Abstract
Gap junction channels connect the cytoplasms of adjacent cells through the end-to-end docking of hexameric hemichannels called connexons. Each connexon is formed by a ring of 24 α-helices that are staggered by 30° with respect to those in the apposed connexon. Current evidence suggests that the connexons are docked by interdigitated, anti-parallel β strands across the extracellular gap. The second extracellular loop, E2, guides selectivity in docking between connexons formed by different isoforms. There is considerably more sequence variability of the N-terminal portion of E2, suggesting that this region dictates connexon coupling. Mutagenesis, biochemical, dye-transfer and electrophysiological data, combined with computational studies, have suggested possible assignments for the 4 transmembrane α-helices. Most current models assign M3 as the major pore-lining helix. Mapping of human mutations onto a Cα model suggested that native helix packing is important for the formation of fully functional channels. Nevertheless, a mutant in which the M4 helix has been replaced with polyalanine is functional, suggesting that M4 is located on the perimeter of the channel. In spite of this substantial progress in understanding the structural biology of gap junction channels, an experimentally determined structure at atomic resolution will be essential to confirm these concepts.
Keywords: gap junction channel, connexon, intercellular communication, electron microscopy, mutagenesis, computational modeling
1 Introduction
Gap junctions are specialized regions of the plasma membrane in which hexameric oligomers, called connexons, dock end-to-end noncovalently across the narrow extracellular gap. These intercellular channels exclude the extracellular environment and allow the exchange of nutrients, metabolites, ions and small molecules of up to ≈1000 Da (Loewenstein, 1981). Intercellular coupling by gap junctions is a fundamental mechanism for cell-to-cell communication within tissues of higher organisms. More than 20 unique connexins have been identified to date in virtually all multicellular organisms, from mesozoa to humans (Bruzzone et al., 1996; Gerido and White, 2004), including a putative invertebrate connexin (Phelan et al., 1998; White et al., 2004).
Each connexon in the channel consists of an annular assembly of six individual connexins, which forms a pore through the plasma membrane. The particular connexins present on the surfaces of adjacent cells may not be identical to each other (reviewed by Kumar and Gilula, 1996). In the simplest homotypic case, the dodecameric channel is formed by a single connexin isoform. In the heterotypic case, a connexon formed by a particular isoform docks with a second connexon formed by a different isoform. Heteromeric connexons also exist in which the hexamer is formed by more than one isoform. The expression of multiple connexins in the same cell type, the multiplicity of isoforms, and the ability to form homomeric and heteromeric connexons, as well as homotypic and heterotypic channels, likely provides exquisite “functional tuning” of this family of membrane channels.
The primary biophysical tools for structure analysis of gap junction channels have included electron microscopy and image analysis, X-ray diffraction, nuclear magnetic resonance (NMR) spectroscopy, and atomic force microscopy (AFM). These techniques have been used in combination with mutagenic, biochemical and in vivo functional studies to explore the structural and functional properties of gap junction channels. In this review we will focus on some current molecular modeling studies based mainly on maps derived by electron cryo-crystallography and structurally focused mutagenesis studies. More comprehensive reviews by Yeager and Nicholson (2000), Harris (2001) and Sosinsky and Nicholson (2005) include more detailed reference lists.
2 Gap junction connexons are formed by a ring of 24 helices
Hydropathy analyses of the sequences of various connexin family members suggests the presence of four hydrophobic domains each comprised of 20–28 residues, referred to as M1, M2, M3 and M4, proceeding from the N- to the C-terminus (Milks et al., 1988). Connecting the transmembrane (TM) domains are two extracellular loops (E1 and E2), each containing 3 cysteines, and one cytoplasmic M2–M3 loop. Both the N- and C-termini reside in the cytoplasm (Milks et al., 1988; Yancey et al., 1989; Yeager and Gilula, 1992). The four TM domains, and particularly the cysteine-containing extracellular loops (E1 that connects M1 to M2 and E2 that connects M3 to M4) represent the most conserved regions of the family (Beyer et al., 1987; Kumar and Gilula, 1986). The most variable domains, both in length and sequence, are the C-terminal domain and the cytoplasmic hydrophilic loop connecting M2 to M3.
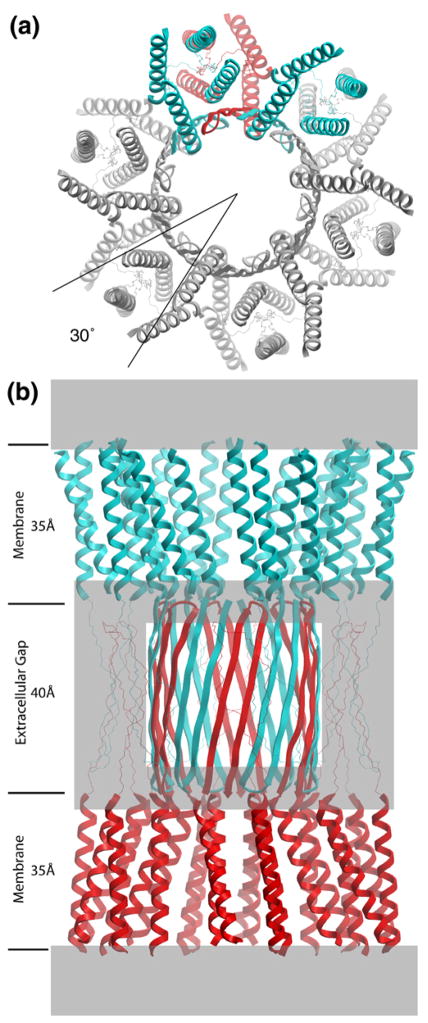
The general higher-order assembly of gap junction channels (Figure 1) and the α-helical structure of the TM domains was first revealed by electron cryo-microscopy and diffraction analysis of two-dimensional crystals (Unwin and Ennis, 1984; Unger et al., 1997, 1999). Each dodecameric channel is formed by the end-to-end docking of two hemi-channels, called connexons, which are rotationally staggered by 30° around the 6-fold symmetry axis (Unger et al., 1997; Perkins et al., 1998) (Figure 1a). The 3D map at 7.5 Å in-plane resolution showed that each connexon contains 24 rod-like densities interpreted as TM α-helices, based on their length, diameter and packing (Unger et al., 1999). The primary sequence identity of each TM helix could not be assigned at this resolution, and they were therefore arbitrarily designated A, B, C and D (Figure 2).
Figure 1.
Molecular design of gap junction channels. (a) Top view showing the 30° rotational stagger between docked connexons. Two subunits of the top connexon (in blue) are above one subunit of the bottom connexon (in red). The other subunits have been colored gray for clarity. The molecular boundary is depicted as a 4-helix bundle, but there are other possibilities (Figure 2). (b) Side view. The top connexon is in blue and the bottom one in red. Grayed areas denote parts of the structure that are most uncertain, especially the folding within the density at the boundary between the TM assembly and the extracellular space. Putative β sheets corresponding to E1 (on the perimeter of the extracellular gap) are drawn with thin lines to emphasize this ambiguity. The E2 loops are depicted as an interdigitating β barrel. Refer to Figures 2 and 3 in Unger et al. (1999) for the corresponding views of the 3D density map derived by electron cryo-crystallography.
