Abstract
Lignin is the second most abundant plant biopolymer mainly present in the secondary walls of tracheary elements and fibers in wood. Understanding how lignin is biosynthesized has long been an interest to plant biologists and will have a significant impact on tree biotechnology. Lignin is polymerized from monolignols that are synthesized through the lignin biosynthetic pathway. To make lignin, all the genes in the lignin biosynthetic pathway need to be coordinately turned on. It has been shown that a common cis-element, namely the AC element, is present in the majority of the lignin biosynthetic genes and required for their expression in lignifying cells. Important progress has been made in the identification of transcription factors that bind to the AC elements and are potentially involved in the coordinated regulation of lignin biosynthesis. The Arabidopsis MYB58 and MYB63 as well as their poplar ortholog PtrMYB28 are transcriptional activators of the lignin biosynthetic pathway, whereas the eucalyptus egMYB2 and pine PtMYB4 transcription factors are likely Arabidopsis MYB46 orthologs involved in the regulation of the entire secondary wall biosynthetic program. It was found that the transcriptional regulation of lignin biosynthesis is under the control of the same transcriptional network regulating the biosynthesis of other secondary wall components, including cellulose and xylan. The identification of transcription factors directly activating lignin biosynthetic genes provides unprecedented tools to potentially manipulate the amount of lignin in wood and other plant products based on our needs.
Key words: lignin, lignin biosynthesis, MYB, secondary wall biosynthesis, transcription factor, transcriptional regulation
Vascular plants appeared on earth during the Silurian period around 430 million years ago and one of the features important for their conquest on dry land is the evolution of vascular tissues that solved the problem of fluid transport throughout the plant body.1 Accompanying the evolution of vascular tissues was the evolution of the ability for vascular plants to synthesize lignin, which provides mechanical strength and hydrophobicity to the water-conducting tracheary elements. The importance of lignin in the functioning of tracheary elements has been unequivocally proven in transgenic plants and mutants with defective lignin deposition showing the deformation of vessels due to the inability of the weakened walls to resist the negative pressure generated during transpiration.2–4 The lignin polymer is mainly deposited in the secondary walls of tracheary elements and fibers, the principal cell types of wood, and its impregnation into the cellulose and hemicellulose network strengthens the secondary walls as well as renders them waterproof due to its hydrophobicity. The lignin polymer is very inert, which confers stable and protective coatings to protect the secondary walls from physical and biological attacks. The protective function of lignin and other phenolic compounds contribute to the resistance of decay of wood; wood in some living bristlecone pine trees has been dated back to ∼4,800 years ago.1 Although lignin is mainly deposited in tracheary elements and fibers, it is also found in other cell types or tissues, such as the endodermis, periderm and epidermis of some plant species, to provide rigidity and impermeability. In addition to the developmental regulation of its deposition, the lignin polymer accumulates in response to many environmental stresses, such as wounding, ultraviolet light irradiation and pathogen attack, which may constitute the first line of defense against biotic and abiotic damages.5
Although lignin is essential for normal plant growth and development, its presence in plant tissues may hinder the uses of plant products by humans. For example, wood is widely used for paper-making, but lignin has to be chemically removed during the pulping process. Chemical removal of lignin not only increases the cost of pulping but also leads to production of chemical wastes that cause severe adverse effect on the environment.5 Lignocellulosic biomass has also recently been considered to be an alternative source of biofuel production. One of the main obstacles in the conversion of lignocellulose into glucose for bioethanol production is the presence of lignin that impedes the enzymatic digestion of cellulose.6 In addition, lignin present in animal forage decreases digestibility. Due to its importance in plant survival and its relevance to human uses of plant products, lignin has been subjected to intensive studies for its biosynthesis and function in the hope of engineering its content and composition in trees and forage crops based on our needs. Currently, the biosynthetic pathway of lignin has been well defined and a number of recent reviews cover the topic of lignin biosynthesis and its genetic modification.7,8 In contrast, our understanding on the transcriptional regulation of the lignin biosynthetic pathway is still limited. This review focuses on recent molecular and genetic dissection of transcription factors involved in the coordinated activation of lignin biosynthetic genes.
