Abstract
For decades after the discovery that a contractile ring made of actin filaments and myosin II produces the force to constrict the cleavage furrow of animal cells, the complexity of cytokinesis has slowed progress in understanding the mechanism. Mechanistic insights, however, have been obtained by genetic, biochemical, microscopic and mathematical modelling approaches in the fission yeast Schizosaccharomyces pombe. Many features that have been identified in fission yeast are probably shared with animal cells, as both inherited many cytokinesis genes from their common ancestor about one billion years ago.
Cell division by cytokinesis completes the cell cycle for every cell. Cytokinesis in fungal, amoeboid and animal cells takes place in four steps (FIG. 1). The process begins when the cell marks the site of the future cleavage furrow relative to the sister chromatids, which are separated by the mitotic apparatus. Accurate placement of the cleavage plane is important because the aim of cytokinesis is to create two cells, each with its own nucleus. At the selected site the cell assembles a contractile ring, which comprises the motor protein myosin II, actin filaments and many other proteins and is attached to the plasma membrane. As in muscle contraction, interactions of myosin II with actin filaments produce force to constrict the contractile ring and to form a cleavage furrow in the plasma membrane. Remarkably, the contractile ring disassembles as it constricts, so it does not become thicker like contracting muscle does. Finally, proteins that bring together membranes for their fusion promote the reorganization of the plasma membrane at the base of the furrow and the separation of the two daughter cells.
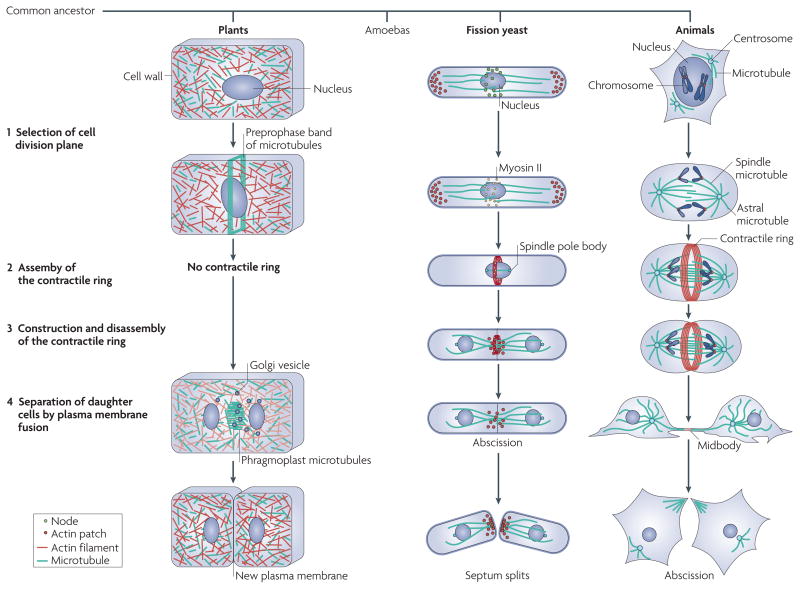
Figure 1. Strategies for cytokinesis used by plant, fission yeast and animal cells.
Plants, amoebas, fungi and animals all arose from a common ancestor, branching off as shown. In plants, the cell division plane is selected by the nucleus specifying the position of a preprophase band of microtubules around the equator. Plants lack key proteins to make a contractile ring, so they depend on membrane fusion to separate the two daughter cells. Phragmoplast microtubules transport Golgi vesicles to the midplane to form the new plasma membrane. Amoebas divide much like animal cells and are not illustrated. In fission yeast, the cell division plane is selected by the nucleus specifying the position of nodes around the equator, whereas in animals, spindle and astral microtubules specify the position of the contractile ring. Fission yeast and animal cells assemble a contractile ring of actin filaments and myosin II around the equator of the cell between the chromosomes, which are separated by microtubules of the mitotic apparatus. The ring constricts and the daughter cells separate by membrane fusion. See also Fig. 2 for more details on the process in fission yeast.
The mechanism of cytokinesis through a contractile ring appeared about one billion years ago in the common ancestor of amoebas, fungi and animals. These organisms share most of the genes used for cytokinesis by the fission yeast Schizosaccharomyces pombe, so lessons learned about mechanisms in fission yeast should apply to animals. The membrane fusion machinery is older than the contractile ring, as plant cells fuse membrane vesicles to build new plasma membrane between the daughter nuclei rather than constricting a furrow1. Little is known about cytokinesis in eukaryotes that branched before algae and plants (for example, Giardia spp. and trypanosomes) except that their genomes also lack myosin II, so the mechanism of cytokinesis in these eukaryotes is different from that in amoebas, fungi and animals.