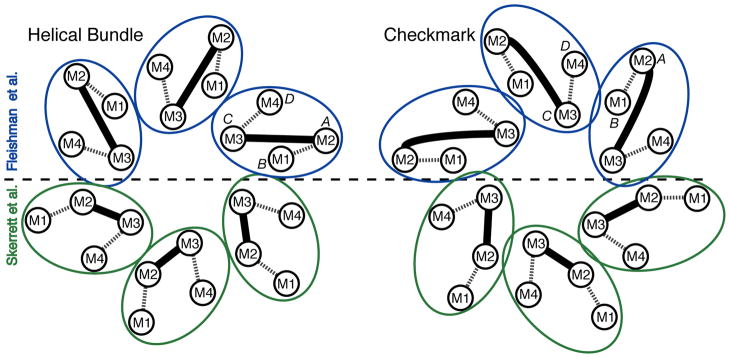
Figure 2.
Possible molecular boundaries for the connexin subunit include a helical bundle (left) and a “checkmark” (right) (following the naming of Unger et al., 1999). Each shows two assignments for the 4 TM α-helices within each subunit, according to Fleishman et al. (2004) (top, blue) and Skerrett et al. (2002) (bottom, green). Dashed lines denote the extracellular loops, E1 and E2, and the solid lines denote the M2–M3 cytoplasmic loops. The α-helical rods designated A, B, C and D in the 3D density map derived by electron cryo-crystallography (Unger et al., 1999) are also indicated.
Milks et al. (1988) first proposed that M3 was the pore-lining helix because it contained a hydrophilic stripe that was presumed to line the aqueous pore. The first experimental evidence for the accessibility of M3 to the aqueous pore was provided by experiments using the substituted cysteine accessibility method (SCAM) in gap junction channels formed by C×46 connexons paired with chimeric C×46, in which the E1 loop was derived from C×32 (Zhou et al., 1997). Two residues in M1, and three in M3, were accessible to aqueous sulfhydryl reagents, leading to a partial block of the channel. Surprisingly, the greatest effect was observed for two adjacent residues centrally located in M1. SCAM studies of C×46 hemichannels have also suggested that residues in M1 are accessible to labeling (Kronengold et al., 2003). Indeed, models have been proposed in which M1 is the major pore-lining α-helix (Oshima et al., 2003).
By combining the results of an improved cryo-EM map (with an in-plane resolution of 5.7 Å and a vertical resolution of 19.8 Å) with biochemical and biophysical evidence, a Cα model for the TM domains within a connexon was proposed (Fleishman et al., 2004). For membrane proteins, evolutionarily conserved amino acids are more likely to mediate protein packing interactions, and variable residues are more likely to face the lipid (Baldwin, 1993). On the basis of the potential relative spatial location of conserved and variable residues within the connexin family, as well as SCAM analysis, the primary sequence of TM segments M1–M4 was assigned to the observed α-helices in the map (A=M2, B=M1, C=M3, D=M4) (Figure 2). The relative rotation angles of the α-helices fitted into the density map were estimated by analysis of evolutionary conservation and hydrophobicity of amino acid residues. Although this is the most well-defined model for the TM domains of gap junctions at the time of this writing, the conformation of the amino acid side chains remains undetermined. In addition, the α-helical rods in the cryo-EM density map display curvature not reflected in the idealized Cα model of Fleishman et al. (2004).
Experimental validation of the model proposed by Fleishman et al. (2004) was provided by expression of connexins with compensatory mutations based upon naturally occurring pathological point substitutions (Fleishman et al., 2006). Competent assembly was assayed by fluorescent labeling of connexins in the plasma membrane. In addition, an attempt was made to identify, in each case, a compensatory mutation for each deleterious mutation that restored plasma membrane targeting. For example, function was restored with the salt-bridge swaps Arg32 with Glu146 and Glu209 with Lys22, and with the mutation of a single packing pair (Ser139 with Asn206). In addition to supporting the above TM assignment, this work is a good example of the tight linkage between structure and the biological function of the molecule.
We note that the helical assignment in Fleishman et al. (2004) differs from a previous assignment (Skerrett et al., 2002), based upon SCAM analysis, in which the assignments of M1 and M2 were reversed (A=M1, B=M2). This discrepancy might be attributed to methodological differences or to possible differences in conformation, such as the latter representing an open conformation and the former a closed conformation. Another possibility is the presence of conformational flexibility or “breathing” that would create transient solvent crevices between α-helices that would allow labeling by water soluble reagents, as has been observed for K+ channels (Simoes et al., 2002).
While progress has been made regarding the assignment of the helices, there still remains ambiguity as to the exact molecular boundary of the individual monomers, since the connecting loops between helices were not visible due to the limited resolution of the 3D cryo-EM map and possible disorder of the cytoplasmic regions of the connexins. The packing of the 24 α-helices within the 6-fold symmetric connexon can accommodate several possible molecular boundaries. Scrutiny of the density map and exclusion of models that require crossovers of the E1 and E2 loops suggests that the most likely molecular boundaries are a closely-packed 4-helix bundle or a more loosely packed “checkmark” arrangement (Unger et al., 1999, Figure 4), shown in Figure 2. For each of these possibilities, we also show the helical assignments of Fleishman et al. (2004) (blue) and Skerrett et al. (2002) (green).
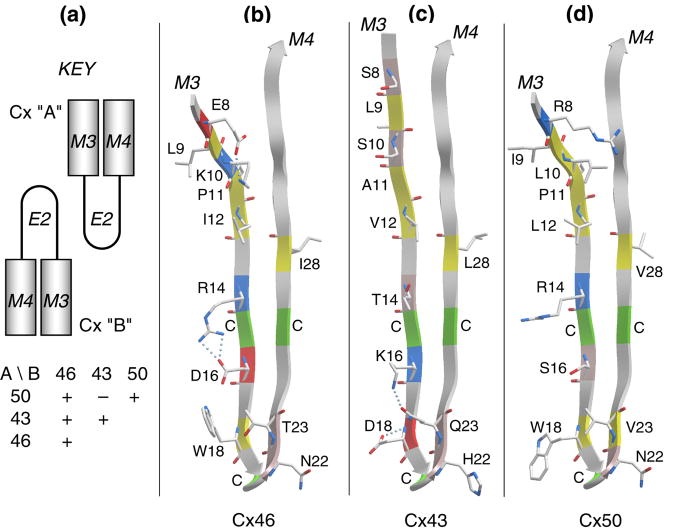
Figure 4.