Lignin Structure and Biosynthesis
Lignin is a complex phenylpropanoid polymer formed through dehydrogenative polymerization of three monolignols, including p-coumaryl alcohol, coniferyl alcohol and sinapyl alcohol.5 Lignin from gymnosperms is composed of guaiacyl unit polymerized from coniferyl alcohol, whereas that from angiosperms typically consists of both guaiacyl and syringyl units polymerized from coniferyl alcohol and sinapyl alcohol, respectively. Lignin from grasses also contains p-hydroxyphenyl unit polymerized from p-coumaryl alcohol in addition to guaiacyl and syringyl units. The biosynthetic pathway of monolignols starts with the general phenylpropanoid pathway leading to the production of hydroxycinnamoyl CoA esters, which are the common precursors of diverse groups of chemical compounds, such as flavonoids, suberin, coumarins, quinones, phytoalexins, acetosyringone and lignin. For production of monolignols, two successive reductive steps catalyzed by cinnamoyl CoA reductase and cinnamyl alcohol dehydrogenase convert hydroxycinnamoyl CoA esters into the three monolignols. Monolignols are then transported, via an unknown mechanism, into the cell wall, where they are oxidized by oxidases such as peroxidases and laccases for polymerization.5
To make lignin, it is conceivable that all the genes involved in the biosynthesis, transport and oxidation of monolignols need to be coordinately expressed. Dissection of the molecular switches controlling the coordinated activation of lignin biosynthetic genes is of importance in understanding the molecular mechanisms underlying tissue-specific deposition of lignin. Currently, little is known about the signals and the master transcription factors that coordinate the activation of the entire lignin biosynthetic pathway. Identification of the molecular switches controlling lignin biosynthesis could allow us to alter the lignin biosynthetic program and thereby genetically engineer trees and biomass crops with altered lignin content.
Cis-Elements Involved in the Activation of Lignin Biosynthetic Genes
Genes involved in the biosynthesis of monolignols have all been identified and characterized in the past three decades.7,8 Promoter deletion-GUS reporter gene analysis has revealed specific regions in the promoters of lignin biosynthetic genes responsible for their expression in lignifying tissues.9–15 Among the lignin gene promoters analyzed, the promoter of phenylalanine ammonia lyase (PAL) that catalyzes the first step of phenylpropanoid pathway has been best characterized. It was shown that multiple regions of the bean PAL2 promoter were able to drive the GUS reporter gene expression in lignifying xylem tissues in transgenic tobacco and that another region was involved in suppression of expression in phloem.11 A similar organization of multiple regions responsible for xylem-specific expression is also present in the promoter of the parsley 4-coumarate CoA ligase-1 gene.10 Detailed analyses of the PAL2 promoter by footprinting and electrophoretic mobility shift assay identified three AC elements together with a G-box involved in xylem-specific expression.16 Mutations of either the AC-I (ACCTACC) or AC-II (ACCAACC) element resulted in a decrease of xylem-associated expression but a gain of phloem-associated expression, whereas simultaneous mutations of both the AC-I and AC-II elements led to a complete loss of xylem-specific expression. Mutation of the AC-III (ACCTAAC) element caused a slight decrease of xylem-associated expression. It was concluded that the combined activity of AC elements determines xylem-specific expression.16 The AC elements, also known as PAL-box and H-box, were first identified in the promoter of parsley PAL1 as one of the two motifs involved in the response to UV light and elicitor treatments.17 The hypothesis that AC elements are required and sufficient for directing xylem-specific expression was further supported by the finding that the AC-II heptamer linked with the cauliflower mosaic virus 35S minimal promoter was able to drive the specific expression of the GUS reporter gene in xylem.18
The discovery that the AC elements specify the expression of lignin biosynthetic genes in lignifying cells was a breakthrough in understanding how lignin biosynthesis is coordinately regulated. Bioinformatic analysis of the promoters of all the lignin biosynthetic genes in Arabidopsis identified the presence of the AC elements in the majority of them.19 The only exceptions are the promoters of cinnamate 4-hydroxylase, ferulate 5-hydroxylase and caffeic acid-O-methyltransferase, which do not have apparent AC elements. However, it was suggested that these genes might have more degenerative AC elements that may not be picked up by the bioinformatic analysis19 or have AC elements present in other regions of the genes instead of their promoters. Together, these analyses indicate that AC elements serve as common cis regulatory elements driving the coordinated expression of lignin biosynthetic genes in lignifying tissues.