Of all the commonly used model organisms, fission yeast offer many advantages for studying cytokinesis, so an active community of investigators is making rapid progress in deciphering the mechanisms of cytokinesis in fission yeast. Like in animal cells, the position of the mitotic apparatus (inside the nucleus in fungi) determines the position of the contractile ring in fission yeast. The fission yeast genome encodes ~ 4,940 proteins. Classical and reverse genetics have produced the best inventory of more than 130 cytokinesis genes (see Supplementary information S1 (table)) and conditional mutations for many of these genes2,3. TABLE 1 lists the proteins that are relevant to the contractile ring. Deletion strains for 98% of the ~ 4,900 of the total genes are available (Bioneer). The size and shape of the cells are ideal for quantitative microscopy and many cytokinesis proteins have been tagged with fluorescent proteins in the genome. These features made it possible to chart a high-resolution time line of cytokinetic events4 and to quantitate the intracellular distributions of key cytokinesis proteins5. In a field dominated by genetics, biochemical analysis has been limited but is growing. Enough quantitative data are available to formulate and test mathematical models for some steps in cytokinesis6,7. This article discusses advances being made through research on fission yeast, with a focus on conserved features that are likely to be used by other cells.
Table 1.
Identified cytokinesis proteins in fission yeast
| Generic name | Schizosaccharomyces pombe name* |
|---|---|
| Cleavage furrow placement proteins | |
| GIN4 family kinase | Cdr2 |
| Ser/Thr kinase | Kin1 |
| Anillin-like protein | Mid1 (Dmf1) |
| γ-Tubulin complex subunits | Mto1 (Mbo1) and Mto2 |
| Polo kinase | Plo1 |
| DYRK kinase | Pom1 |
| Contractile ring assembly and maturation proteins | |
| Capping protein | Acp1 and Acp2 |
| Actin | Act1 (cps8) |
| Cofilin | Cof1 (Adf1) |
| α-Actinin | Ain1 |
| Aurora B kinase | Ark1 (Aim1) |
| Microtubule cross-linking factor | Ase1 |
| Survivin | Bir1 (Cut17) |
| Novel protein | Blt1 (SPBC1A4.05)‡ |
| Formin | Cdc12 |
| F-BAR domain containing protein | Cdc15 |
| Profilin | Cdc3 |
| Myosin II light chain | Cdc4 |
| Tropomyosin | Cdc8 |
| Cdc14 phosphatase | Clp1 (Flp1) |
| Fimbrin | Fim1 |
| Rho GEF | Gef2‡ |
| Hsp90 chaperone | Hsp90 (Swo1) |
| Kinesin | Klp8‡ |
| Myosin II heavy chains | Myo2 (Rng5) and Myp2 (Myo3) |
| Borealin | Nbl1 |
| Phosphoinositide-dependent kinase | Pdk1 (Ppk21) |
| Inner centromere protein (INCENP) | Pic1 |
| GEF for Rho1 | Rgf3 (Lad1) |
| Myosin II regulatory light chain | Rlc1 |
| IQGAP family protein | Rng2 |
| UCS domain-containing protein | Rng3 |
Acp, F-actin-capping protein; Act, actin; Ain1, α-actinin-like protein 1; Ase1, anaphase spindle elongation protein 1; Cdc, cell division control; Clp1, Cdc14-like phosphatase 1; GEF, guanine nucleotide exchange factor; Hsp, heat shock protein; Klp8, kinesin-like protein 8; Myo2, myosin II; Myp2, unconventional myosin II; Rlc1, regulatory light chain 1; Rng, ring assembly.
Alternative protein names are provided in brackets.
The role needs to be further tested.
Main pathway of cytokinesis
The advantages of fission yeast (discussed above) made it possible to show that events during cytokinesis occur like clockwork (see Supplementary information S2 (movie)). Events can be followed with a precision of one or two minutes on an absolute time-scale anchored to time zero, when the spindle pole bodies separate4. Spindle pole bodies are the structures that anchor micro-tubules of the mitotic spindle in fungi. Use of this timescale (FIG. 2) allows for comparisons of observations between laboratories and for detecting subtle problems, such as changes in timing that result from experimental manipulations. The following account summarizes our views regarding contractile ring assembly in fission yeast, but we want the reader to appreciate that questions remain regarding every step in the process.
