Abstract
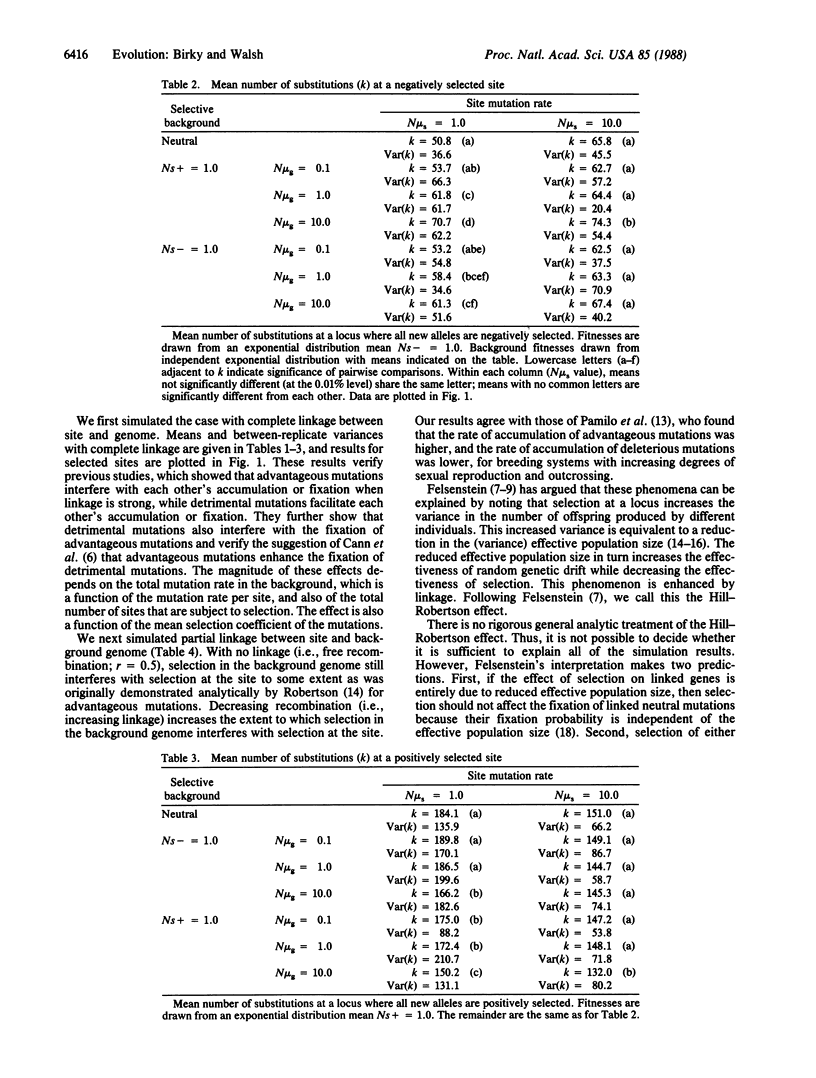
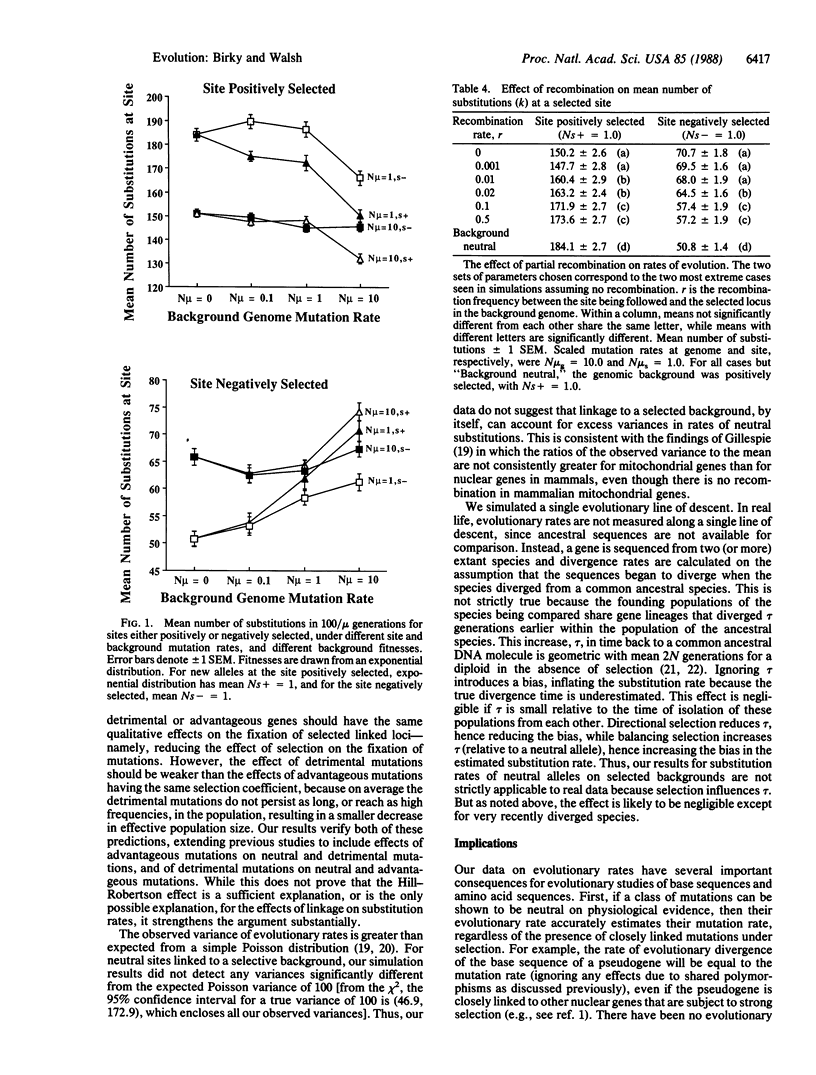
When an advantageous mutation is fixed in a population by selection, a closely linked selectively neutral or mildly detrimental mutation may "hitchhike" to fixation along with it. It has been suggested that hitchhiking might increase the rate of molecular evolution. Computer simulations and a mathematical argument show that complete linkage to either advantageous or deleterious mutations does not affect the substitution of selectively neutral mutations. However, the simulations show that linkage to selected background mutations decreases the rate of fixation of advantageous mutations and increases the rate of fixation of detrimental mutations. This is true whether the linked background mutations are advantageous or detrimental, and it verifies and extends previous observations that linkage tends to reduce the effects of selection on evolution. These results can be interpreted in terms of the Hill-Robertson effect: a locus linked to another locus under selection experiences a reduction in effective population size. The interpretation of differences in evolutionary rates between different genomes or different regions of a genome may be confounded by the effects of strong linkage and selection. Recombination is expected to reduce the overall rate of molecular evolution while enhancing the rate of adaptive evolution.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birky C. W., Jr Relaxed cellular controls and organelle heredity. Science. 1983 Nov 4;222(4623):468–475. doi: 10.1126/science.6353578. [DOI] [PubMed] [Google Scholar]
- Cann R. L., Brown W. M., Wilson A. C. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics. 1984 Mar;106(3):479–499. doi: 10.1093/genetics/106.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974 Oct;78(2):737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., Langley C. H. Are evolutionary rates really variable? J Mol Evol. 1979 Jun 8;13(1):27–34. doi: 10.1007/BF01732751. [DOI] [PubMed] [Google Scholar]
- Gillespie J. H. Variability of evolutionary rates of DNA. Genetics. 1986 Aug;113(4):1077–1091. doi: 10.1093/genetics/113.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966 Dec;8(3):269–294. [PubMed] [Google Scholar]