Transcription Factors Regulating the Expression of Lignin Biosynthetic Genes
The AC element sequences uncovered in the lignin biosynthetic genes are similar to the binding site (CCT/AACC) identified through binding site selection for the maize MYB protein P.20 In addition, the Antirrhinum MYB305 was found to be able to bind to the AC elements and activate the expression of the AC element-containing bean PAL2 promoter-driven reporter gene.21,22 Therefore, it was reasoned that transcription factors that bind to the AC elements in lignin biosynthetic genes are also MYB proteins. The first line of genetic evidence on the possible involvement of MYBs in the regulation of lignin biosynthesis came from the study of two MYB proteins, AmMYB308 and AmMYB330, from Antirrhinum (Table 1).23 Overexpression of the Antirrhinum MYB proteins in transgenic tobacco plants caused a reduction in the expression of several lignin biosynthetic genes and a decrease in lignin content, suggesting that the Antirrhinum MYBs are able to regulate the expression of lignin biosynthetic genes and thereby affect lignin biosynthesis. Since then, several MYBs from Arabidopsis24 and grapes25 have been shown to alter the expression of phenylpropanoid biosynthetic genes and lignin biosynthesis when overexpressed. However, none of these MYBs have been proven to bind to the AC elements, nor have they been demonstrated to be expressed in lignifying tissues. The latter is especially important because developmental regulators of lignin biosynthetic genes should be expressed in cells undergoing lignification. Thus, it is uncertain whether these MYBs are indeed regulators of lignin biosynthesis or their effects on lignin biosynthesis observed in the overexpressors are indirect. In fact, one of these MYBs, PAP1, has been demonstrated to be a regulator of anthocyanin biosynthesis, and the increased accumulation of lignin caused by PAP1 overexpression is due to the elevation of the common hydroxycinnamoyl CoA esters shared by the biosynthetic pathways of both lignin and anthocyanin.24
Table 1.
MYB transcription factors affecting lignin biosynthesis
| Genes | Tissue level expression pattern | Bind to AC elements | Overexpression phenotypes | Downregulation phenotypes | Reference |
| Antirrhinum | N/D | N/D | Reduced lignin | N/D | 23 |
| AmMYB308 | |||||
| AmMYB330 | |||||
| Arabidopsis PAP1 | N/D | N/D | Increased lignin | N/D | 24 |
| Maize | Lignifying tissues | N/D | Reduced lignin | N/D | 50 |
| ZmMYB31 | |||||
| ZmMYB42 | |||||
| Pine PtMYB4 | Secondary xylem | Yes | Ectopic lignin deposition | N/D | 26 |
| Pine | Secondary xylem | N/D | Ectopic lignin deposition | N/D | 29 |
| PtMYB1 | |||||
| PtMYB8 | |||||
| Eucalyptus EgMYB2 | Secondary xylem | Yes | Increased wall thickening and Lignin metabolism | N/D | 28 |
| Arabidopsis | Lignifying tissues | Yes | Ectopic lignin deposition | Reduced wall thickening and lignin deposition | 34 |
| MYB58 | |||||
| MYB63 | |||||
| Arabidopsis MYB85 | Lignifying tissues | N/D | Ectopic lignin deposition | Reduced wall thickening | 35 |
N/D, not determined.