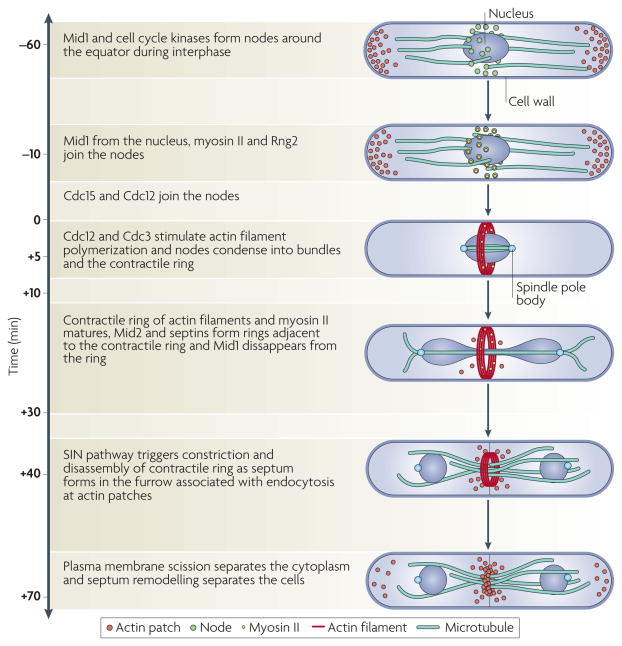
Figure 2. Time course of cytokinesis in fission yeast.
Time zero is defined as the time when the spindle pole bodies separate. Starting at time −60 minutes, interphase nodes containing the anillin-like protein Mid1 (also known as Dmf1) and cell cycle kinases form near the plasma membrane. Negative signals from the ends of the cell position these nodes around the equator. Dynamic microtubules push the nucleus to the centre of the cell. At time −10 minutes, interphase nodes begin to mature into cytokinesis nodes by the addition of myosin II (Myo2), ring assembly protein 2 (Rng2; a member of the IQGAP family), the F-BAR domain-containing protein cell division control protein 15 (Cdc15) and the formin Cdc12. Following spindle pole body separation, Cdc12 and Cdc3 (also known as profilin) stimulate the polymerization of actin filaments that bind tropomyosin and cross-linking proteins. During anaphase A (at time +5 minutes), interactions of myosin II with actin filaments condense nodes into a contractile ring, which matures by adding more Cdc15, capping protein, unconventional myosin II (Myp2; also known as Myo3) and other proteins. In anaphase B (at time +10 to +30 minutes) the mitotic spindle elongates and the anillin-like protein Mid2 and septins form double rings adjacent to the contractile ring. Mid1 disappears from the ring at the onset of its constriction. At the end of anaphase, a signalling pathway consisting of a GTPase and three protein kinases (the septation initiation network (SIN)) triggers constriction of the contractile ring (at time +40 minutes), membrane invagination and synthesis of a new cell wall to form a septum. Constriction ends at time +70 minutes. After another 30 minutes, scission of the plasma membrane separates the cytoplasm and septum remodelling separates the cells.
Assembly of interphase nodes
Protein assemblies called nodes are precursors of the contractile ring8–11. The adaptor anillin-like protein, the product of the mid1 (also known as dmf1) gene in fission yeast, appears in interphase nodes around the equator of interphase cells more than 1 hour before time zero4,10, along with the kinases Cdr1, Cdr2 and Wee1 and other proteins (Blt1 (also known as SPBC1A4.05), kinesin-like protein 8 (Klp8) and Rho guanine nucleotide exchange factor Gef2)12–14. Assembly of these interphase nodes depends on the presence of kinase-active Cdr2, which interacts with Mid112,14 and might be anchored to the cell cortex through its carboxyl terminus15. Cdr1 and Cdr2 phosphorylate and inhibit Wee1, a kinase that holds the cell in G2 phase by phosphorylating the master cell cycle kinase cell division control protein 2 (Cdc2; also known as Cdk1), which controls cell cycle progression13.
The location of nodes during interphase depends on the kinase Pom1 and another unknown inhibitor6,16–18. Pom1 concentrates at both ends of the cell and restricts interphase nodes containing Cdr2 and Mid1 to the middle of the cell. As a cell grows longer, the inhibitory activities decline in the middle of the cell, allowing Cdr1 and Cdr2 to phosphorylate and inhibit Wee1. This releases the kinase Cdc2 to trigger the transition into mitosis, thus coupling growth to the cell cycle12,13. In addition to the ‘negative regulation’ that excludes interphase nodes from cell tips, the polo kinase Plo1 releases Mid1 from the nucleus before mitosis19 and seems to prepare the ~ 65 interphase nodes around the equator of the cell for cytokinesis before the onset of mitosis. The position of the nucleus determines the location of these maturing nodes and, therefore, the position of the contractile ring by a mechanism that is still under investigation.
Maturation of Mid1 interphase nodes
Starting 10 minutes before spindle pole body separation, nodes containing ~ 25 copies of Mid1 mature by sequentially adding, from cytoplasmic pools, ~ 25 molecules of myosin II (each composed of 2 heavy chains and 4 light chains) and ~ 25 ring assembly protein 2 (Rng2; a member of the IQGAP family) molecules, followed by ~ 25 copies of the F-BAR domain-containing protein Cdc15 and ~ 2 dimers of the formin Cdc12, which makes the node competent for actin assembly5. We call these mature nodes cytokinesis nodes. Detecting the formin Cdc12 is difficult20,21 because it is the least abundant known cytokinesis node protein5 and because it arrives last, so most (younger) nodes have no detectable Cdc12 (REF. 4.) Some interphase node proteins (Cdr2, Blt1, Klp8 and Gef2) remain with Mid1 during the formation of the contractile ring, but Cdr2 leaves the ring during anaphase B12,13.