The E2 loop is a primary determinant of isoform compatibility. (a) A schematic key to show the compatibilities in pairing of heterotypic channels formed by C×46, C×43 and C×50. (b), (c) and (d) show a comparison of the E2 loops of C×46, C×43 and C×50, respectively. Only amino acids that differ in at least one of the three isoforms are shown in a stick representation. Cysteines are indicated with C and colored green on the ribbon. Residue coloring is: yellow, hydrophobic; pink, polar; red, acidic; blue, basic. Note that the C-terminal halves of the loops are highly conserved, suggesting that selectivity is dictated by the N-terminal regions of the loops. As discussed in the text, the hydrogen-bonding pattern near the β hairpin of C×43, along with its H22, may account for its incompatibility with C×50. This analysis was based on the 3 localized regions that are predicted to be β strands and did not depend on the complete β-turn-β fold depicted in the figure.
3 Helical structure within the N- and C-terminal and cytoplasmic loops
There is limited information on the structure of the N- and C- termini and the M2–M3 cytoplasmic loop. These regions of the protein are critical in channel regulation. The N-terminus participates in voltage sensing (Verselis et al., 1994; Oh et al., 2004), the M2–M3 loop participates in pH-mediated gating (Duffy et al., 2002; Morley et al., 1996), and the C-tail participates in phosphorylation-dependent gating (Warn-Cramer et al., 1996; Zhou et al., 1999) and in pH-dependent gating (Ek-Vitorín et al., 1996; Morley et al., 1996). The C-tail of C×43 also binds v-SRC (Loo et al., 1995; Kanemitsu et al., 1997) and the second PDZ domains of zonula occludens-1 and -2 (Sorgen et al., 2004; Singh et al., 2005).
NMR spectroscopy of the isolated C-terminus of C×43 indicates that the polypeptide is mostly disordered, with the exception of two short non-associating helical stretches (amino acids 315–326 and 340–348) (Sorgen et al., 2004). These authors proposed that these helical regions may in fact dimerize and be important in gating.
A peptide corresponding to the N-terminus of C×26 has also been characterized using NMR spectroscopy (Purnick et al., 2000). The isolated 13-amino-acid peptide consists of a two-turn α-helix, which then unravels into a flexible loop-like structure. It was hypothesized that this short amino-terminal helix is oriented parallel to the TM helices lining the entrance to the pore, thus forming part of the conduction path and contributing to the voltage dependence of the channel. The structural mechanism by which regulation occurs and the overall arrangement of these domains in the complete channel awaits more detailed structural and/or biochemical studies.
NMR studies of a 26 residue peptide in the carboxyl portion of theM2–M3 cytoplasmic loop suggest the formation of two short α-helices under acidic conditions (Duffy et al., 2002). These regions were proposed to bind to the C-tail of C×43 and thereby mediate pH-dependent gating (Duffy et al., 2002). We also note that the rod of density assigned to M3 in the cryoEM map extends into the cytoplasm, suggesting continuation of the N-terminal portion of the loop as an α-helix (Unger et al., 1999, Figure 3a).
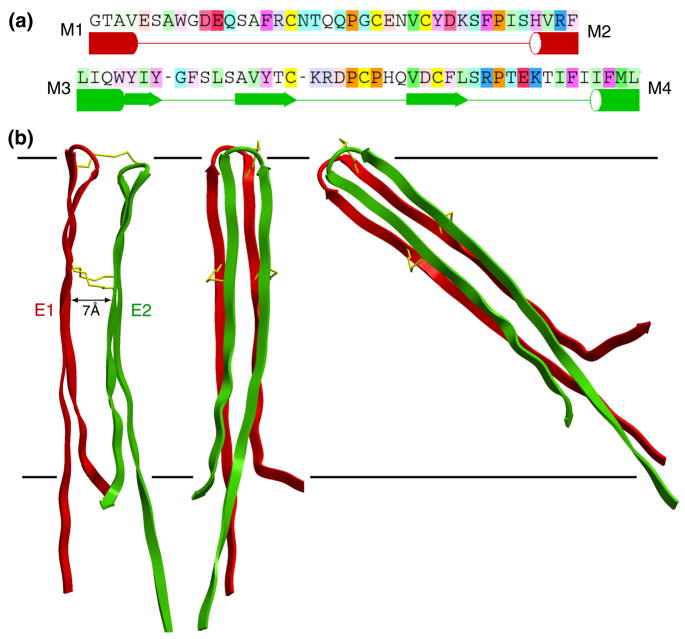
Figure 3.
(a) Sequences of loops E1 (top) and E2 and a statistical secondary-structure prediction with ICM software (in preparation). Tubes and arrows are predicted to be TM helices and β-sheet secondary structure, respectively. Note that the prediction for E2 shows partial β-strand structure. (The residue coloring comes from the alignment, and depends on both the conservation and the residue type as in Figure 4.) (b) Loops E1 (red) and E2 (green) belonging to a single connexin subunit. The left side shows a side view, while the middle and right sides are pore views. The separation between the loops is ~7 Å. The right side shows that the 35 amino acids of the complete loops would have to be tilted by ~50° to be accommodated within the 40 Å extracellular gap (represented by horizontal lines), if one assumes that they form extended β strands. The cysteines forming disulfide bonds are indicated by gold sticks.
4 A portion of the extracellular loops pack as anti-parallel β-pleated sheets
Data are sparse as to the topological location of E1 and E2 in the extracellular gap. A careful electrophysiological and SCAM study of C×46 hemichannels suggested that at least a portion of E1 is accessible for labeling (Kronengold et al., 2003). If M3 is the major pore-lining helix, then we would expect that at least the N-terminal portion of E2 is accessible to the pore. The cryo-EM density map displays a continuous ring of density centered at a diameter of ~34 Å and 6 arcs of density at a diameter of ~52 Å. These features suggest that the polypeptide in the gap is highly ordered and that the apposed connexins have extensive contact area, which is needed to form a tight seal separating the channel from the extracellular environment. However, these features do not allow confident assignment of the secondary structure of the protein within the extracellular gap. Foote et al. (1998) proposed, based on the pattern of disulfide bonds between E1 and E2, a double-β-barrel model for the extracellular region of the channel. Each barrel would be formed by a dodecameric, antiparallel interdigitation of β-turn-β projections from each connexon. In order to assess the plausibility of this proposition, we built a double β-barrel model, and Figure 1 shows the inner layer, using the diameter of ~34 Å in the cryoEM map (Unger et al., 1999). Although this may appear to be reasonable, the distance between loops belonging to adjacent connexins is shorter than ideal for a β barrel, and as a consequence, only some of the inter-loop hydrogen bonds would be geometrically favored in our computational model. Also, note that this was also the case even for a virtually vertical orientation of the β strands (Figure 3b). Usually, β strands in known β-barrel structures are more tilted with respect to the membrane plane (Schirmer, 1998), which would worsen the packing issue. We estimate that a tilt of about 50° would be needed in order for the full β-turn-β loops (about 35 amino acids) to pack within the accepted vertical gap space of ~40 Å (Figure 3b). We therefore hypothesize that the extracellular loops near the surface of the lipid bilayer break from a β conformation. Presumably this region of the loops (indicated with a shaded horizontal box in Figure 1) has a more complicated conformation so as to fit the 40 Å gap. A plausible variation could be that the β sheets of each connexin do not lie with their planes perpendicular to the radius vectors, but are somewhat rotated, which would also help accommodate the inner barrel within the ~34 Å diameter seen in the map. The outer arcs of density in the extracellular gap, observed at a diameter of ~52 Å, imply that the β strands cannot be packed as a continuous β barrel (since the same number of strands as in the inner barrel would span a larger diameter). Therefore, the continuous cylinder of density adjacent to the pore likely functions as the primary seal with the extracellular space.