- Keightley P. D., Hill W. G. Directional selection and variation in finite populations. Genetics. 1987 Nov;117(3):573–582. doi: 10.1093/genetics/117.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Hill W. G. Effects of linkage on response to directional selection from new mutations. Genet Res. 1983 Oct;42(2):193–206. doi: 10.1017/s0016672300021650. [DOI] [PubMed] [Google Scholar]
- Martin C. H., Meyerowitz E. M. Characterization of the boundaries between adjacent rapidly and slowly evolving genomic regions in Drosophila. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8654–8658. doi: 10.1073/pnas.83.22.8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckelmann N., Li K., Wade R. P., Shuster R., Wallace D. C. cDNA sequence of a human skeletal muscle ADP/ATP translocator: lack of a leader peptide, divergence from a fibroblast translocator cDNA, and coevolution with mitochondrial DNA genes. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7580–7584. doi: 10.1073/pnas.84.21.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamilo P., Nei M., Li W. H. Accumulation of mutations in sexual and asexual populations. Genet Res. 1987 Apr;49(2):135–146. doi: 10.1017/s0016672300026938. [DOI] [PubMed] [Google Scholar]
- Rice W. R. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics. 1987 May;116(1):161–167. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satta Y., Ishiwa H., Chigusa S. I. Analysis of nucleotide substitutions of mitochondrial DNAs in Drosophila melanogaster and its sibling species. Mol Biol Evol. 1987 Nov;4(6):638–650. doi: 10.1093/oxfordjournals.molbev.a040464. [DOI] [PubMed] [Google Scholar]
- Schaeffer S. W., Aquadro C. F. Nucleotide sequence of the Adh gene region of Drosophila pseudoobscura: evolutionary change and evidence for an ancient gene duplication. Genetics. 1987 Sep;117(1):61–73. doi: 10.1093/genetics/117.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N. On the overdispersed molecular clock. Genetics. 1987 May;116(1):169–179. doi: 10.1093/genetics/116.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C., Ye J. H., Neckelmann S. N., Singh G., Webster K. A., Greenberg B. D. Sequence analysis of cDNAs for the human and bovine ATP synthase beta subunit: mitochondrial DNA genes sustain seventeen times more mutations. Curr Genet. 1987;12(2):81–90. doi: 10.1007/BF00434661. [DOI] [PubMed] [Google Scholar]
- Wilson A. C., Carlson S. S., White T. J. Biochemical evolution. Annu Rev Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]