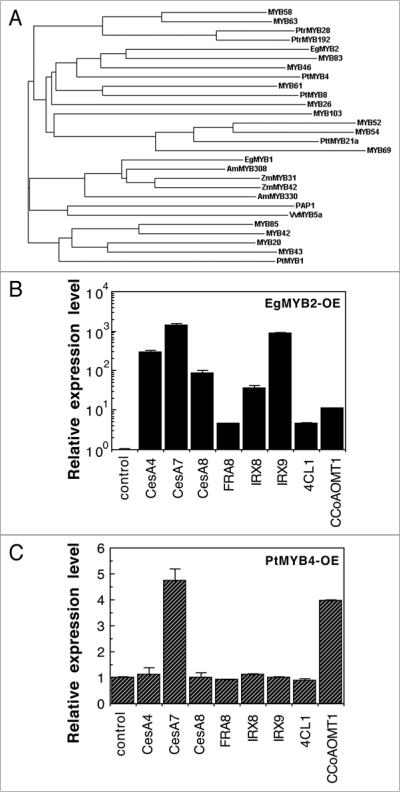
Several MYBs from pine and eucalyptus have been shown to be strong candidates as regulators of lignin biosynthesis (Table 1). The pine PtMYB1 and PtMYB426,27 and the eucalyptus EgMYB228 bind to the AC elements and their genes are expressed in developing wood that undergoes secondary wall thickening and lignin biosynthesis. When overexpressed in tobacco plants, PtMYB4 and EgMYB2 induced the expression of some of the lignin biosynthetic genes and led to ectopic lignin deposition or increased secondary wall thickening. In addition, the wood-associated pine PtMYB1 and PtMYB8 also caused ectopic lignin deposition and wall thickening when overexpressed in spruce.29 PtMYB8 is a close homolog of the Arabidopsis MYB61 (Fig. 1A) whose overexpression could cause ectopic lignin deposition but its exact functions remain to be studied.30–32 It was concluded that these pine and eucalyptus MYBs are involved in regulation of lignin biosynthesis during wood formation. However, PtMYB4 and EgMYB2 are phylogenetically grouped together with the Arabidopsis MYB46 (Fig. 1A), which has been shown to be a key regulator of the biosynthesis of all the three major secondary wall components, including cellulose, xylan and lignin.33 Overexpression of EgMYB2 in Arabidopsis protoplasts was found to be able to activate the expression of the biosynthetic genes of cellulose, xylan and lignin (Fig. 1B). Overexpression of PtMYB4 could induce the expression of a cellulose synthase gene and a lignin biosynthetic gene (Fig. 1C). These findings indicate that EgMYB2 and perhaps also PtMYB4 are orthologs of MYB46 and they regulate the entire secondary wall biosynthetic program during wood formation.
Figure 1.
(A) Phylogenetic analysis of MYB genes that are involved in regulation of secondary wall biosynthesis and/or lignification. The phylogenetic relationship of MYBs was analyzed with ClustalW.54 (B and C) Activation of secondary wall biosynthetic genes by overexpression of eucalyptus EgMYB2 and pine PtMYB4. EgMYB2 or PtMYB4 cDNAs driven by the cauliflower mosaic virus 35S promoter were transfected into Arabidopsis protoplasts.35 rNAs from the transfected protoplasts were used for quantitative PCR analysis of gene expression.
Studies of secondary wall-associated transcription factors in Arabidopsis led to the identification of two MYBs, MYB58 and MYB63, that are transcriptional activators of lignin biosynthetic genes.34 MYB58 and MYB63 are specifically expressed in cells undergoing lignification. Their overexpression was found to induce ectopic deposition of lignin but not cellulose and xylan, whereas their dominant repression resulted in a reduction in secondary wall thickening and lignin deposition, indicating that MYB58 and MYB63 are specifically involved in the regulation of lignin biosynthesis. The hypothesis that MYB58 and MYB63 are transcriptional activators of lignin biosynthesis was further supported by the findings that they bind to the AC elements and directly activate the expression of lignin biosynthetic genes. In addition to MYB58 and MYB63, another MYB transcription factor, MYB85, was also found to activate lignin biosynthetic genes and cause ectopic lignin deposition when overexpressed.35
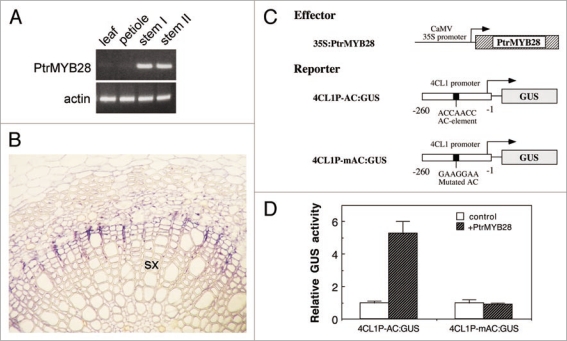
Orthologs of MYB58 and MYB63 are present in tree species, such as poplar (Fig. 1A). One of these orthologs, the poplar PtrMYB28, is predominantly expressed in developing wood undergoing secondary wall thickening and lignification (Fig. 2A and B). Expression of PtrMYB28 in Arabidopsis protoplasts effectively induced the expression of the lignin biosynthetic gene 4CL1 and this induction was dependent on the presence of the AC element (Fig. 2C and D), indicating that the mechanisms underlying the transcriptional regulation of lignin biosynthesis are conserved between herbaceous Arabidopsis and tree species.