We know little about the assembly, architecture or attachment of nodes to the cortex in either interphase or mitosis, and they have not been seen by electron microscopy. Not much quantitative biochemical data are available on interactions among the large, poorly soluble, multi-domain node proteins. Mutation of Mid1 in a region called the amphipathic a-helix compromises its association with the cortex22, but this element is part of a much larger insoluble domain of unknown architecture. Like other anillins, Mid1 has a C-terminal PH domain, but it is not required for association with the cortex10. Lateral diffusion of nodes in the plane of the membrane is very slow (20 nm2 s−1) up to time +2 minutes. Anchoring by actin filaments might constrain their movements, but they do not diffuse faster in the absence of actin filaments7.
Actin filament assembly at time +2 minutes
The assembly of actin filaments for the contractile ring depends on the formin Cdc1220,23,24 and a profilin — a small protein that can interact with actin monomers and sequences of multiple proline residues found in other proteins25. Formins nucleate actin filaments from free actin monomers26 and cooperate with profilin to elongate the filament26–29. Profilin–actin complexes bind to multiple polyproline sequences in the Cdc12 formin homology 1 (FH1) domain20,23,26 and transfer rapidly onto the fast growing barbed end of the filament26,28,29, whereas the FH2 domain moves processively on the growing end without dissociating26,28. Thus, Cdc12 might anchor actin filaments to the cytokinesis nodes.
Cdc12 cannot elongate actin filaments using actin fused to green fluorescent protein (GFP)5, so contractile ring actin filaments can only be labelled indirectly with fluorescent phalloidin in fixed cells30 or with GFP fused to an actin filament-binding domain, such as a calponin homology domain, or Lifeact in live cells21,31. GFP–calponin homology domains decorate transient linear connections between cytokinesis nodes that might be single actin filaments7, although this has not yet been verified by other methods. These putative actin filaments grow from cytokinesis nodes in random directions close to the plasma membrane at about 80 subunits per second and contact neighbouring cytokinesis nodes. The endoplasmic reticulum is closely opposed to the plasma membrane and might help to restrict these filaments to the plane close to the plasma membrane. Cdc8 (also known as tropomyosin) is an a-helical coiled-coil protein that binds along the actin filament helix and increases the rate of elongation of filaments by Cdc12 but can also dissociate Cdc12 from the barbed end or trap Cdc12 between two annealed filaments32. It is not known how cells control the association of Cdc12 with nodes or the nucleation activity of Cdc12.
Cytokinesis nodes condense into a contractile ring
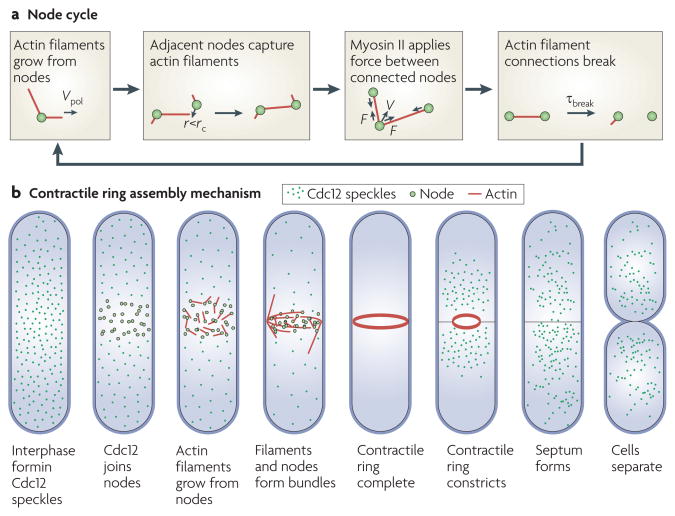
As soon as actin filaments appear in the cortex around the equator at time +2 minutes, cytokinesis nodes start moving at about 30 nm s−1 in short bursts of ~ 20 seconds. Over 10 minutes these stochastic movements condense the nodes into a nearly continuous ring around the equator7. Our hypothesis to explain these intermittent movements is that myosin II (Myo2) in cytokinesis nodes captures the actin filaments growing from neighbouring nodes. The nodes are then pulled together transiently until the actin filament connection is broken by the dissociation of myosin II from the filament, thereby severing the filament (probably by the actin filament severing the protein cofilin), or by Cdc12 turnover (FIG. 3). Monte Carlo simulations (a stochastic simulation method based on the probability of each reaction) of a simple search, capture, pull and release model, with parameters similar to those measured in live cells, reproduce the assembly of the contractile ring in the correct time of 10 minutes, providing that the mechanism includes frequent breaks in the connections between the cytokinesis nodes7.