5 Molecular modeling of the extracellular loops
We performed a statistical secondary-structure prediction of the E1 and E2 extracellular loops using ICM software (Abagyan et al., 1994). For E1, there was no preferred secondary structure, i.e., appearing to favor random coil. For the E2 loop, the prediction yielded three segments of β strand which spanned about 40% of the length of the loop, consistent with the above model.
By assuming a β-sheet secondary structure and carefully examining the sequence of the extracellular loops, particularly the positions of conserved prolines and cysteines, a potential spatial relationship between the extracellular loops (E1 and E2) can be defined. We therefore built a stacked β sheet model of E1 and E2 to see if the proposed disulfide bonding pattern of Foote et al. (1998) could be constructed with correct covalent geometry and stereochemistry. This was actually the case (Figure 3b). This model was built using the sequences of the C×43 loops and placing the centrally located proline of each loop at the center of the β hairpin. The cysteine residues align very well with good geometry, allowing the disulfide bonds to form correctly, even though the central one is shifted by one residue between E1 and E2. (This model was then inserted into that of the whole channel shown in Figure 1.)
However, the distance between the loops provided by this model, about 7 Å, is less than that seen between the inner ring and outer arcs in the cryo-EM map (9–10 Å). A possible explanation, as was suggested by others (Foote et al., 1998; Sosinsky and Nicholson, 2005), could be that the 7 Å separation holds only for undocked connexons, but upon docking together, the disulfide bonds are exchanged to form intermolecular crosslinks between the cysteines that reside in the middle of the β strands (i.e., not the β hairpin cysteines). In order for these disulfides top form, the distance between the β hairpins would have to be ~30 Å, as opposed to the ~40 Å as suggested in Figure 3. The resulting gap of about 9 Å might presumably be occupied by the winding portion of the neighboring connexin’s loops.
6 Selectivity of heterotypic interactions may be guided by the N-terminal portion of the E2 loops
Most homotypic couplings between connexons result in functional channels, but many heterotypic channels do not. An extensive summary of these compatibilities was presented in Yeager and Nicholson (2000, pp. 47ff). In particular, it has been shown that C×46 may be functionally paired with either C×43 or C×50, whereas C×50 does not form functional channels with C×43 (White et al., 1994). These electrophysiological experiments in paired oocytes suggested that the E2 loop was a major determinant of isoform selectivity. We analyzed this particular triplet (C×46, C×43 and C×50) in an attempt to elucidate a plausible set of rules dictating this selectivity behavior based on the E2 sequence difference between these isoforms (Figure 4). Non-conserved residues are labeled and colored by polarity. Note that there is substantially more conservation in sequence in the C-terminal halves of the E2 loops, suggesting that selectivity is dictated by the more variable N-terminal regions. We observe notable differences in certain key positions (arbitrarily numbered 8, 10, 16, 18 and 23), which could potentially participate in hydrogen-bonds and/or salt-bridges (indicated by small spheres) that are quite different between the isoforms. The only disallowed pairing presented here, according to White et al. (1994), is C×43/C×50. We suspect that this incompatibility is mainly the result of an Arg at position 8 in C×50, conflicting with the nature of side chains near the β hairpin (residues 16–23) of the paired connexon. For C×43, these residues could form H-bonds amongst themselves, leaving the side chain of H22 free, which together with the free side chain of R8 of C×50, might obstruct the channel, or result in a steric clash. In contrast, when C×50 is paired with C×46, residues D16, N22 or T23 of the latter would be available to form H-bonds with the side chain of R8 of C×50, constraining its conformation so as to avoid an obstruction or clash. Homotypic C×50 channels would therefore be allowed, for the same reason (N22 H-bonded to R8). By these criteria, the pairs C×46/C×43, C×46/C×46 and C×43/C×43 present no obstacle.
Of course, this most likely represents an overly simplistic view based on a very limited sample size. Nevertheless, it appears that the potential packing of a simple secondary-structure model of E2 might begin to provide a working hypothesis for understanding the origin of pairing preferences amongst connexins.
7 Replacement of the TM segments alters packing and functionality
The role of individual residues in transmembrane helices has been traditionally addressed by scanning mutagenesis (generally Cys- or Ala-scanning mutagenesis), where the TM residues are substituted, one at a time, and the functional and/or structural consequences of the substitutions are investigated (Kaback et al., 2001; Frillingos et al., 1998; Lu et al., 2001; Uveges et al., 2002). In the case of one of the better studied membrane proteins, the lactose permease of E. coli (12 TMs), it has been established that only 6 of its 417 amino acids are essential for function and cannot be replaced with any other residue (Kaback et al., 2001; Abramson et al., 2003). The recent discovery of an archaebacterial protein, Hsmr, with TMs composed of >40% Ala and Val (Ninio and Schuldiner, 2003) supports the notion that a large number of amino acid variations are tolerated in functional molecules. However, we note that Beahm et al. (2006) described a related experiment whereby the mutation of a conserved threonine in M3 eliminates channel function, without preventing its formation.
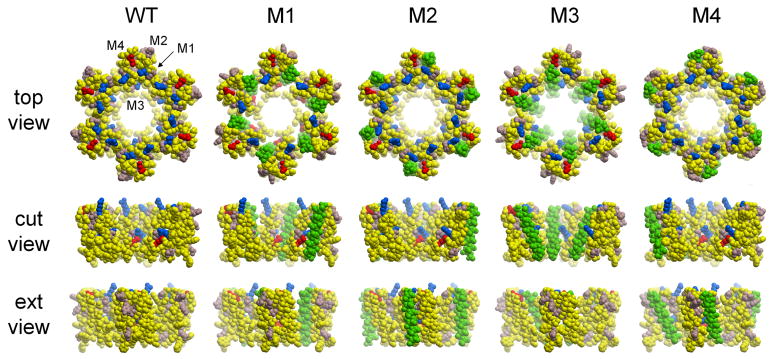
Multiple simultaneous mutations are potentially very informative (Ninio and Schuldiner, 2003), but it is generally thought that they are difficult to design because of the complex interactions of membrane-protein residues. With the possibility that amino-acid side chains within transmembrane α-helices are not necessary to generate and maintain the native folding of the membrane protein (Abramson et al., 2003), and that non-polar inter-helix interactions are the most frequent packing interactions in membrane proteins (Lee, 2002), Altenberg and his colleagues have replaced complete TMs with poly-Ala sequences (Miller et al., 2001, 2002) without homology to the wild-type amino-acid sequence (Figure 5). A priori, this approach looks risky because of potential effects of complete helix replacement on protein expression, targeting, folding, oligomerization and function. However, individual replacement of each of the C×43 TMs with poly-Ala sequences yielded mutant hemichannels that were expressed at the plasma membrane (Figure 6B) and exhibited dye transfer (Figure 6C).
