Abstract
FR-900098 is a potent chemotherapeutic agent for the treatment of malaria. Here we report the heterologous production of this compound in E. coli by re-constructing the entire biosynthetic pathway using a three plasmid system. Based on this system, whole cell feeding assays in combination with in vitro enzymatic activity assays reveal an unprecedented functional role of nucleotide conjugation and lead to the complete elucidation of the previously unassigned late biosynthetic steps. These studies also suggest a biosynthetic route to a second phosphonate antibiotic, FR-33289. A thorough understanding of the FR-900098 biosynthetic pathway now opens the possibilities for metabolic engineering in E. coli to increase production of the antimalarial antibiotic and combinatorial biosynthesis to generate novel derivatives of FR-900098.
Keywords: antimalarial, phosphonic acids, synthetic biology, biosynthesis
Introduction
Malaria remains one of the most common life-threatening infections in developing countries with up to 500 million infections and over 1 million deaths per year (Snow et al., 2005). Current antimalarial treatments include chloroquine and sulfadoxin pyrimethamine, but the rapid emergence of antibiotic resistance strains is rendering these current therapies futile, creating an urgent need for new effective drugs (Ridley, 2002; Wiesner et al., 2003). FR-900098 and fosmidomycin represent a new class of antimalarial compounds. These compounds were first isolated from Streptomyces rubellomurinus and Streptomyces lavendulae, respectively (Okuhara et al., 1980a). Studies have shown that these two compounds bind in the active site and effectively inhibit 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR), the first step in the nonmevalonate pathway for isoprenoid biosynthesis (Mac Sweeney et al., 2005). Genomic analyses indicate that the genes encoding the nonmevalonate pathway are present in a wide variety of Gram-negative and Gram-positive bacteria, plants, and the parasite causing the most virulent form of malaria, Plasmodium falciparum (Boucher and Doolittle, 2000; Lange et al., 2000). Mammals produce isoprenoids via the mevalonate pathway; therefore, FR-900098 and fosmidomycin should not be detrimental to humans (Beytia and Porter, 1976). Both compounds are currently undergoing clinical trials and have shown great promise as malaria treatments (Borrmann et al., 2004; Borrmann et al., 2006). Earlier work revealed that FR-900098 is twice more effective than fosmidomycin against various strains of Plasmodium falciparumin vitro and the closely related Plasmodium vinckei in mice (Jomaa et al., 1999). Progress has been made in the development of chemical synthesis routes for fosmidomycin (Hemmi et al., 1982) and FR-900098 (Fokin et al., 2007; Hemmi et al., 1982), but we chose to pursue a biological route for a more cost-effective way to produce these antimalarial compounds.
Heterologous production of FR-900098 in Streptomyces lividans and a proposed biosynthetic pathway were reported by Eliot et al. (Eliot et al., 2008). The first step involved in FR-900098 biosynthesis, a phosphoenolpyruvate (PEP) mutase reaction catalyzed by FrbD, is common in almost all other known phosphonate biosyntheses. Currently, only three other complete phosphonate biosynthetic pathways have been identified (2-aminoethylphosphonate (AEP) (Barry et al., 1988), fosfomycin (Hidaka et al., 1995; Woodyer et al., 2006), and bialaphos (Blodgett et al., 2005; Schwartz et al., 2004)); all of which have a common second step. In all of these cases, an irreversible thiamine pyrophosphate-dependent decarboxylation of phosphonopyruvate (PnPy) to form phosphonoacetaldehyde is required to provide a sufficient thermodynamic driving force to overcome the unfavorable equilibrium created by PEP mutase. However, the FR-900098 biosynthetic cluster does not contain a PnPy decarboxylase. FrbC, a homocitrate synthase homolog, catalyzes the formation of 2-phosphonomethylmalate, thus creating a novel route for phosphonate biosynthesis (Eliot et al., 2008).
