translational inhibition. This led to the prediction that cells chronically exposed to low levels of inflammatory signals may have significantly enhanced responses to stress stimuli that inhibit protein synthesis because of signaling crosstalk: increased constitutive IKKβ activity results in increased turnover of the NF-κB-bound IκBα, and inhibition of protein synthesis rapidly depletes the pool of the intrinsically unstable free IκBα. This prediction was tested with low doses of inflammatory signals that only marginally enhance IKK and NF-κB activities, but when combined with UV or endopiasmic reticulum (ER) stress agents that inhibit protein synthesis (via phosphorylation of the translation initiation factor eIF2α), they produce a highly synergistic increase in NF-κB activity.23 These studies suggest that the homeostatic state of the cell, with altered IκB turnover due to inflammatory signals, predetermines the responsiveness of NF-κB to metabolic stress stimuli. Other signaling crosstalk studies indicated that combined treatment of UV irradiation and the inflammatory cytokine IL-1 synergistically enhances not only NF-κB activation but also apoptosis. The latter was proposed to be mediated by enhanced NF-κB responsive TNFα expression.30,31
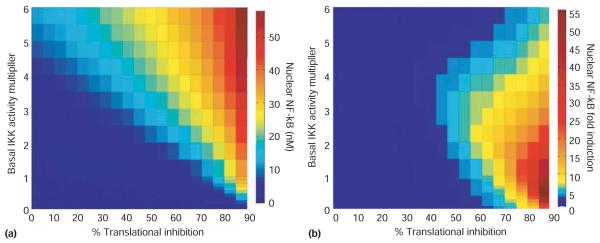
To chart the effect of varying both basal IKK activity and induced translational inhibition, a three-dimensional heat map of NF-κB activity is useful. Using the IKK-IκB-NF-κB mathematical model, NF-κB activity after 8 h of varied doses of translational inhibition was simulated, and each simulation was run at different constitutive levels of IKK activity. Absolute NF-κB activity levels were plotted on the heat map (Figure 5(a)) and, as expected, higher preexisting or constitutive IKK activity levels provided for more NF-κB activity in response to all doses of translational inhibition. However, when the fold induction (NF-κB activity at 8 h over NF-κB activity at 0 h), rather than absolute value, of NF-κB activity is plotted, it becomes evident that the highest constitutive IKK activity level is not the most optimal for NF-κB responsiveness to translational inhibition. Instead, we find that the optimal NF-κB responsiveness occurs at specific IKK activity values that are dependent on the level of translational inhibition. At 90% translational inhibition, NF-κB responsiveness is maximum when IKK is about 1 nM; at 50% inhibition it is maximum at about 3 nM (Figure 5(b)).
FIGURE 5.
NF-κB responses to metabolic stresses. NF-κB activation in response to metabolic stresses that cause translational inhibition was computationally simulated as a function of the level of basal/constitutive IKK activity. (a) Simulations of nuclear NF-κB (nM in color scale) after 8 hours of the indicated degree of translational repression (% on the x-axis) at indicated levels of constitutive IKK activity (y-axis). (b) Simulations of nuclear NF-κB fold-induction (nuclear NF-κB at 8 hours over nuclear basal NF-κB) after 8 hours of the indicated degree of translational inhibition (% on the x-axis) at indicated levels of constitutive IKK activity (y-axis). The IKK activity multiplier is the degree to which the wild-type level of constitutive IKK activity (1% of the IKK is active in the “wild-type model”) was multiplied by during the equilibration phase, prior to the induction of translational repression and held constant throughout each simulation.
As different cells may have different homeostatic states, the degree of NF-κB responsiveness to common assaults is different. For example, high levels of constitutive IKK activity are associated with cancer cells that have elevated NF-κB activity, which may promote survival, tumorigenesis, angiogenesis, or chemoresistance. Pharmacological inhibition of IKK activity may lower basal NF-κB activity, but based on these computational predictions, it may also result in increased NF-κB responsiveness to metabolic stress associated with chemotherapeutic agents, diminishing the efficacy of such combination therapy.