Figure 5.
Helix-scanning mutagenesis of connexon43. Each of the four helices was substituted by a poly-alanine helix (shown in green). The coloring scheme is the same as in Figure 4. The M4 substitution is the only one which preserved channel function (Bao et al., 2005), consistent with the assignment of Fleishman et al. (2004) that M4 is on the perimeter of the channel.
Figure 6.
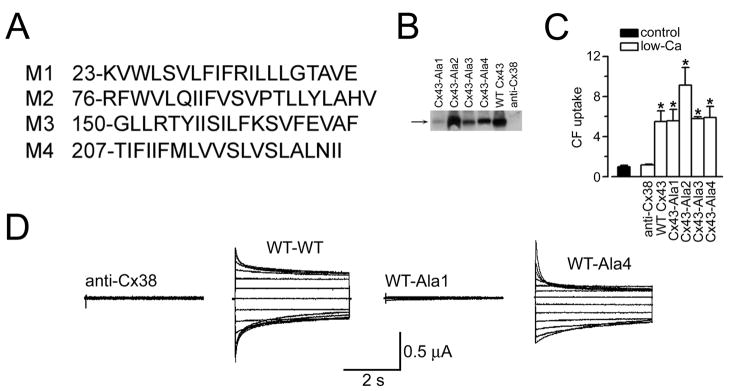
Formation of regulated gap junction channels and hemichannels by C×43 poly-Ala mutants (adapted from Bao et al., 2005). (A) Sequences of wild-type C×43 TM helices replaced with poly-Ala sequences. The position of the first residue of each sequence is indicated. (B) Immunoblots of plasma membranes from frog oocytes injected with anti-C×38 antisense oligonucleotide alone (anti-C×38) or in combination with cRNA coding for WT rat C×43 (WT C×43) or the C×43 poly-Ala mutants (C×43-Ala1 to C×43-Ala4). Data representative of 3 similar blots. (C) Uptake of CF via hemichannels. Data were normalized to the uptake in low-[Ca2+] solution in oocytes expressing WT C×43. Data are means ±SEM of 30–40 oocytes per condition. *P<0.05 compared to the control value, in the presence of 1.8 mM Ca2+. Units in the vertical axis are relative fluorescence values. (D) Formation of gap junction channels. Typical gap junction currents. Oocytes injected with anti-C×38 antisense oligonucleotide (anti-C×38), or expressing WT C×43 or poly-Ala mutants, were paired, and the junctional currents (Ij) were measured upon 4-s transjunctional voltage (Vj) steps between −120 and 120 mV, applied in 20-mV increments from a holding voltage of −60 mV. Four sets of records are shown, which Ij correspond to anti-C×38-anti-C×38, WT-WT, WT-Ala1 and WT-Ala4 oocyte pairs. The recordings are representative of 6–10 experiments per condition, obtained using the dual-electrode voltage-clamp technique (Bao et al., 2004b).
In the experiments of Bao et al. (2004c); Bao et al. (2004b); Bao et al. (2004a); Bao, and Chen et al. (2005), functional C×43 mutant hemichannels were defined as those capable of forming a plasma-membrane-regulated pore permeable to large hydrophilic solutes. Heterologous expression of recombinant C×43 in single frog oocytes resulted in uptake of the 376-Da molecular weight hydrophilic probe carboxyfluorescein (CF) via C×43 hemichannels (Bao et al., 2004a, 2004b). Figure 6C shows that CF uptake is very low with normal extracellular [Ca2+], due to specific hemichannel block (Bao et al., 2004a; Gomez-Hernandez et al., 2003; Muller et al., 2002; Pfahnl and Dahl, 1999), and that exposure to a low-[Ca2+] medium resulted in a significant increase in CF uptake in the WT and poly-Ala mutants. Two known blockers of gap junction channels and hemichannels, 18-β-glycyrrhetinic acid and octanol, (Bao et al., 2004a; Eskandari et al., 2002) also blocked this CF uptake (Bao et al., 2005). In normal [Ca2+], CF uptake is increased by the PKC inhibitor calphostin C because of dephosphorylation of Ser368 (Bao et al., 2004a, 2004b, 2004c). Calphostin C also increased CF uptake in oocytes expressing a C×43 poly-Ala mutant (Bao et al., 2005). For these experiments, expression of endogeneous Cx was suppressed by injecting the oocytes with C×38 antisense oligonucleotides.
Now, as to the whole channels, paired oocytes expressing poly-Ala mutants had no detectable gap junction currents, but C×43-Ala4 formed gap junction channels with wild-type C×43 (Figure 6D, WT-Ala4 trace on the right). Although there was some asymmetry in the time dependency of the junctional currents elicited by transjunctional voltage pulses in the WT-Ala4 channels, the voltage dependency of the current was generally conserved (i.e., slow inactivation at large voltages). Figure 6D (WT-Ala1 trace) also shows the absence of gap junction currents between an oocyte expressing WT C×43 and another expressing C×43-Ala1. Junctional currents were also absent between oocyte pairs consisting of one oocyte expressing WT C×43 and the other one C×43-Ala2 or C×43-Ala3 (Bao et al., 2005).
As described above, gap junction channel formation by docking of connexons of different connexin composition (heterotypic channels) occurs only between certain isoforms, i.e., is highly selective (Yeager and Nicholson, 2000; Harris, 2001). A change in the conformation of the extracellular loops has been shown to prevent hemichannel docking (Harris, 2001; Foote et al., 1998), and it is therefore likely that a similar conformational change occurs in the poly-Ala mutants due to alterations in the packing or membrane-exit conformation of the TM helices. Changes in helix packing of the poly-Ala mutants are expected because small residues such as Ala favor closer helix packing in membrane proteins and the substitution of native polar residues can affect helix-helix interactions (Curran and Engelman, 2003; Eilers et al., 2000; Javadpour et al., 1999; Jiang and Vakser, 2004). In any case, the results indicate that poly-Ala helix mutants form hemichannels whose permeability is increased by lowering [Ca2+] and by blocking PKC, both known regulatory mechanisms of C×43 gap junction channels and hemichannels. In addition, at least one of the C×43 mutants (C×43-Ala4) also formed functional gap junction channels.