The subsequent steps parallel the tricarboxylic (TCA) cycle, resulting in the formation of 2-oxo-4-phosphonobutyrate, an analog of α-ketoglutarate. The final steps were proposed based only on sequence homology of the remaining genes, without any definitive assignment of the order or function. A complete understanding of FR-900098 pathway could provide significant insight into the biosynthesis of all phosphonates. More importantly, heterologous expression inside a more genetically tractable host will act as a platform towards creating novel and potentially more potent derivatives of FR-900098 and increasing production of the antimalarial compound to industrially desirable levels. Here we report the heterologous production of FR-900098 in E. coli and deciphering of the late steps in FR-900098 biosynthesis, verified through whole cell feeding experiments and in vitro assays with purified enzymes.
Results and Discussion
Heterologous Production of FR-900098 in E. coli
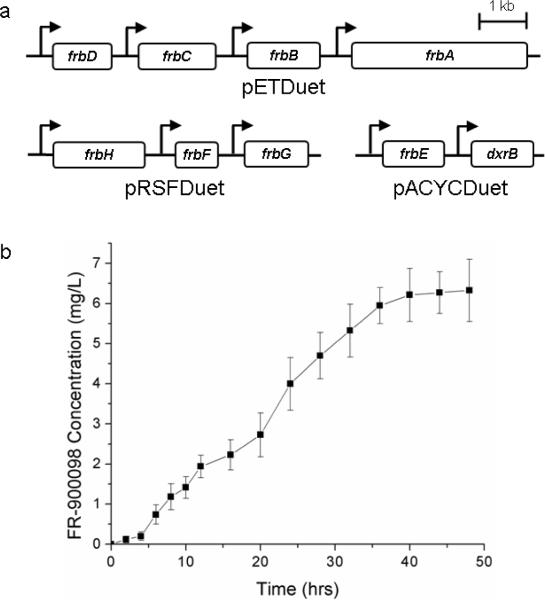
To achieve heterologous production of FR-900098 in E. coli, each of the genes in the biosynthetic gene cluster were first checked for soluble expression inside the new host. Each of the eleven genes were cloned, expressed, and then analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot. All of the proteins had high expression and showed clear overexpression bands in E. coli, except for FrbE and DxrB, which demonstrated low levels of soluble expression detected only by Western blot analysis (data not shown). To reconstitute the FR-900098 biosynthetic pathway in E. coli, each of the eleven genes in the biosynthetic cluster were cloned into a set of three compatible Duet expression vectors (pETDuet, pRSFDuet and pACYCDuet). Each individual gene is under the transcriptional control of a strong T7 promoter and expression is inducible by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG). The constructed plasmids were transformed into the heterologous host E. coli to create the production strain. The heterologous production of FR-900098 was confirmed using liquid chromatography/mass spectrometry (LC-MS) analysis as described in Eliot et al. (Eliot et al., 2008).
Deletion analysis was utilized to determine which genes were required for heterologous production of the FR-900098. Different strains of E. coli were created which lacked a single gene from the pathway and each was tested for its ability to produce the compound. The strain lacking dxrB grew very slowly upon protein induction and although the strain eventually grew to saturation, it produced significantly lower amounts of FR-900098. As mentioned earlier, the dxrB gene encodes a homolog of 1-deoxy-D-xylulose-5-phosphate (DXP) reductoisomerase, the known target of the antibiotic. This provided evidence for the necessary inclusion of the dxrB gene in the E. coli FR-900098 producing strains as means of resistance from increasing concentrations of the compound. The genes frbI and frbJ were not essential for FR-900098 production in E. coli; the overexpression of these two proteins more than likely results in the unnecessary consumption of cellular resources, which results in lower FR-900098 production. The final production strain (pETDuet-frbABCD, pRSFDuet-frbHFG, pACYCDuet-frbE-dxrB) did not include frbI or frbJ, resulting in the highest production of FR-900098, 6.3 mg/L (Fig. 1). This is the first known heterologous production of a phosphonate antibiotic in E. coli.
Figure 1.
Heterologous Production of FR-900098 in E. coli. (a) Scheme for the three plasmid system in FR-900098 production strain. Each gene is under transcriptional control by strong T7 promoter indicated by arrow. (b) Time course production of FR-900098 in E. coli.
