Abstract
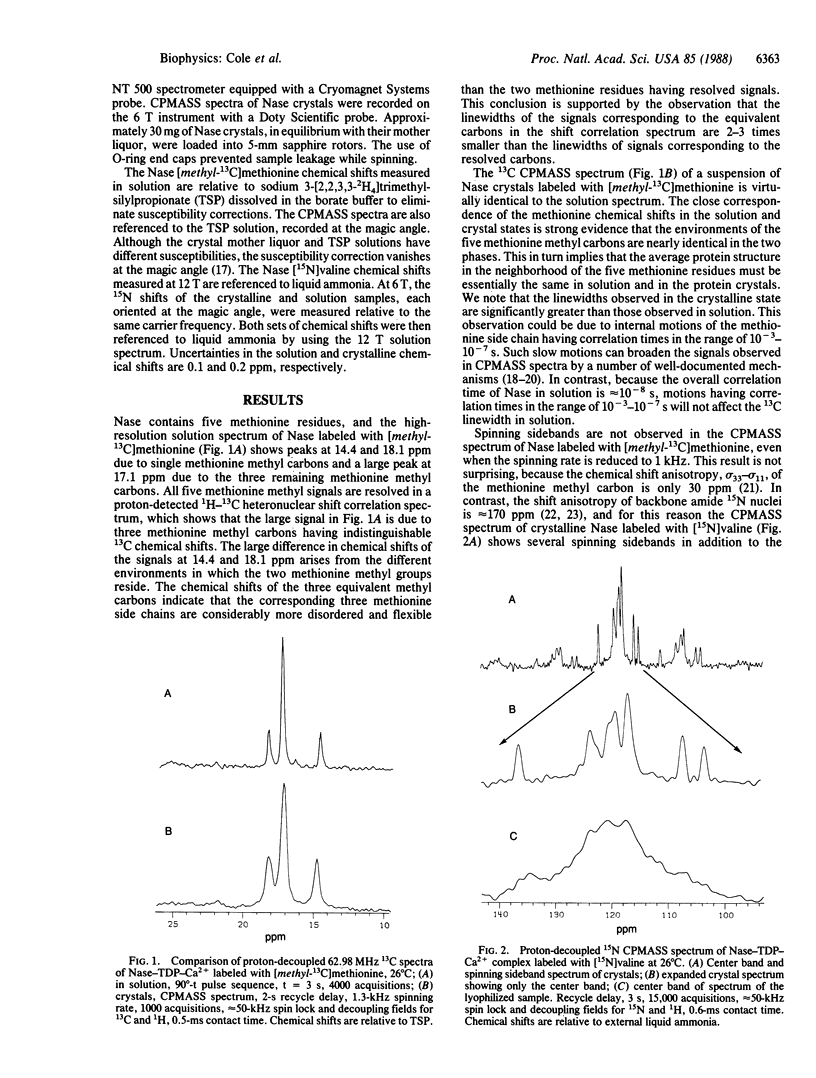
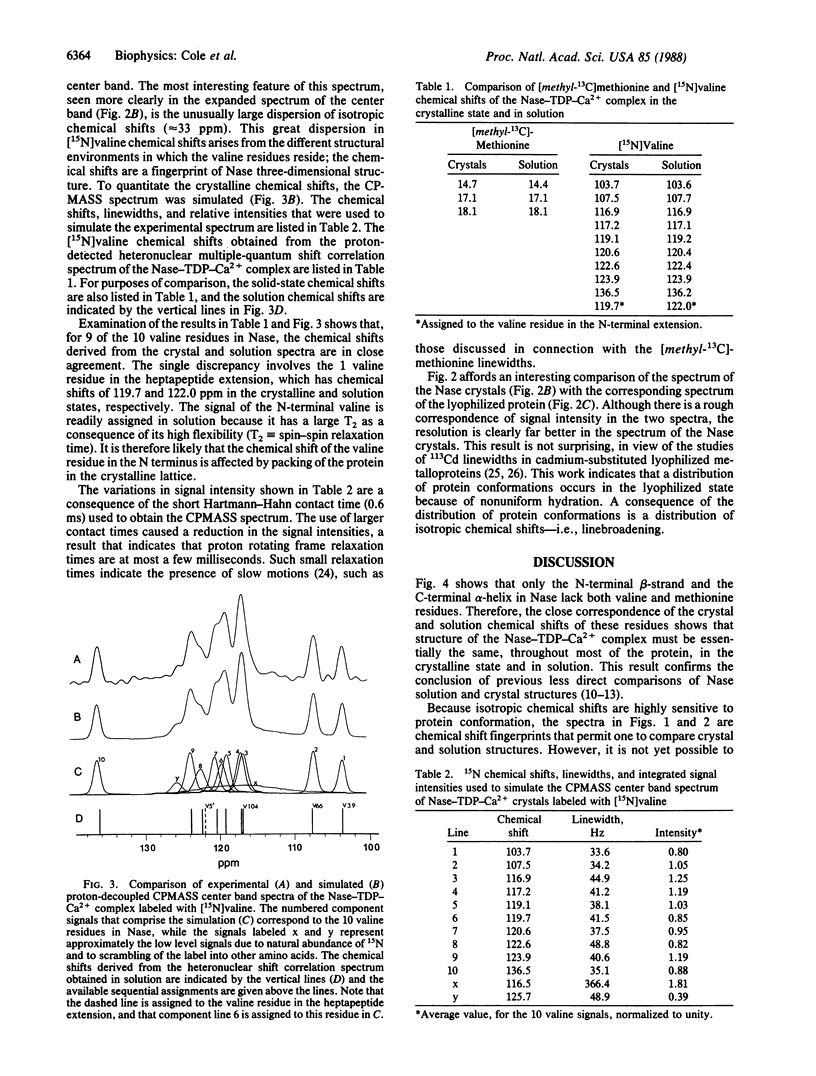
We report high-resolution 13C and 15N NMR spectra of crystalline staphylococcal nuclease (Nase) complexed to thymidine 3',5'-diphosphate and Ca2+. High sensitivity and resolution are obtained by applying solid-state NMR techniques--high power proton decoupling and cross-polarization magic angle sample spinning (CPMASS)--to protein samples that have been efficiently synthesized and labeled by an overproducing strain of Escherichia coli. A comparison of CPMASS and solution spectra of Nase labeled with either [methyl-13C]methionine or [15N]valine shows that the chemical shifts in the crystalline and solution states are virtually identical. This result is strong evidence that the protein conformations in the solution and crystalline states are nearly the same. Because of the close correspondence of the crystal and solution chemical shifts, sequential assignments obtained in solution apply to the crystal spectra. It should therefore be possible to study the molecular structure and dynamics of many sequentially assigned atomic sites in Nase crystals. Similar experiments are applicable to the growing number of proteins that can be obtained from efficient expression systems.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnone A., Bier C. J., Cotton F. A., Hazen E. E., Jr, Richardson D. C., Richardson J. S. The extracellular nuclease of Staphylococcus aureus: structures of the native enzyme and an enzyme-inhibitor complex at 4 A resolution. Proc Natl Acad Sci U S A. 1969 Oct;64(2):420–427. doi: 10.1073/pnas.64.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon R. O., Stolowich N. J., Gerlt J. A., Sturtevant J. M. Thermal denaturation of staphylococcal nuclease. Biochemistry. 1985 Oct 22;24(22):6044–6049. doi: 10.1021/bi00343a004. [DOI] [PubMed] [Google Scholar]
- Cotton F. A., Hazen E. E., Jr, Richardson D. C. Crystalline extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1966 Oct 10;241(19):4389–4390. [PubMed] [Google Scholar]
- Fox R. O., Evans P. A., Dobson C. M. Multiple conformations of a protein demonstrated by magnetization transfer NMR spectroscopy. Nature. 1986 Mar 13;320(6058):192–194. doi: 10.1038/320192a0. [DOI] [PubMed] [Google Scholar]