Figure 2.
Expression and transactivation analyses of Poplar PtrMYB28. (A) Reverse transcription PCR analysis of expression of PtrMYB28 in different organs. Stem I and Stem II are from elongating and non-elongating stems, respectively. (B) In situ mRNA hybridization showing expression of PtrMYB28 in the developing secondary xylem (sx) in a poplar (Populus tricocarpa) stem. (C) Diagram of the effector and reporter constructs used for transactivation analysis. The native or mutated AC element in the 4CL1 promoter is marked. (D) Transactivation analysis in Arabidopsis protoplasts showing that PtrMYB28 activates the expression of the GUS reporter gene driven by the native Arabidopsis 4CL1 promoter but not by the 4CL1 promoter with the mutated AC element.
Besides MYBs, two transcription factors, NtLIM1 and ACBF, belonging to other families have been isolated based on their ability to bind to the AC elements.18,36 The tobacco NtLIM1 protein shows sequence similarity to members of the LIM protein family and is able to activate the AC element-driven GUS reporter gene expression in tobacco protoplasts.36 Antisense inhibition of NtLIM1 expression in transgenic tobacco plants caused a reduction in lignin content in stems, indicating that it is required for normal lignin biosynthesis. However, it remains to be determined whether NtLIM1 directly regulates the expression of lignin biosynthetic genes. The other AC element-binding transcription factor, ACBF, has not been investigated for its possible role in regulation of lignin biosynthesis.18
Transcriptional Network Regulating Secondary Wall Biosynthesis and Lignification
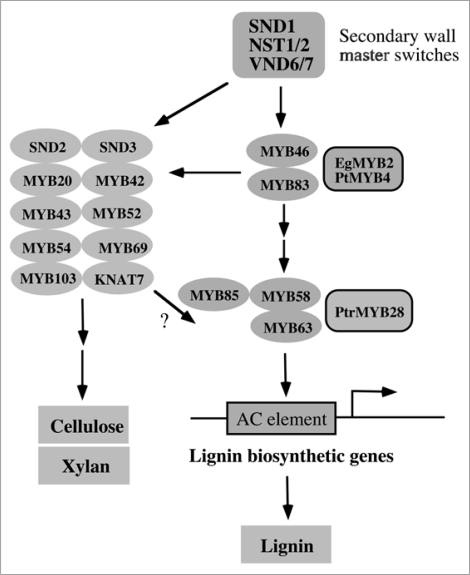
Recent studies on the transcriptional regulation of secondary wall biosynthesis in Arabidopsis have revealed that the transcriptional regulation of lignin biosynthesis is under the control of the common transcriptional network activating the entire secondary wall biosynthetic program (Fig. 3). In this network, the NAC domain transcription factors, SND1 and its close homologs, NST1, NST2, VND6 and VND7, act as master switches leading to the activation of the biosynthetic genes of cellulose, xylan and lignin.35,37–44 Simultaneous knockout of SND1 and NST1 completely blocks secondary wall thickening and lignin deposition in fibers,38,41,42 indicating that the biosynthesis of lignin together with cellulose and xylan is under the transcriptional control of the same master switches SND1 and NST1. SND1 and its homologs regulate a number of downstream transcription factors involved in secondary wall biosynthesis.35 Among them, MYB46, SND3, MYB103 and KNAT7 were found to be SND1 direct targets, and MYB46 acts as another level of master switch able to activate the entire secondary wall biosynthetic program.33 Therefore, SND1 and MYB46 function at the top of the transcriptional network leading to regulation of biosynthesis of lignin together with cellulose and xylan. The finding that the direct transcriptional activators of lignin biosynthesis, MYB58 and MYB63, are downstream targets of SND1 and MYB46,34 further indicates that transcription factors directly regulating lignin biosynthesis are part of the SND1- and MYB46-mediated transcriptional network regulating secondary wall biosynthesis.
Figure 3.
Diagram of the transcriptional network regulating secondary wall biosynthesis and lignification. In this network, SND1 and its close homologs are master switches of the secondary wall biosynthetic program. MYB46 and its tree orthologs EgMYB2 and PtMYB4 function downstream of the NAC master switches. MYB58 and MYB63 as well as poplar PtrMYB28 directly activate the expression of lignin biosynthetic genes through binding to the AC elements.