Whole Cell Feeding Studies
To identify possible intermediates in the late steps of FR-900098 biosynthesis, a series of whole cell feeding experiments were conducted. Resting cells of E. coli were fed with 2-amino-4-phosphonobutyrate (2APn), a glutamate analog which is efficiently taken up by E. coli and also shown to cross-feed into S. lividans deletion mutants (data not shown). Culture supernatants were analyzed for possible phosphonate intermediates using a coupled LC-MS approach. Cells expressing FrbH were found to accumulate CMP-5'-3-aminopropylphosphonate (CMP-5'-3APn) and cells expressing FrbH, FrbF, and FrbG had detectable levels of CMP-5'-N-acetyl-3-aminopropylphosphonate (CMP-5'-Ac3APn), CMP-5'-FR-900098, and FR-900098. Thus, these initial whole cell feeding studies suggested that cytidine-5'-monophosphate (CMP) conjugated phosphonate intermediates are present during the late steps of FR-900098 biosynthesis. These studies also showed that FrbG is an essential component in the late steps of FR-900098 biosynthesis as definitive assignment of the function of the enzyme was never made (Eliot et al., 2008). It is also interesting to note that 2-oxo-4-phosphonobutyrate was detected in samples containing a negative control (empty vector), providing evidence that a ubiquitous cellular transaminase is able to carry out the transamination of 2OPn to form 2APn.
Bifunctional Activity of FrbH
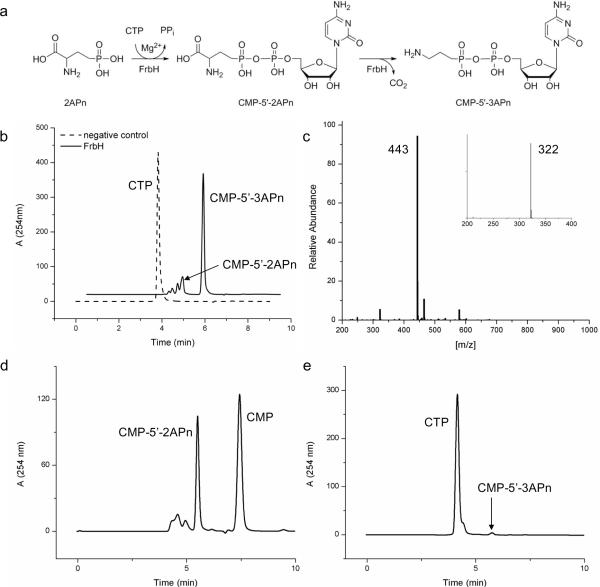
The role of frbH was investigated after overexpression and purification of a histidine-tagged FrbH fusion from E. coli. Based on sequence homology to other known PLP-dependent enzymes, the protein was originally believed to catalyze the decarboxylation of 2APn to yield 3APn (Eliot et al., 2008). However, repeated attempts to demonstrate this in vitro reaction failed. A BLAST search of full-length FrbH revealed two distinct domains: a nucleotide transferase domain followed by a PLP-dependent aminotransfease/decarboxylase domain. Only the second domain was originally believed to be active, but based on the whole cell feeding studies with 2APn, we began to investigate whether FrbH is bifunctional (Fig. 2a). An in vitro reaction mixture containing both 2APn and cytidine-5'-triphosphate (CTP) showed nearly complete conversion of CTP to several compounds, including cytidine-5'-monophosphate (CMP), cytidine-5'-diphosphate (CDP), CMP-5'-2APn and predominantly CMP-5'-3APn (Fig. 2b). LC-MS of the products CMP-5'-2APn and CMP-5'-3APn revealed the masses m/z 487 and 443, respectively (Fig. 2c). Furthermore, fragmentation of these two products in negative mode showed the characteristic fragment ion of m/z 322, indicating the CMP moiety on both of these compounds. These results suggest FrbH condenses 2APn and CTP to form CMP-5'-2APn and then decarboxylates CMP-5'-2APn to yield CMP-5'-3APn.