8 Folding of connexins and functional implications
It is generally considered that the mechanisms of folding of membrane proteins are simpler than those of soluble proteins because of the constraints established by the lipid bilayer environment. In membrane proteins, the folding is viewed as a two-stage process (Popot and Engelman, 2000). First, the α-helices insert into the lipid bilayer with the correct topology, and second, the packing of these helices results in the final, functional, folded structure. Reality may be more complex (Engelman et al., 2003), but this simple two-stage model explains a substantial fraction of experimental observations. The fact that the primary sequence of the C×43 TM helices is not essential to form functional and regulated hemichannels suggests that the hydrophobic interactions between TM helices may not be the exclusive guide for the folding of connexins. Since the C-terminal domain of C×43 is not required for the formation of hemichannels, either the remaining cytoplasmic loops (N-terminal, M2–M3) and/or extracellular loops (E1 and E2) may participate in directing the folding and assembly of the connexon. In this regard, the extracellular loops may contribute appreciabledriving force because they are highly structured compared with the N- and C-termini and the M2–M3 loop. Although conserved Cys residues in these loops are required for the formation of functional channels (Foote et al., 1998), they are not needed for the assembly of hemichannels, as demonstrated with a C×43 mutant that lacks these conserved Cys residues (Bao et al., 2004b, a nd unpublished observations).
The formation of functional hemichannels by all C×43-Ala mutants suggests that none of the helices contains specific residues needed for the formation of a large pore that is permeable to hydrophilic solutes. Even though dye transfer is maintained in the C×43 poly-Ala mutant hemichannels, variations in the primary sequence of the helices likely tune function by alterations in helix packing and/or electrostatics. For example, the absence of larger wild-type side chains by the poly-Ala replacements of M1 and M3 could directly result in a larger effective pore for hydrophilic compounds, with an increase in permeability and size cut-off. The replacements can also reduce hydrophobic interactions of some permeants (e.g., fluorescent probes) with the pore (Harris, 2001; Weber et al., 2004), with no easily predictable effects of permeability. The effects of replacements of M2 and M4 may depend mostly on the effects on helix packing. In summary, the above results clearly indicate that fairly drastic changes in the primary sequence of the C×43 TM helices do not interfere with the assembly of connexons. Nevertheless, the primary sequence of different connexin isoforms will determine size and substrate selectivity, which are critical for the function and regulation of gap junction channels.
9 Conclusions
In the last decade, there has been impressive progress in the analysis of several classes of membrane proteins, including reaction centers, porins, ligand-gated channels, voltage-gated channels, transporters and aquaporins (Raman et al., 2006). By comparison, the tempo of discovery in the gap junction channel field has been slower. Possible reasons include difficulties with expression of engineered connexins with sufficient stability and quantity to allow detailed biochemical and biophysical analysis, difficulties in performing electrophysiological studies on a channel that spans two membranes, and lack of a repertoire of pharmacological agents to probe channel function. Nevertheless, recent electrophysiological, biochemical and biophysical studies, and analysis of engineered and pathological mutations have yielded a consensus model for the general molecular design of gap junction channels, which we have summarized in this review. There is currently no information on amino-acid, side-chain conformations, which will be essential to understand the molecular basis of (1) the stability and selectivity in docking of connexons, (2) mechanisms of gating (pH, divalent cations, phosphorylation, membrane active agents), and (3) the ability to form homomeric and heteromeric, as well as homotypic and heterotypic channels. Structural data at atomic resolution are required to gain insight into these unique functional properties of gap junction channels.
Acknowledgments
This work was supported by NIH grants 2 R01 HL048908-11 (M.Y.), 1 R01 GM071872-01 (R.A.) and R21 DC007150 (G.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abagyan R, Totrov M, Kuznetsov D. ICM: a new method for structure modeling and design: Applications to docking and structure prediction from the distorted native conformation. J Comput Chem. 1994;15:488–506. [Google Scholar]
- Abramson J, Smirnova I, Kasho V, Verner G, Iwata S, Kaback HR. The lactose permease of Escherichia coli: overall structure, the sugar-binding site and the alternating access model for transport. FEBS Letters. 2003;555:96–101. doi: 10.1016/s0014-5793(03)01087-1. [DOI] [PubMed] [Google Scholar]
- Baldwin JM. The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 1993;12:1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Altenberg GA, Reuss L. Mechanism of regulation of the gap junction protein connexin 43 by protein kinase C-mediated phosphorylation. Amer J Physiol – Cell Physiol. 2004a;286:C647–C654. doi: 10.1152/ajpcell.00295.2003. [DOI] [PubMed] [Google Scholar]
- Bao X, Chen Y, Lee SH, Lee SC, Reuss L, Altenberg GA. Membrane transport proteins with complete replacement of transmembrane helices with polyalanine sequences remain functional. J Biol Chem. 2005;280:8647–8650. doi: 10.1074/jbc.M413536200. [DOI] [PubMed] [Google Scholar]
- Bao X, Chen Y, Reuss L, Altenberg GA. Functional expression in Xenopus oocytes of gap-junctional hemichannels formed by a cysteine-less connexin 43. J Biol Chem. 2004b;279:9689–9692. doi: 10.1074/jbc.M311438200. [DOI] [PubMed] [Google Scholar]
- Bao X, Reuss L, Altenberg GA. Regulation of purified and reconstituted Connexin 43 hemichannels by protein kinase C-mediated phosphorylation of serine 368. J Biol Chem. 2004c;279:20058–20066. doi: 10.1074/jbc.M311137200. [DOI] [PubMed] [Google Scholar]
- Beahm DL, Oshima A, Gaietta GM, Hand GM, Smock AE, Zucker SN, Toloue MM, Chandrasekhar A, Nicholson BJ, Sosinsky GE. Mutation of a conserved threonine in the third transmembrane helix of α- and β-connexins creates a dominant-negative closed gap junction channel. J Biol Chem. 2006;281:7994–8009. doi: 10.1074/jbc.M506533200. [DOI] [PubMed] [Google Scholar]