Figure 3. Mechanism of contractile ring assembly in fission yeast.
a. | the cycle of reactions hypothesized in the search, capture, pull and release mechanism of contractile ring assembly. Cytokinesis nodes contain the anillin-like protein Mid1 (also known as Dmf1), myosin II (Myo2) and the formin cell division control protein 12 (Cdc12). Actin filaments grow in random directions from nodes by adding subunits at rate Vpol (the rate of actin polymerization). If a filament approaches a node within distance r, less than a defined distance rc (capture radius), myosin II can capture the filament. Myosin II applies force on attached filaments and moves nodes at velocity V. Actin filament connections between nodes break owing to severing or other reactions, with a time constant of τbreak, after which the cycle is repeated. b | An overview of contractile ring assembly and constriction in fission yeast. During interphase, Cdc12 is distributed throughout the cell in small clusters called speckles. At time zero minutes, Cdc12 joins nodes around the equator and nucleates actin filaments. The actin filaments form bundles as myosin II pulls the cytokinesis nodes into a ring. As the contractile ring constricts, the septum forms between the daughter cells, which separate by membrane fusion.
Bundles of actin filaments form naturally during the condensation of cytokinesis nodes into a contractile ring and several groups have proposed that contractile rings form by cross-linking filaments in a ‘leading cable’ that is nucleated from a single ‘spot’ containing Cdc1220,23,33–35. This was a reasonable idea, as Cdc12 is more obvious after actin filaments and nodes form bundles than during the short interval between time zero and the onset of cytokinesis node condensation, when each node contains just a few Cdc12 molecules. Similarly, bundles of actin filaments are much easier to image than the thin connections between dispersed cytokinesis nodes early in the process. The existence of spots that promote the formation of leading cables depends on the actin filament cross-linking protein α-actinin-like 1 (Ain1), but normal contractile rings can form without Ain1 or the spot20,21. Electron micrographs of permeabilized anaphase cells treated with the myosin head domain were interpreted to show that contractile rings consist of two bundles of actin filaments with opposite polarities that originate from a single source35. However, some of the filaments in these bundles appear to us to be anti-parallel, consistent with mechanisms other than the leading cable model, such as the search and capture mechanism. Electron micrographs of filaments early in the assembly process might help to explain the relationship between dispersed networks of cytokinesis nodes connected by short filaments and the bundled filaments in more mature contractile rings.
Many mechanistic questions remain about contractile ring assembly. We do not know whether formin-dependent polymerization of actin filaments suffices to explain the onset of cytokinesis node condensation, or whether cells must also regulate the activity of Myo2 with the UCS domain-containing protein Rng3 (REF. 36), phosphorylation of myosin II heavy chains11,37 or light chains37,38, or other mechanisms.
Maturation of the contractile ring
Contractile rings do not change in size or shape between time +11 and +35 minutes, but many proteins are exchanged with others from the cytoplasmic pool20,39,40. During this time the ring acquires other proteins, such as capping protein, the unconventional myosin II (Myp2; also known as Myo3) and the F-BAR domain-containing protein Imp2, and loses Mid1. Contractile rings can form without input from the septation initiation network (SIN) signalling pathway, which consists of a GTPase and three protein kinases41, but the polo kinase Plo1 and the SIN pathway are required for maturation of a compact contractile ring42. A network of proteins including the F-BAR domain-containing proteins Cdc15 and Imp2, the C2 domain-containing protein Fic1 and paxillin-related protein 1 (Pxl1) also stabilize the ring40,43,44. The number of proteins in the mature ring is known, but how they are organized and how the ring is attached to the plasma membrane are unknown. Adjacent to the contractile ring, the anillin-like protein Mid2 and four GTP-binding proteins called septins polymerize to form two rings that remain after the contractile ring has constricted4,45–47.
Contractile ring constriction and disassembly
Starting at time +35 minutes, the ring constricts circumferentially down to a small spot at about 5nm s−1 (REFS 4,39,48) — a linear rate that is almost 50 times slower than the large contractile rings of sea urchin eggs49 and nematode embryos50, following the trend that constriction rate is proportional to the circumference of the ring50. Constriction is presumed to occur by a sliding filament mechanism, similar to that in striated muscles, but the details are unknown. As in sea urchin eggs51 and nematode embryos50, the ring loses actin filaments and actin-binding proteins, by an uncharacterized mechanism, in proportion to the decline in circumference4,5. The actin filament concentration is therefore constant. In contrast to nematode embryos50, fission yeast contractile rings concentrate myosin II as they constrict4.