Figure 2.
FrbH reaction. (a) Proposed order of reaction where CMP attachment occurs before decarboxylation. (b) HPLC analysis of FrbH reaction product. (c) MS/MS data of CMP-5'-3APn. (d) HPLC analysis of FrbH-K466A mutant, which should remove decarboxylase activity. (e) HPLC analysis of FrbH-K38A mutant, which should remove nucleotide transferase activity.
Single point mutations were designed to eliminate the activity of each of the individual domains of FrbH to verify the proposed order of reaction. PLP-dependent enzymes are known to bind to the cofactor with a conserved lysine residue. A sequence alignment with CobD from Salmonella typhirium revealed the PLP-binding motif and the lysine at position 466 (Brushaber et al., 1998). A K466A mutant was constructed, expressed, and purified, which should be unable to bind the PLP and thus perform the decarboxylation. A second mutant was designed to disrupt nucleotide transferase activity of FrbH. A sequence alignment with IspD (an enzyme in the nonmevalonate pathway for isoprenoid biosynthesis) from E. coli revealed a common lysine residue (K38 in FrbH) that eliminated enzymatic activity (Richard et al., 2004). Under the same in vitro reaction conditions as the wild type FrbH, the K466A mutant produced only CMP-5'-2APn (Fig. 2d). Although unable to totally abolish nucleotide transferase activity, the K38A mutant had a mostly unreacted CTP peak with a small peak corresponding to CMP-5'-3APn (Fig. 2e). These results are conclusive evidence for FrbH catalyzing the CMP attachment to 2APn followed by decarboxylation, yielding CMP-5'-3APn (Fig. 2a).
N-Acetyltransferase Activity of FrbF and N-Hydroxylase Activity of FrbG
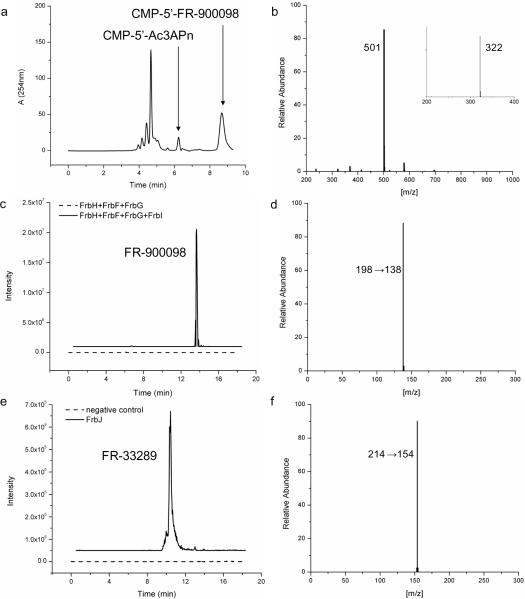
To complete the biosynthesis of FR-900098, the amino group of CMP-5'-3APn requires both hydroxylation and acetylation. The order and identity of the enzymes catalyzing these two steps was never definitively assigned in the original pathway (Eliot et al., 2008). FrbF has strong homology to known N-acetyltransferases and it was originally believed that FrbJ may be involved in the hydroxylation step. However, based on the results of the whole cell feeding assays and deletion studies described above, FrbG, a FAD/NADPH dependent monooxygenase, was now thought to catalyze the hydroxylation step instead of FrbJ. The whole cell feeding studies also suggest the order of the two steps as acetylation followed by hydroxylation, since CMP-5'-Ac3APn was seen and not N-hydroxy-3-aminopropylphosphonate (CMP-5'-H3APn). Coupled in vitro reactions with FrbF and FrbG also supported this order of reaction as there was always a CMP-5'-Ac3APn peak present, but little to no CMP-5'-H3APn monitored (Fig. 3ab). To determine the order of the acetylation and hydroxylation steps, each of the potential substrates was purified and kinetic parameters for FrbF and FrbG were obtained.
Figure 3.