- Beyer EC, Paul DL, Goodenough DA. Connexin43: A protein from rat heart homologous to a gap junction protein from liver. J Cell Biol. 1987;105:2621–2629. doi: 10.1083/jcb.105.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone R, White TW, Paul DL. Connections with connexins: the molecular basis of direct intercellular signaling. Eur J Biochem. 1996;238:1–27. doi: 10.1111/j.1432-1033.1996.0001q.x. [DOI] [PubMed] [Google Scholar]
- Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- Duffy HS, Sorgen PL, Girvin ME, O’Donnell P, Coombs W, Taffet SM, Delmar M, Spray DC. pH-dependent intramolecular binding and structure involving C×43 cytoplasmic domains. J Biol Chem. 2002;277:36706–36714. doi: 10.1074/jbc.M207016200. [DOI] [PubMed] [Google Scholar]
- Eilers M, Shekar SC, Shieh T, Smith SO, Fleming PJ. Internal packing of helical membrane proteins. Proc Natl Acad Sci USA. 2000;97:5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek-Vitorín JF, Calero G, Morley GE, Coombs W, Taffet SM, Delmar M. pH regulation of connexin43: Molecular analysis of the gating particle. Biophys J. 1996;71:1273–1284. doi: 10.1016/S0006-3495(96)79328-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman DM, Chen Y, Chin CN, Curran AR, Dixon AM, Dupuy AD, Lee AS, Lehnert U, Matthews EE, Reshetnyak YK, Senes A, Popot JL. Membrane protein folding: beyond the two stage model. FEBS Letters. 2003;555:122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- Eskandari S, Zampighi GA, Leung DW, Wright EM, Loo DDF. Inhibition of gap junction hemichannels by chloride channel blockers. J Membrane Biol. 2002;185:93–102. doi: 10.1007/s00232-001-0115-0. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, Sabag AD, Ophir E, Avraham KB, Ben-Tal N. The structural context of disease-causing mutations in gap junctions. J Biol Chem. 2006;281:28958–28963. doi: 10.1074/jbc.M605764200. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, Unger VM, Yeager M, Ben-Tal N. A Cα model for the transmembrane α helices of gap junction intercellular channels. Mol Cell. 2004;15:879–888. doi: 10.1016/j.molcel.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Foote CI, Zhou L, Zhu X, Nicholson BJ. The pattern of disulfide linkages in the extracellular loop regions of connexin 32 suggests a model for the docking interface of gap junctions. J Cell Biol. 1998;140:1187–1197. doi: 10.1083/jcb.140.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frillingos S, Sahin-Tóth M, Wu J, Kaback HR. Cys-scanning mutagenesis: a novel approach to structure-function relationships in polytopic membrane proteins. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- Gerido DA, White TW. Connexin disorders of the ear, skin, and lens. Biochim Biophys Acta. 2004;1662:159–170. doi: 10.1016/j.bbamem.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Gómez-Hernández JM, de Miguel M, Larrosa B, González D, Barrio LC. Molecular basis of calcium regulation in connexin-32 hemichannels. Proc Natl Acad Sci USA. 2003;100:16030–16035. doi: 10.1073/pnas.2530348100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AL. Emerging issues of connexin channels: biophysics fills the gap. Quart Rev Biophys. 2001;34:325–472. doi: 10.1017/s0033583501003705. [DOI] [PubMed] [Google Scholar]
- Javadpour MM, Eilers M, Groesbeek M, Smith SO. Helix packing in polytopic membrane proteins: Role of glycine in transmembrane helix association. Biophys J. 1999;77:1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Vakser IAA. Shorter side chains optimize helix-helix packing. Protein Sci. 2004;13:1426–1429. doi: 10.1110/ps.03505804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback HR, Sahin-Tóth M, Weinglass AB. The kamikaze approach to membrane transport. Nature Rev Mol Cell Biol. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- Kanemitsu MY, Loo LWM, Simon S, Lau AF, Eckhart W. Tyrosine phosphorylation of connexin 43 by v-Src is mediated by SH2 and SH3 domain interactions. J Biol Chem. 1997;272:22824–22831. doi: 10.1074/jbc.272.36.22824. [DOI] [PubMed] [Google Scholar]
- Kronengold J, Trexler EB, Bukauskas FF, Bargiello TA, Verselis VK. Single-channel SCAM identifies pore-lining residues in the first extracellular loop and first transmembrane domains of C×46 hemichannels. J Gen Physiol. 2003;122:389–405. doi: 10.1085/jgp.200308861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar NM, Gilula NB. Cloning and characterization of human and rat liver cDNAs coding for a gap junction protein. J Cell Biol. 1986;103:767–776. doi: 10.1083/jcb.103.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar NM, Gilula NB. The gap junction communication channel. Cell. 1996;84:381–388. doi: 10.1016/s0092-8674(00)81282-9. [DOI] [PubMed] [Google Scholar]
- Lee AG. Ca2+-ATPase structure in the E1 and E2 conformations: mechanism, helix-helix and helix-lipid interactions. Biochim Biophys Acta. 2002;1565:246–266. doi: 10.1016/s0005-2736(02)00573-4. [DOI] [PubMed] [Google Scholar]
- Loewenstein WR. Junctional intercellular communication: the cell-to-cell membrane channel. Physiol Rev. 1981;61:829–913. doi: 10.1152/physrev.1981.61.4.829. [DOI] [PubMed] [Google Scholar]
- Loo LWM, Berestecky JM, Kanemitsu MY, Lau AF. pp60src-mediated phosphorylation of connexin 43, a gap junction protein. J Biol Chem. 1995;270:12751–12761. doi: 10.1074/jbc.270.21.12751. [DOI] [PubMed] [Google Scholar]
- Lu ZL, Saldanha JW, Hulme EC. Transmembrane domains 4 and 7 of the M1 muscarinic acetylcholine receptor are critical for ligand binding and the receptor activation switch. J Biol Chem. 2001;276:34098–34104. doi: 10.1074/jbc.M104217200. [DOI] [PubMed] [Google Scholar]
- Milks LC, Kumar NM, Houghten R, Unwin N, Gilula NB. Topology of the 32-kD liver gap junction protein determined by site-directed antibody localizations. EMBO J. 1988;7:2967–2975. doi: 10.1002/j.1460-2075.1988.tb03159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JS, Kennedy RJ, Kemp DS. Short, solubilized polyalanines are conformational chameleons: Exceptionally helical if N- and C-capped with helix stabilizers, weakly to moderately helical if capped with rigid spacers. Biochemistry. 2001;40:305–309. doi: 10.1021/bi0019500. [DOI] [PubMed] [Google Scholar]
- Miller JS, Kennedy RJ, Kemp DS. Solubilized, spaced polyalanines: A context-free system for determining amino acid α-helix propensities. J Amer Chem Soc. 2002;124:945–962. doi: 10.1021/ja011726d. [DOI] [PubMed] [Google Scholar]
- Morley GE, Taffet SM, Delmar M. Intramolecular interactions mediate pH regulation of connexin43 channels. Biophys J. 1996;70:1294–1302. doi: 10.1016/S0006-3495(96)79686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller DJ, Hand GM, Engel A, Sosinsky GE. Conformational changes in surface structures of isolated connexin 26 gap junctions. EMBO J. 2002;21:3598–3607. doi: 10.1093/emboj/cdf365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio S, Schuldiner S. Characterization of an archaeal multidrug transporter with a unique amino acid composition. J Biol Chem. 2003;278:12000–12005. doi: 10.1074/jbc.M213119200. [DOI] [PubMed] [Google Scholar]