The SIN pathway is required for constriction and disassembly of the contractile ring, but the mechanisms of these processes are not known owing to limited information about the substrates of the SIN pathway proteins52, except for the Sid2 kinase. Sid2 phosphorylates Cdc14-like phosphatase 1 (Clp1), creating a binding site for the 14-3-3 serine phosphate-binding protein Rad24, which retains Clp1 in the cytoplasm53. Mid1 anchors Clp1 in the contractile ring54, where it dephosphorylates Cdc15 and contributes to the stability of the ring.
Backup pathway for ring assembly
The ability of fission yeast cells to form contractile rings and divide without Mid1 or cytokinesis nodes4,8,11,42,55–57 revealed a mechanism that corrects defects in the normal assembly pathway31,43 (see BOX 1 for the different views of geneticists and biophysicists on what it means to be essential). During mitosis, cells without Mid1 form strands of contractile ring proteins (including Myo2, actin, Cdc12 and Cdc15) scattered over the cortex. These proteins can therefore assemble without Mid1 as a scaffold. In some cells, these strands become connected into rings oriented at random angles and positions relative to the long axis of the cell. These oblique rings can slowly constrict the plasma membrane and direct the formation of a septum, but are usually off-centre, so they do not reliably separate the two daughter nuclei.
Box 1. On being ‘essential’.
What does it mean to say that a gene or protein is ‘essential’ for a complicated process such as cytokinesis? Geneticists and biophysics use the word essential in different ways. From the genetics perspective, a gene or protein is essential if the viability of the organism depends on it and ‘non-essential’ if the organism can survive without it, even if survival is a struggle. From the biophysics perspective, a gene or protein is essential if the system does not work normally in its absence, showing that the component is required for normal timing and/or fidelity. Some genes such as the anillin-like mid1 (also known as dmf1) are not essential in the sense that strains lacking mid1 are viable. However, cells lacking the Mid1 protein do not grow well and cytokinesis is far from normal, so the protein is absolutely required for the ‘normal’ process of cytokinesis. The fact that some cells lacking Mid1 manage to cleave in two illustrates the important point that cells have mechanisms to correct serious defects that occur along the normal pathway.
This backup pathway depends on the SIN pathway42 and activation of this pathway in interphase may produce contractile rings by this mechanism58. This alternative pathway works better in cells with a temperature sensitive mutation of 1,3β glucan synthase component 1 (Bgs1; also known as Cps1), by allowing oblique contractile rings to slide into a normal orientation that is perpendicular to the long axis of the cell57,59. Nothing is known about how the mechanisms of this pathway relate to the normal search, capture, pull and release pathway, or to the normal disappearance of Mid1 before constriction.
Septation and membrane scission
Cells use the SIN pathway to coordinate the formation of a specialized cell wall, called the septum, with ring constriction and fusion and scission of the plasma membrane41. Supplementary information S1 (table) lists more than 70 genes, including septins, an anillin (Mid2), enzymes, GTPases and membrane trafficking machinery, that contribute to septum formation and membrane fusion and scission. The role of Bgs1 in restricting the motion of contractile rings57,59 suggests that the contractile ring might be physically connected to the enzymes that make the septum.
Major open questions
We understand the assembly, constriction and disassembly of contractile rings in fission yeast better than in any other organism, but our understanding is incomplete and much work is yet to be done. The identification of the long list of cytokinesis genes in Supplementary information S1 (table) is a remarkable achievement for this small field. One might hope that the list is close to completion, but it has doubled in 10 years, so some cytokinesis genes are probably yet to be discovered. The less complete inventories of cytokinesis proteins in animals overlap with the proteins used by fission yeast. The participation of anillin, myosin II, actin and formins, as well as the general order of events, indicate that mechanisms of cytokinesis are likely to be similar in fission yeast and animals. More detailed studies on animals will be required to document the extent of these similarities and any fundamental differences. Punctate contractile ring precursors containing myosin II and anillin have been seen in animal cells, and condensation of these precursors into a contractile ring depends on the formin cytokinesis defect protein 1 (CYK-1) in Caenorhabditis elegans embryos60. More work is required to learn how similar the mechanism is to that in fission yeast.
Learning how well the molecular mechanisms of cytokinesis are conserved will depend on better inventories of cytokinesis genes from the main branches of the phylogenetic tree and a better understanding of the reactions at the system level. The current state of knowledge suggests that most new cytokinesis genes that appeared during evolution were conserved in subsequent branches of the tree. Actin is the most ancient component, having arisen in the common ancestor of all forms of life. Prokaryotes use actin for many interesting processes, but apparently not for cytokinesis. After algae and plants diverged, primitive amoeboid eukaryotes evolved myosin II and the contractile ring mechanism based on actin filaments. Amoebas, fungi and animals retained this mechanism for reliable cell division. The genes for membrane traffic and scission are more ancient than the genes that encode myosin II and are still used by all higher eukaryotes.