FrbG, FrbF, FrbI and FrbJ reactions. (a) HPLC analysis of FrbG and FrbF combined reaction product. (b) MS/MS data of CMP-5'-FR-900098. (c) HPLC analysis of FrbI reaction product. (d) MS/MS data of FR-900098. (e) HPLC analysis of FrbJ reaction product, and (f) MS/MS data of FR-33289.
Although CMP-5'-Ac3APn was believed to be an intermediate in the pathway instead of CMP-5'-H3APn, no activity was observed for FrbG towards this substrate. FrbF was able to accept either of the two potential substrates, meaning CMP-5'-Ac3APn is actually a dead end in the FR-900098 biosynthetic pathway rather than a true intermediate (Table 1). Kinetic analysis of these two enzymes indicate that FrbF and FrbG compete for the same intermediate, CMP-5'-3APn, as they both prefer this compound compared to the alternative (CMP-H3APn for FrbF and CMP-5'-Ac3APn for FrbG). However, FrbG has 2.3-fold lower KM for CMP-5'-3APn in comparison to FrbF, so the enzyme can effectively direct the flux away from the dead end. The reason for observing CMP-5'-Ac3APn rather than CMP-5'-H3APn throughout the feeding experiments is due to the buildup of the dead end intermediate CMP-5'-Ac3APn, whereas the true intermediate, CMP-5'-H3APn, is prevented from accumulating because it is further processed by FrbF. These results demonstrate that the flux of the pathway must go through N-hydroxylation by FrbG followed by N-acetylation by FrbF and subsequently the more electrophilic CMP-5'-H3APn intermediate.
Table 1.
Kinetic parameters for FrbF and FrbG reactions with each of the potential substrates.
| Enzyme | Substrate | kcat (min−1) (mean ± SD) | KM (μM) (mean ± SD) | kcat / KM (mM−1 min−1) |
|---|---|---|---|---|
| FrbF | CMP-5'-3APn | 3.46 ± 0.16 | 35.2 ± 3.2 | 98.0 |
| CMP-5'-H3APn | 2.04 ± 0.43 | 52.3 ± 3.0 | 39.0 | |
| FrbG | CMP-5'-3APn | 5.92 ± 0.38 | 15.1 ± 1.5 | 391 |
| CMP-5'-Ac3APn | ND | |||
ND: no detectable activity; SD: standard deviation.
Nucleotide Hydrolase Activity of FrbI
The last step in the biosynthesis of FR-900098 was evident based on the presence of the intermediate CMP-5'-FR-900098. Removal of the CMP moiety from CMP-5'-FR-900098 is necessary in order to produce the final compound FR-900098. Nucleotide hydrolases are known to catalyze the hydrolytic breakdown of a wide variety of nucleotide derivatives (McLennan, 2006). FrbI has high homology to this family of enzymes and could likely catalyze the hydrolysis of CMP-5'-FR-900098 to produce FR-900098 and CMP. In order to perform in vitro enzymatic studies, the enzyme FrbI was overexpressed in E. coli and purified to homogeneity. An in vitro reaction involving FrbI was tested and found to produce a significant amount of FR-900098, whereas only a very small amount of FR-900098 was present in the absence of FrbI, likely due to the instability of CMP-5'-FR-900098 (Fig. 3cd). Thus, it is clear that FrbI catalyzes the hydrolysis of CMP-5'-FR-900098 to form FR-900098 and CMP. Interestingly, FrbI also catalyzed the hydrolysis of all the other CMP-containing intermediates within the pathway and even showed a small amount of activity toward CTP. This is not surprising given that nucleotide hydrolases typically have broad substrate specificity (Bessman et al., 1996). This promiscuous behavior is evident from the gene deletion analysis previously mentioned. The gene frbI is not required for FR-900098 biosynthesis in E. coli since a ubiquitous nucleotide hydrolase is able to cleave the CMP, yielding the final compound FR-900098.