- Oh S, Rivkin S, Tang Q, Verselis VK, Bargiello TA. Determinants of gating polarity of a connexin 32 hemichannel. Biophys J. 2004;87:912–928. doi: 10.1529/biophysj.103.038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima A, Doi T, Mitsuoka K, Maeda S, Fujiyoshi Y. Roles of Met-34, Cys-64, and Arg-75 in the assembly of human connexin 26. J Biol Chem. 2003;278:1807–1816. doi: 10.1074/jbc.M207713200. [DOI] [PubMed] [Google Scholar]
- Perkins GA, Goodenough DA, Sosinsky GE. Formation of the gap junction intercellular channel requires a 30° rotation for interdigitating two apposing connexons. J Mol Biol. 1998;277:171–177. doi: 10.1006/jmbi.1997.1580. [DOI] [PubMed] [Google Scholar]
- Pfahnl A, Dahl G. Gating of C×46 gap junction hemichannels by calcium and voltage. Pflügers Archiv – Eur J Physiol. 1999;437:345–353. doi: 10.1007/s004240050788. [DOI] [PubMed] [Google Scholar]
- Phelan P, Stebbings LA, Baines RA, Bacon JP, Davies JA, Ford C. Drosophila shaking-B protein forms gap junctions in paired Xenopus oocytes. Nature. 1998;391:181–184. doi: 10.1038/34426. [DOI] [PubMed] [Google Scholar]
- Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Ann Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- Purnick PEM, Benjamin DC, Verselis VK, Bargiello TA, Dowd TL. Structure of the amino terminus of a gap junction protein. Arch Biochem Biophys. 2000;381:181–190. doi: 10.1006/abbi.2000.1989. [DOI] [PubMed] [Google Scholar]
- Raman P, Cherezov V, Caffrey M. The membrane protein data bank. Cell Mol Life Sci. 2006;63:36–51. doi: 10.1007/s00018-005-5350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer T. General and specific porins from bacterial outer membranes. J Struct Biol. 1998;121:101–109. doi: 10.1006/jsbi.1997.3946. [DOI] [PubMed] [Google Scholar]
- Simoes M, Garneau L, Klein H, Banderali U, Hobeila F, Roux B, Parent L, Sauvé R. Cysteine mutagenesis and computer modeling of the S6 region of an intermediate conductance IKCa channel. J Gen Physiol. 2002;120:99–116. doi: 10.1085/jgp.20028586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Solan JL, Taffet SM, Javier R, Lampe PD. Connexin 43 interacts with Zona Occludens-1 and -2 proteins in a cell cycle stage-specific manner. J Biol Chem. 2005;280:30416–30421. doi: 10.1074/jbc.M506799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerrett IM, Aronowitz J, Shin JH, Cymes G, Kasperek E, Cao FL, Nicholson BJ. Identification of amino acid residues lining the pore of a gap junction channel. J Cell Biol. 2002;159:349–359. doi: 10.1083/jcb.200207060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgen PL, Duffy HS, Sahoo P, Coombs W, Delmar M, Spray DC. Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and Zonula Occludens-1. J Biol Chem. 2004;279:54695–54701. doi: 10.1074/jbc.M409552200. [DOI] [PubMed] [Google Scholar]
- Sosinsky GE, Nicholson BJ. Structural organization of gap junction channels. Biochim Biophys Acta. 2005;1711:99–125. doi: 10.1016/j.bbamem.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Unger VM, Kumar NM, Gilula NB, Yeager M. Projection structure of a gap junction membrane channel at 7 Å resolution. Nature Struct Biol. 1997;4:39–43. doi: 10.1038/nsb0197-39. [DOI] [PubMed] [Google Scholar]
- Unger VM, Kumar NM, Gilula NB, Yeager M. Three-dimensional structure of a recombinant gap junction membrane channel. Science. 1999;283:1176–1180. doi: 10.1126/science.283.5405.1176. [DOI] [PubMed] [Google Scholar]
- Unwin PNT, Ennis PD. Two configurations of a channel-forming membrane protein. Nature. 1984;307:609–613. doi: 10.1038/307609a0. [DOI] [PubMed] [Google Scholar]
- Uveges AJ, Kowal D, Zhang Y, Spangler TB, Dunlop J, Semus S, Jones PG. The role of transmembrane helix 5 in agonist binding to the human H3 receptor. J Pharm Experimental Therapeutics. 2002;301:451–458. doi: 10.1124/jpet.301.2.451. [DOI] [PubMed] [Google Scholar]
- Verselis VK, Ginter CS, Bargiello TA. Opposite voltage gating polarities of two closely related connexins. Nature. 1994;368:348–351. doi: 10.1038/368348a0. [DOI] [PubMed] [Google Scholar]
- Warn-Cramer BJ, Lampe PD, Kurata WE, Kanemitsu MY, Loo LWM, Eckhart W, Lau AF. Characterization of the mitogen-activated protein kinase phosphorylation sites on the connexin-43 gap junction protein. J Biol Chem. 1996;271:3779–3786. doi: 10.1074/jbc.271.7.3779. [DOI] [PubMed] [Google Scholar]
- Weber PA, Chang HC, Spaeth KE, Nitsche JM, Nicholson BJ. The permeability of gap junction channels to probes of different size is dependent on connexin composition and permeant-pore affinities. Biophys J. 2004;87:958–973. doi: 10.1529/biophysj.103.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TW, Bruzzone R, Wolfram S, Paul DL, Goodenough DA. Selective interactions among the multiple connexin proteins expressed in the vertebrate lens: the second extracellular domain is a determinant of compatibility between connexins. J Cell Biol. 1994;125:879–892. doi: 10.1083/jcb.125.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TW, Wang H, Mui R, Litteral J, Brink PR. Cloning and functional expression of invertebrate connexins from Halocynthia pyriformis. FEBS Letters. 2004;577:42–48. doi: 10.1016/j.febslet.2004.09.071. [DOI] [PubMed] [Google Scholar]
- Yancey SB, John SA, Lal R, Austin BJ, Revel JP. The 43-kD polypeptide of heart gap junctions: Immunolocalization (I), topology (II), and functional domains (III) J Cell Biol. 1989;108:2241–2254. doi: 10.1083/jcb.108.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M, Gilula NB. Membrane topology and quaternary structure of cardiac gap junction ion channels. J Mol Biol. 1992;223:929–948. doi: 10.1016/0022-2836(92)90253-g. [DOI] [PubMed] [Google Scholar]
- Yeager M, Nicholson BJ. Structure and biochemistry of gap junctions. In: Hertzberg E, editor. Gap Junctions. Vol. 30 of Advances in Molecular and Cell Biology. JAI Press; Stamford, CT: 2000. pp. 31–98. [Google Scholar]
- Zhou L, Kasperek EM, Nicholson BJ. Dissection of the molecular basis of pp60v-src induced gating of connexin 43 gap junction channels. J Cell Biol. 1999;144:1033–1045. doi: 10.1083/jcb.144.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XW, Pfahnl A, Werner R, Hudder A, Llanes A, Luebke A, Dahl G. Identification of a pore lining segment in gap junction hemichannels. Biophys J. 1997;72:1946–1953. doi: 10.1016/S0006-3495(97)78840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]