- Griffey R. H., Redfield A. G. Proton-detected heteronuclear edited and correlated nuclear magnetic resonance and nuclear Overhauser effect in solution. Q Rev Biophys. 1987 Feb;19(1-2):51–82. doi: 10.1017/s0033583500004029. [DOI] [PubMed] [Google Scholar]
- Hibler D. W., Stolowich N. J., Reynolds M. A., Gerlt J. A., Wilde J. A., Bolton P. H. Site-directed mutants of staphylococcal nuclease. Detection and localization by 1H NMR spectroscopy of conformational changes accompanying substitutions for glutamic acid-43. Biochemistry. 1987 Sep 22;26(19):6278–6286. doi: 10.1021/bi00393a048. [DOI] [PubMed] [Google Scholar]
- Jelinski L. W., Torchia D. A. 13C/1H high power double magnetic resonance investigation of collagen backbone motion in fibrils and in solution. J Mol Biol. 1979 Sep 5;133(1):45–65. doi: 10.1016/0022-2836(79)90250-x. [DOI] [PubMed] [Google Scholar]
- Keniry M. A., Rothgeb T. M., Smith R. L., Gutowsky H. S., Oldfield E. Nuclear magnetic resonance studies of amino acids and proteins. Side-chain mobility of methionine in the crystalline amino acid and in crystalline sperm whale (Physeter catodon) myoglobin. Biochemistry. 1983 Apr 12;22(8):1917–1926. doi: 10.1021/bi00277a028. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983 May-Jun;22(2-3):181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- Tucker P. W., Hazen E. E., Jr, Cotton F. A. Staphylococcal nuclease reviewed: a prototypic study in contemporary enzymology. I. Isolation; physical and enzymatic properties. Mol Cell Biochem. 1978 Dec 22;22(2-3):67–77. doi: 10.1007/BF00496235. [DOI] [PubMed] [Google Scholar]
- Tucker P. W., Hazen E. E., Jr, Cotton F. A. Staphylococcal nuclease reviewed: a prototypic study in contemporary enzymology. II. Solution studies of the nucleotide binding site and the effects of nucleotide binding. Mol Cell Biochem. 1979 Jan 15;23(1):3–16. doi: 10.1007/BF00226675. [DOI] [PubMed] [Google Scholar]
- Tucker P. W., Hazen E. E., Jr, Cotton F. A. Staphylococcal nuclease reviewed: a prototypic study in contemporary enzymology. III. Correlation of the three-dimensional structure with the mechanisms of enzymatic action. Mol Cell Biochem. 1979 Jan 26;23(2):67–86. doi: 10.1007/BF00226229. [DOI] [PubMed] [Google Scholar]
- Tucker P. W., Hazen E. E., Jr, Cotton F. A. Staphylococcal nuclease reviewed: a prototypic study in contemporary enzymology. IV. The nuclease as a model for protein folding. Mol Cell Biochem. 1979 Feb 9;23(3):131–141. doi: 10.1007/BF00219452. [DOI] [PubMed] [Google Scholar]