Monooxygenase Activity of FrbJ
FrbJ is homologous to α-ketoglutarate dependent monooxygenases capable of functionalizing an inactivated C-H bond. Although not required for FR-900098 biosynthesis, we believed the enzyme may be involved in the synthesis of another phosphonate antibiotic, FR-33289. Production of this compound was observed in both the native Streptomyces rubellomurinus and heterologous Streptomyces lividans 4G7 host (data not shown). FR-33289 has similar antibacterial properties compared to FR-900098 and could also be another effective antimalarial compound (Okuhara et al., 1980b). An in vitro reaction mixture containing active FrbJ was able to convert FR-900098 to FR-33289 (Fig. 3ef). Expression of frbJ along with the remaining genes in the pathway could provide a means for production of FR-33289. In summary, we have heterologously produced FR-900098 in E. coli and demonstrated that the late steps of FR-900098 biosynthesis follow a series of novel intermediates (Fig. 4). Of special note, the nucleotide attachment to the intermediates in FR-900098 biosynthesis seems to play an unprecedented role. It is traditionally believed that attachment of the nucleotide serves only as an activator for otherwise challenging chemistry in natural product biosyntheses containing nucleotide conjugated intermediates. For example, in the biosynthesis of the phosphinothricin tripeptide (PTT), PhpF catalyzes the condensation of CTP with phosphonoformate to form CMP-5'-phosphonoformate (CMP-5'-PF). This reaction was required so that the activated intermediate would be primed for transfer of the phosphonoformate group to the appropriate acceptor molecule (Blodgett et al., 2007). As part of the nonmevalonate pathway, IspD has been shown to catalyze the formation of 4-diphosphocytidyl-2-C-methyl-D-erythritol (CDP-ME) from 2-C-methyl-D-erythritol 4-phosphate (MEP) and CTP, which facilitates the subsequent formation of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (Rohdich et al., 1999). However, in FR-900098 biosynthesis, nucleotide attachment clearly does not activate the bond for subsequent chemical reactions. Conjugation to CMP likely functions as a recognition mechanism to help distinguish among other metabolites within the cell and more importantly, a requirement for enzymatic activity. This hypothesis was substantiated by our recent determination of the co-crystal structure of FrbG with the cofactor NADPH (data not shown). Examination of this structure reveals that there is no room for the binding of the substrate in the active site. Thus, we proposed a novel reaction mechanism for the N-hydroxylation catalyzed by FrbG. Briefly, the substrate and NADPH cofactor compete for the same binding pocket in the enzyme, and the nucleotide helps dock the substrate in the correct position. The remaining enzymes in the pathway likely evolved to accept substrates conjugated to CMP rather than those proposed in the original pathway.
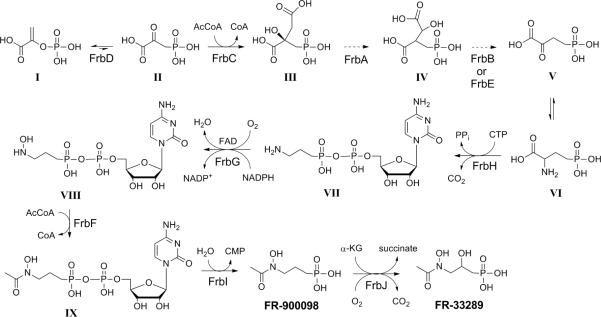
Figure 4.
Revised FR-900098 pathway. Each of the final steps in the FR-900098 biosynthetic pathway has been demonstrated in vitro using purified enzymes. Intermediates in the FR-900098 biosynthetic pathway are as follows: I, phosphoenolpyruvate; II, phosphonopyruvate; III, 2-phosphonomethylmalate; IV, 3-phosphonomethylmalate; V, 2-oxo-4-phosphonobutyrate; VI, 2-amino-4-phosphonobutyrate; VII, CMP-5'-3-aminopropylphosphonate; VIII, CMP-5'-N-hydroxy-3-aminopropylphosphonate; IX. CMP-5'-FR-900098.