Certain aspects of cytokinesis are better understood in systems other than fission yeast. For example, we understand how mitotic spindle microtubules, Rho family GTPases and chromosomal passenger proteins control contractile ring positioning, assembly and constriction better in animals than in fission yeast61. Numerous examples show that organisms adapted ancient proteins for novel cytokinesis strategies, illustrating how evolution can stumble upon any number of strategies based on available materials. However, we are more impressed by the conservation of basic mechanisms, so deciphering elements of the system in a favourable organism should provide insights into cytokinesis in other cells.
Beyond an account of the participating molecules, understanding the mechanisms will depend on better ideas, more information about the participating molecules, high quality quantitative measurements in live cells and mathematical models. Better ideas are needed because some fundamental concepts are missing, such as strategies to assemble contractile ring precursors. Our understanding of the mechanisms of most cytokinesis proteins (IQGAPs, for example) is still primitive. Even where characterization is advanced, such as for actin, myosin II and formins, gaps remain, such as in understanding the mechanisms regulating the activity of Cdc12. GFP fusion proteins and spectacular new microscopes enable the precise measurement of cellular events, but this work is in its infancy. Mathematical models have been useful for formulating and testing some ideas about cytokinesis, and we expect them to be the indispensable gold standard for designing and interpreting experiments in the future.
Supplementary Material
Acknowledgments
The work in the T.D.P. laboratory is supported by the National Institutes of Health (NIH) research grant GM-026338 and the work in the J.-Q.W. laboratory is supported by an American Cancer Society Ohio Pilot research grant, American Heart Association Great Rivers Affiliate, Ohio Cancer Research Associates, Basil O’Connor Starter Scholar research award from the March of Dimes Foundation and the NIH research grant GM-086546. The authors thank Z. Cande, F. Chang, Q. Chen, J. Moseley and S. Saha for comments on the manuscript.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Contributor Information
Thomas D. Pollard, Department of Molecular Cellular and Developmental Biology, and the Departments of Molecular Biophysics and Biochemistry and of Cell Biology, Yale University, PO BOX 208103, New Haven, Connecticut 06520-8103, USA
Jian-Qiu Wu, Department of Molecular Genetics, and the Department of Molecular and Cellular Biochemistry, The Ohio State University, Columbus, Ohio 43210, USA.
References
- 1.Otegui MS, Verbrugghe KJ, Skop AR. Midbodies and phragmoplasts: anallogous structures involved in cytokinesis. Trends Cell Biol. 2005;15:404–413. doi: 10.1016/j.tcb.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- 3.Guertin DA, Trautmann S, McCollum D. Cytokinesis in eukaryotes. Microbiol Mol Biol Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 5.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 6.Padte NN, Martin SG, Howard M, Chang F. The cell-end factor Pom1p inhibits Mid1p in specification of the cell division plane in fission yeast. Curr Biol. 2006;16:2480–2487. doi: 10.1016/j.cub.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 8.Bähler J, et al. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998;143:1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motegi F, Nakano K, Mabuchi I. Molecular mechanism of myosin-II assembly at the division site in Schizosaccharomyces pombe. J Cell Sci. 2000;113:1813–1825. doi: 10.1242/jcs.113.10.1813. [DOI] [PubMed] [Google Scholar]
- 10.Paoletti A, Chang F. Analysis of Mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol Biol Cell. 2000;11:2757–2773. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motegi F, Mishra M, Balasubramanian MK, Mabuchi I. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J Cell Biol. 2004;165:685–695. doi: 10.1083/jcb.200402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 13.Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- 14.Almonacid M, et al. Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Curr Biol. 2009;19:961–966. doi: 10.1016/j.cub.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Morrell JL, Nichols CB, Gould KL. The GIN4 family kinase, Cdr2p, acts independently of septins in fission yeast. J Cell Sci. 2004;117:5293–5302. doi: 10.1242/jcs.01409. [DOI] [PubMed] [Google Scholar]
- 16.Moseley JB, Nurse P. Cdk1 and cell morphology: connections and directions. Curr Opin Cell Biol. 2009;21:82–88. doi: 10.1016/j.ceb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Celton-Marizur S, Racine V, Sibarita JB, Paoletti A. Pom1 kinase links division plane position to cell polarity by regulating Mid1p cortical distribution. J Cell Sci. 2006;119:4710–4718. doi: 10.1242/jcs.03261. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y, Chew TG, Ge W, Balasubramanian MK. Polarity determinants Tea1p, Tea4p, and Pom1p inhibit division-septum assembly at cell ends in fission yeast. Dev Cell. 2007;12:987–996. doi: 10.1016/j.devcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Bähler J, Pringle JR. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 1998;12:1356–1370. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yonetani A, et al. Regulation and targeting of the fission yeast formin Cdc12p in cytokinesis. Mol Biol Cell. 2008;19:2208–2219. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coffman VC, Nile AH, Lee IJ, Liu H, Wu JQ. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Celton-Marizur S, Bordes N, Fraisier V, Tran P, Paoletti A. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Mol Cell Biol. 2004;24:10621–10635. doi: 10.1128/MCB.24.24.10621-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang F, Drubin D, Nurse P. Cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu J, Pollard TD. Profilin binding to poly-L-proline and actin monomers along with ability to catalyse actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero S, et al. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 28.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 29.Vavylonis D, Kovar DR, O’Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marks J, Hyams JS. Localization of F-actin through the cell division cycle of Schizosaccharomyces pombe. Eur J Cell Biol. 1985;39:27–32. [Google Scholar]