Concluding Remarks
Tremendous progress has been made in the last two decades regarding the transcriptional regulation of the biosynthesis of lignin, the second most abundant biopolymer produced by vascular plants. It is established that the AC elements present in the promoters of the lignin biosynthetic genes serve as a common regulatory element driving their expression, and that specific MYB transcription factors bind to the AC elements and thereby regulate the coordinated expression of lignin biosynthetic genes. Recent studies have also provided evidence that the transcriptional regulation of lignin biosynthesis is under the control of the same transcriptional network regulating the biosynthesis of other secondary wall components. Despite these progresses, many outstanding issues regarding the transcriptional regulation of lignin biosynthesis still remain. First, the transcriptional activation of lignin biosynthetic genes is likely mediated through multiple cis-elements and a combinatorial interaction of multiple transcription factors, a scenario similar to the transcriptional regulation of the branch of the phenylpropanoid pathway leading to flavonoid biosynthesis.45 Earlier studies of the promoter activities of lignin biosynthetic genes have indicated the presence of cis elements besides the AC elements and of multiple proteins binding to the promoter sequences.9,12,16,21 In addition, some MYBs have been suggested to function as repressors in fine-tuning the expression level of phenylpropanoid biosynthetic genes.46–50 Identification and characterization of all the cis elements and transcription factors involved in regulation of lignin biosynthesis will be essential for gaining a full picture of the complexity of transcriptional control of lignin biosynthesis. Second, little is known about how lignin heterogeneity is regulated at the transcription level. For example, it is well recognized that the lignin composition varies among different cell types, such as vessels and fibers, within the same plant species, which attributes to the differential expression of genes committed to the biosynthesis of sinapyl alcohol.19 Understanding of the transcriptional regulation of sinapyl alcohol biosynthetic genes will contribute to uncovering the mechanisms underlying the regulation of lignin heterogeneity. Third, lignin biosynthesis is not only developmentally regulated but also induced in response to many environmental stresses, such as wounding, UV light irradiation and pathogen attacks, but little is known about transcriptional activation of stress-induced lignin biosynthesis. Because these environmental stresses typically do not induce secondary wall thickening, it is likely that the transcriptional regulation of stress-induced lignin biosynthesis is different from that of the developmentally activated lignin biosynthesis. Early promoter deletion studies have identified specific regions in the promoters of lignin biosynthetic genes responsible for wounding, UV irradiation and pathogen activations51 and suggest that the AC elements are implicated in response to environmental stresses.17,52 Further investigation on stress-specific cis elements and the corresponding transcription factors will be necessary to understand how stress signals are transduced to activate lignin biosynthesis. Finally, lignin constitutes 15–30% of the biomass of wood and understanding the transcriptional control of lignin biosynthesis during wood formation will have important implications in tree biotechnology. It will be possible to use one or a few transcription factors to down- or up-regulate the entire lignin biosynthetic pathway and thereby alter lignin content in wood based on our needs. Recently, it has been reported that conversion of biomass into electricity instead of to biofuel for automobile propulsion captures more biomass energy.53 Since lignin has higher energy density than other polysaccharides, it would be desirable to genetically engineer biomass crops with higher lignin content for conversion into electricity. With the availability of the ever-increasing molecular and genomic tools, it is expected that a complete picture of the transcriptional regulation of lignin biosynthesis will soon emerge.