Inflammatory signaling determines responsiveness to developmental signals via control of p100 and RelB expression
Inflammation interacts with developmental processes. Lymph node development and homeostasis appears to require not only the developmental regulator LTβ but also the inflammatory cytokine TNFα.32 Conversely, inflammation can derail normal developmental or cellular homeostasis and be a major factor in cancer progression.33 Over the last few years, it has become apparent that the NF-κB signaling system may be a primary integrator of these diverse signals to mediate cross talk in both physiological and pathological processes.
Following the identification of the fourth IκBδ activity that mediates noncanonical signaling, its NF-κB responsive expression was explored as a cross-talk mechanism.7 Because IκBδ is not degraded by canonical IKKβ-mediated signals, inflammatory stimuli increase its abundance, thereby amplifying the RelA:p50 responsiveness to developmental signals to the extent that physiological developmental stimuli may cause inflammatory gene expression. Conversely, long-term RelA:p50 signaling in response to some pathogen-associated substances is terminated by IκBδ at late times, but this inhibition may be relieved by concomitant exposure to developmental signals (V. Shih, S. Basak, and A. Hoffmann, unpublished observations).
Similarly, the extent of RelB NF-κB activity induced by developmental stimuli may be regulated by inflammatory coexposure. However, in this case—as RelB:p52 dimers are generated cotranslationally—it is the continued RelA-responsive transcription of RelB (and to a lesser extent of p100) that controls RelB:p52 formation.34
Finally, recent studies report that the NIK, long associated with the regulation of the noncanonical pathway, does in fact regulate canonical NEMO-associated IKK activity.35 As the short half life of NIK is extended by developmental stimuli inhibiting the TNFR associated factor 3 (TRAF3) ubiquitin ligase, NIK accumulates in these conditions and may amplify canonical IKK activation. As such, NIK may mediate potent signaling cross talk between developmental and inflammatory stimuli, whose physiological or pathological importance remains to be explored more fully.
REFERENCES
- 1.Sullivan JC, Kalaitzidis D, Gilmore TD, Finnerty JR. Rel homology domain-containing transcription factors in the cnidarian Nematostella vectensis. Dev Genes Evol. 2007;217:63–72. doi: 10.1007/s00427-006-0111-6. [DOI] [PubMed] [Google Scholar]
- 2.Graef IA, Gastier JM, Francke U, Crabtree GR. Evolutionary relationships among Rel domains indicate functional diversification by recombination. Proc Natl Acad Sci U S A. 2001;98:5740–5745. doi: 10.1073/pnas.101602398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Courtois G, Gilmore TD. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 4.Naugler WE, Karin M. NF-kappaB and cancer-identifying targets and mechanisms. Curr Opin Genet Dev. 2008;18:19–26. doi: 10.1016/j.gde.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Courtois G, Gilmore TD. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 7.Basak S, Kim H, Kearns JD, Tergaonkar V, O'Dea E, et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. Embo J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto M, Takeda K. Role of nuclear IkappaB proteins in the regulation of host immune responses. J Infect Chemother. 2008;14:265–269. doi: 10.1007/s10156-008-0619-y. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 2004;430:218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- 11.Trinh DV, Zhu N, Farhang G, Kim BJ, Huxford T. The nuclear IkappaB protein IkappaBzeta specifically binds NF-kappaB p50 homodimers and forms a ternary complex on kappaB DNA. J Mol Biol. 2008;379:122–135. doi: 10.1016/j.jmb.2008.03.060. [DOI] [PubMed] [Google Scholar]
- 12.Bundy DL, McKeithan TW. Diverse effects of BCL3 phosphorylation on its modulation of NF-kappaB p52 homodimer binding to DNA. J Biol Chem. 1997;272:33132–33139. doi: 10.1074/jbc.272.52.33132. [DOI] [PubMed] [Google Scholar]
- 13.Hirotani T, Lee PY, Kuwata H, Yamamoto M, Matsumoto M, et al. The nuclear IkappaB protein IkappaBNS selectively inhibits lipopolysaccharide-induced IL-6 production in macrophages of the colonic lamina propria. J Immunol. 2005;174:3650–3657. doi: 10.4049/jimmunol.174.6.3650. [DOI] [PubMed] [Google Scholar]