- 31.Karagiannis J, Bimbo A, Rajagopalan S, Liu J, Balasubramanian M. The nuclear kinase Lsk1p positively regulates the septation initiation network and promotes the successful completion of cytokinesis in response to perturbation of the actomyosin ring in Schizosaccharomyes pombe. Mol Biol Cell. 2005;16:358–371. doi: 10.1091/mbc.E04-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arai R, Mabuchi I. F-actin ring formation and the role of F-actin cables in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 2002;115:887–898. doi: 10.1242/jcs.115.5.887. [DOI] [PubMed] [Google Scholar]
- 34.Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J Cell Biol. 2003;162:851–862. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamasaki T, Osumi M, Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol. 2007;178:765–771. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lord M, Pollard TD. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J Cell Biol. 2004;167:315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sladewski TE, Previs M, Lord M. Regulation of fission yeast myosin-II function and contractile ring dynamics by regulatory light chain and heavy chain phosphorylation. Mol Biol Cell. 2009;20:3941–3952. doi: 10.1091/mbc.E09-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loo TH, Balasubramanian M. Schizosaccharomyces pombe Pak-related protein, Pak1p/Orb2p, phosphorylates myosin regulatory light chain to inhibit cytokinesis. J Cell Biol. 2008;183:785–793. doi: 10.1083/jcb.200806127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelham RJ, Jr, Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- 40.Roberts-Galbraith RH, Chen JS, Wang J, Gould KL. The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J Cell Biol. 2009;184:113–127. doi: 10.1083/jcb.200806044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krapp A, Simanis V. An overview of the fission yeast septation initiation network (SIN) Biochem Soc Trans. 2008;36:411–415. doi: 10.1042/BST0360411. [DOI] [PubMed] [Google Scholar]
- 42.Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wachtler V, Huang Y, Karagiannis J, Balasubramanian M. Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:3254–3266. doi: 10.1091/mbc.E05-11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge W, Balasubramanian M. Pxl1p, a paxillin-related protein, stabilizes the actomyosin ring during cytokinesis in fission yeast. Mol Biol Cell. 2008;19:1680–1692. doi: 10.1091/mbc.E07-07-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berlin A, Paoletti A, Chang F. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J Cell Biol. 2003;160:1083–1092. doi: 10.1083/jcb.200212016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tasto JJ, Morrell JL, Gould KL. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J Cell Biol. 2003;160:1093–1103. doi: 10.1083/jcb.200211126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An H, Morrell JL, Jennings JL, Link AJ, Gould KL. Requirements of fission yeast septins for complex formation, localization and function. Mol Biol Cell. 2004;15:5551–5564. doi: 10.1091/mbc.E04-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakano K, Mabuchi I. Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol Biol Cell. 2006;17:1933–1945. doi: 10.1091/mbc.E05-09-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bement WM, Benink HA, von Dassow G. A microtubule-dependent zone of active RhoA during cleavage plane specification. J Cell Biol. 2005;170:91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carvalho A, Desai A, Oegema K. Structural memory in the contractile ring makes the duration of cyotkinesis independent of cell size. Cell. 2009;137:926–937. doi: 10.1016/j.cell.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 51.Schroeder TE. The contractile ring. II Determining its brief existence, volumetric changes, and vital role in cleaving Arbacia eggs. J Cell Biol. 1972;53:419–434. doi: 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts-Galbraith RH, Gould KL. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 2008;22:3082–3088. doi: 10.1101/gad.1748908. [DOI] [PubMed] [Google Scholar]
- 53.Chen CT, et al. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr Biol. 2008;18:1509–1594. doi: 10.1016/j.cub.2008.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clifford DM, et al. The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J Cell Biol. 2008;181:79–88. doi: 10.1083/jcb.200709060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- 56.Chang F, Wollard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- 57.Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997;11:1519–1534. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- 59.Pardo M, Nurse P. Equatorial retention of the contractile actin ring by microtubules during cytokinesis. Science. 2003;300:1569–1574. doi: 10.1126/science.1084671. [DOI] [PubMed] [Google Scholar]
- 60.Werner M, Munro E, Glotzer M. Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr Biol. 2007;15:1286–1297. doi: 10.1016/j.cub.2007.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nature Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.