Acknowledgements
Work in the authors' lab was supported by a grant from National Science Foundation.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9875
References
- 1.Raven PH, Evert RF, Eichhorn SE. Biology of plants. 6th ed. New York, NY: WH Freeman and Company; 1999. [Google Scholar]
- 2.Zhong R, Morrison WH, Negrel J, Ye Z-H. Dual methylation pathways in lignin biosynthesis. Plant Cell. 1998;10:2033–2046. doi: 10.1105/tpc.10.12.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones L, Ennos AR, Turner SR. Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J. 2001;26:205–216. doi: 10.1046/j.1365-313x.2001.01021.x. [DOI] [PubMed] [Google Scholar]
- 4.Goujon T, Ferret V, Mila I, Pollet B, Ruel K, Burlat V, et al. Downregulation of the AtCCR1 gene in Arabidopsis thaliana: effects on phenotype, lignins and cell wall degradability. Planta. 2003;217:218–228. doi: 10.1007/s00425-003-0987-6. [DOI] [PubMed] [Google Scholar]
- 5.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 6.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Weng JK, Chapple C. Improvement of biomass through lignin modification. Plant J. 2008;54:569–581. doi: 10.1111/j.1365-313X.2008.03457.x. [DOI] [PubMed] [Google Scholar]
- 8.Vanholme R, Morreel K, Ralph J, Boerjan W. Lignin engineering. Curr Opin Plant Biol. 2008;11:278–285. doi: 10.1016/j.pbi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Hauffe KD, Lee SP, Subramaniam R, Douglas CJ. Combinatorial interactions between positive and negative cis-acting elements control spatial patterns of 4CL-1 expression in transgenic tobacco. Plant J. 1993;4:235–253. doi: 10.1046/j.1365-313x.1993.04020235.x. [DOI] [PubMed] [Google Scholar]
- 10.Hauffe KD, Paszkowski U, Schulze-Lefert P, Hahlbrock K, Dangl JL, Douglas CJ. A parsley 4CL-1 promoter fragment specifies complex expression patterns in transgenic tobacco. Plant Cell. 1991;3:435–443. doi: 10.1105/tpc.3.5.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leyva A, Liang X, Pintor-Toro JA, Dixon RA, Lamb CJ. cis-element combinations determine phenylalanine ammonia-lyase gene tissue-specific expression patterns. Plant Cell. 1992;4:263–271. doi: 10.1105/tpc.4.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacombe E, Doorsselaere JV, Boerjan W, Boudet AM, Grima-Pettenati J. Characterization of cis-elements required for vascular expression of the Cinnamoyl CoA Reductase gene and for protein-DNA complex formation. Plant J. 2000;23:663–676. doi: 10.1046/j.1365-313x.2000.00838.x. [DOI] [PubMed] [Google Scholar]
- 13.Lauvergeat V, Rech P, Jauneau A, Guez C, Coutos-Thevenot P, Grima-Pettenati J. The vascular expression pattern directed by the Eucalyptous gunnii cinnamyl alcohol dehydrogenase EgCAD2 promoter is conserved among woody and herbaceous plant species. Plant Mol Biol. 2002;50:497–509. doi: 10.1023/a:1019817913604. [DOI] [PubMed] [Google Scholar]
- 14.Rogers LA, Campbell MM. The genetic control of lignin deposition during plant growth and development. New Phytol. 2004;164:17–30. doi: 10.1111/j.1469-8137.2004.01143.x. [DOI] [PubMed] [Google Scholar]
- 15.Soltani BM, Ehlting J, Hamberger B, Douglas CJ. Multiple cis-regulatory elements regulate distinct and complex patterns of developmental and woundinduced expression of Arabidopsis thaliana 4CL gene family members. Planta. 2006;224:1226–1238. doi: 10.1007/s00425-006-0296-y. [DOI] [PubMed] [Google Scholar]
- 16.Hatton D, Sablowski R, Yung M-H, Smith C, Schuch W, Bevan M. Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. Plant J. 1995;7:859–876. doi: 10.1046/j.1365-313x.1995.07060859.x. [DOI] [PubMed] [Google Scholar]
- 17.Lois R, Dietrich A, Hahlbrock K, Schulz W. A phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 1989;8:1641–1648. doi: 10.1002/j.1460-2075.1989.tb03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Séguin A, Laible G, Leyva A, Dixon RA, Lamb CJ. Characterization of a gene encoding a DNA-binding protein that interacts in vitro with vascular specific cis elements of the phenylalanine ammonia-lyase promoter. Plant Mol Biol. 1997;35:281–291. doi: 10.1023/a:1005853404242. [DOI] [PubMed] [Google Scholar]
- 19.Raes J, Rohde A, Christensen JH, Peer YV, Boerjan W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003;133:1051–1071. doi: 10.1104/pp.103.026484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grotewold E, Drummond BJ, Bowen B, Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 21.Sablowski RWM, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M. A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 1994;13:128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sablowski RW, Baulcombe DC, Bevan M. Expression of a flower-specific Myb protein in leaf cells using a viral vector causes ectopic activation of a target promoter. Proc Natl Acad Sci USA. 1995;92:6901–6905. doi: 10.1073/pnas.92.15.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell. 1998;10:135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, et al. Characterization of a grapevine R3R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 2006;140:499–511. doi: 10.1104/pp.105.067231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, et al. Characterisation of a pine MYB that regulates lignification. Plant J. 2003;36:743–754. doi: 10.1046/j.1365-313x.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- 27.Patzlaff A, Newman LJ, Dubos C, Whetten RW, Smith C, McInnis S, et al. Characterization of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol Biol. 2003;53:597–608. doi: 10.1023/B:PLAN.0000019066.07933.d6. [DOI] [PubMed] [Google Scholar]