- 14.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-kappaB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 16.Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A. IkappaBepsilon provides negative feedback to control NF-kappaB oscillations, signaling dynamics, and inflammatory gene expression. J Cell Biol. 2006;173:659–664. doi: 10.1083/jcb.200510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 18.O'Dea EL, Barken D, Peralta RQ, Tran KT, Werner SL, et al. A homeostatic model of IkappaB metabolism to control constitutive NF-kappaB activity. Mol Syst Biol. 2007;3:111. doi: 10.1038/msb4100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathes E, O'Dea EL, Hoffmann A, Ghosh G. NF-kappaB dictates the degradation pathway of IkappaBalpha. Embo J. 2008;27:1357–1367. doi: 10.1038/emboj.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Castelao B, Castano JG. Mechanism of direct degradation of IkappaBalpha by 20 S proteasome. FEBS Lett. 2005;579:4797–4802. doi: 10.1016/j.febslet.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 21.Truhlar SM, Torpey JW, Komives EA. Regions of IkappaBalpha that are critical for its inhibition of NF-kappaB.DNA interaction fold upon binding to NF-kappaB. Proc Natl Acad Sci U S A. 2006;103:18951–18956. doi: 10.1073/pnas.0605794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice NR, Ernst MK. In vivo control of NF-kappa B activation by I kappa B alpha. Embo J. 1993;12:4685–4695. doi: 10.1002/j.1460-2075.1993.tb06157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Dea EL, Kearns JD, Hoffmann A. UV as an amplifier rather than inducer of NF-kappaB activity. Mol Cell. 2008;30:632–641. doi: 10.1016/j.molcel.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang TT, Feinberg SL, Suryanarayanan S, Miyamoto S. The zinc finger domain of NEMO is selectively required for NF-kappaB activation by UV radiation and topoisomerase inhibitors. Mol Cell Biol. 2002;22:5813–5825. doi: 10.1128/MCB.22.16.5813-5825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato T, Jr, Delhase M, Hoffmann A, Karin M. CK2 is a C-terminal IkappaB kinase responsible for NF-kappaB activation during the UV response. Mol Cell. 2003;12:829–839. doi: 10.1016/s1097-2765(03)00358-7. [DOI] [PubMed] [Google Scholar]
- 26.Mabb AM, Wuerzberger-Davis SM, Miyamoto S. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat Cell Biol. 2006;8:986–993. doi: 10.1038/ncb1458. [DOI] [PubMed] [Google Scholar]
- 27.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 28.Pomerantz JL, Baltimore D. Two pathways to NFkappaB. Mol Cell. 2002;10:693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 29.Basak S, Hoffmann A. Crosstalk via the NF-kappaB signaling system. Cytokine Growth Factor Rev. 2008;19:187–197. doi: 10.1016/j.cytogfr.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kothny-Wilkes G, Klums D, Luger TA, Kubin M, Scwarz T. Interleukin-1 protects transformed keratinocytes from tumor necrosis factor-related apoptosis-inducing ligand- and CD95-induced apoptosis but not from ultraviolet radiation-induced apoptosis. J Biol Chem. 1999;274:28916–28921. doi: 10.1074/jbc.274.41.28916. [DOI] [PubMed] [Google Scholar]
- 31.Strozyk E, Pöppelmann B, Schwarz T, Klums D. Differential effects of NF-kappaB on apoptosis induced by DNA-damaging agents: the type of DNA damage determines the final outcome. Oncogene. 2006;25:6239–6251. doi: 10.1038/sj.onc.1209655. [DOI] [PubMed] [Google Scholar]
- 32.Rennert PD, James D, Mackay F, Browning JL, Hochman PS. Lymph node genesis is induced by signaling through the lymphotoxin beta receptor. Immunity. 1998;9:71–79. doi: 10.1016/s1074-7613(00)80589-0. [DOI] [PubMed] [Google Scholar]
- 33.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 34.Basak S, Shih VF, Hoffmann A. Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system. Mol Cell Biol. 2008;28:3139–3150. doi: 10.1128/MCB.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarnegar B, Yamazaki S, He JQ, Cheng G. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc Natl Acad Sci U S A. 2008;105:3503–3508. doi: 10.1073/pnas.0707959105. [DOI] [PMC free article] [PubMed] [Google Scholar]