- 28.Goicoechea M, Lacombe E, Legay S, Mihaljevic S, Rech P, Jauneau A, et al. EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 2005;43:553–567. doi: 10.1111/j.1365-313X.2005.02480.x. [DOI] [PubMed] [Google Scholar]
- 29.Bomal C, Bedon F, Caron S, Mansfield SD, Levasseur C, Cooke JE, et al. Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. J Exp Bot. 2008;59:3925–3939. doi: 10.1093/jxb/ern234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell. 2001;13:2777–2791. doi: 10.1105/tpc.010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman LJ, Perazza DE, Juda L, Campbell MM. Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 2004;37:239–250. doi: 10.1046/j.1365-313x.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- 32.Liang Y-K, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol. 2008;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 33.Zhong R, Richardson EA, Ye Z-H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell. 2007;19:2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Chan L, Zhong R, Ye Z-H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. A battery of transcription factors Involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H. Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J. 2000;22:289–301. doi: 10.1046/j.1365-313x.2000.00737.x. [DOI] [PubMed] [Google Scholar]
- 37.Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguch M, Ito J, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulates secondary wall thickening and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ko JH, Yang SH, Park AH, Lerouxel O, Han KH. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 2007;50:1035–1048. doi: 10.1111/j.1365-313X.2007.03109.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhong R, Demura T, Ye Z-H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong R, Richardson EA, Ye Z-H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta. 2007;225:1603–1611. doi: 10.1007/s00425-007-0498-y. [DOI] [PubMed] [Google Scholar]
- 43.Zhong R, Ye Z-H. Regulation of cell wall biosynthesis. Curr Opin Plant Biol. 2007;10:564–572. doi: 10.1016/j.pbi.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi M, Kubo M, Fukuda H, Demura T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008;55:652–664. doi: 10.1111/j.1365-313X.2008.03533.x. [DOI] [PubMed] [Google Scholar]
- 45.Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol. 2005;57:155–171. doi: 10.1007/s11103-004-6910-0. [DOI] [PubMed] [Google Scholar]
- 46.Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, et al. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karpinska B, Karlsson M, Srivastava M, Stenberg A, Schrader J, Sterky F, et al. MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Mol Biol. 2004;56:255–270. doi: 10.1007/s11103-004-3354-5. [DOI] [PubMed] [Google Scholar]
- 48.Fornalé S, Sonbol FM, Maes T, Capellades M, Puigdomínech P, Rigau J, et al. Downregulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol Biol. 2006;62:809–823. doi: 10.1007/s11103-006-9058-2. [DOI] [PubMed] [Google Scholar]
- 49.Legay S, Lacombe E, Goicoechea M, Briere C, Seguin A, Mackay J, et al. Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci. 2007;173:542–549. [Google Scholar]
- 50.Sonbol FM, Fornalé S, Capellades M, Encina A, Touriño S, Torres JL, et al. The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol Biol. 2009;70:283–296. doi: 10.1007/s11103-009-9473-2. [DOI] [PubMed] [Google Scholar]
- 51.Ohl S, Hedrick SA, Chory J, Lamb CJ. Functional properties of a phenylalanine ammonia-lyase promoter from Arabidopsis. Plant Cell. 1990;2:837–848. doi: 10.1105/tpc.2.9.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.da Costa e Silva O, Klein L, Schmelzer E, Trezzini GF, Hahlbrock K. BPF-1, a pathogen-induced DNA-binding protein involved in the plant defense response. Plant J. 1993;4:125–135. doi: 10.1046/j.1365-313x.1993.04010125.x. [DOI] [PubMed] [Google Scholar]
- 53.Ohlrogge J, Allen D, Berguson B, Dellapenna D, Shachar-Hill Y, Stymne S. Driving on biomass. Science. 2009;324:1019–1020. doi: 10.1126/science.1171740. [DOI] [PubMed] [Google Scholar]
- 54.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nuc Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]