Significance
We have successfully produced a potent antimalarial agent, FR-900098 in a recombinant E. coli strain and elucidated the biosynthetic mechanisms of the late steps involved in FR-900098 biosynthesis. Notably, a nucleotide CMP is conjugated with several of the intermediates, which seems to play an unprecedented role in natural product biosynthesis. Instead of activating a chemical bond, the nucleotide conjugation seems to serve as a substrate recognition mechanism. In addition, a biosynthetic route to a second phosphonic acid antibiotic, FR-33289, was discovered, which is embedded in the biosynthetic route to FR-900098, and its production is regulated by a single enzyme, FrbJ. This compound could also be effective in the treatment of malaria. These studies now open the possibilities for metabolic engineering in E. coli to increase production of the antimalarial antibiotic and combinatorial biosynthesis to generate novel derivatives of FR-900098 with more potent antimalarial activity.
Experimental Procedures
Heterologous Production of FR-900098 in E. coli
An overnight culture of the production strain (pETDuet-frbABCD, pRSFDuet-frbHFG, pACYCDuet-frbE-dxrB in E. coli BL21(DE3) cells) were grown at 37 °C in Luria-Bertani (LB) medium broth using the following antibiotic concentrations: 50 μg mL−1 ampicillin, 25 μg mL−1 kanamycin, and 12.5 μg mL−1 chloramphenicol. The cultures were diluted 1:100 in fresh medium and grown at 37 °C to an OD600 of ~0.7. At that point, the expression of the recombinant genes was induced with 0.3 mM IPTG and the culture was incubated at 30 °C with shaking (250 rpm). Samples (1 mL) were then removed from the cultures at specific time points and stored at −80 °C. Frozen samples were thawed at room temperature and centrifuged at maximum speed for 2 minutes. Supernatant from the samples were then analyzed by LC-MS to determine the concentration of FR-900098.
Whole Cell Feeding Studies
E. coli BL21(DE3) cells expressing empty vector (control), FrbH, and cells overexpressing FrbH, FrbF, FrbG, and DxrB were cultured overnight at 37 °C in Terrific Broth (TB) medium with appropriate antibiotics. The cultures were diluted 1:100 in fresh medium and grown at 37 °C to an OD600 of ~0.7. Protein expression was induced by the addition of 0.3 mM IPTG final concentration and cultures were grown at 25 °C for 24 hours. Cultures were then centrifuged at 4,000 rpm for 15 minutes and the supernatant was discarded. The cell pellet was resuspended in 1 mL of 10 mM Tris-HCl buffer pH 7.0. Resuspended cells were mixed with 20 mM final concentration (±)-2-amino-4-phosphonobutyrate (2APn) and incubated at 30 °C for 24 hours. Samples were lysed by freeze/thaw and centrifuged at maximum speed in a microcentrifuge for 10 minutes. The supernatant (1 mL) was mixed with 9 mL of methanol, vortexed, and centrifuged at 4,000 rpm for 15 minutes. The pellet was discarded and the supernatant was rotovaporated down to a volume of 1 mL. Samples were placed in the −80 °C freezer until analyzed by LC-MS using a 7 Tesla LTQ-FT mass spectrometer (Thermo Finnigan, San Jose, CA).
Cloning, Expression, and Purification
The cloning of genes frbH, frbF, frbG, frbI and frbJ from fosmid 4G7 was similar to that of Eliot et al. (14). Briefly, the genes were PCR amplified with NdeI and HindIII sites and cloned into corresponding sites of pET28a, designed to create an N-terminal histidine tag and transformed BL21(DE3) cells. The cells were grown in LB medium supplemented with kanamycin (50 μg mL−1) at 37 °C to OD600~0.7 and then induced at 25 °C by the addition of IPTG to a final concentration of 0.3 mM. During the purification of FrbH, PLP was added to the media for a final concentration of 50 μg mL−1. After 6 to 8 hours, the cells were harvested by centrifugation and resuspended in 20 mM Tris-HCl (pH 7.65 at 4 °C), 15% glycerol and 0.5M NaCl. The sample was freeze-thawed and then lysed further by sonication. After cellular debris removed by centrifugation, the N-His6-tagged proteins were purified using immobilized metal affinity chromatography (IMAC) with TALON Superflow Co2+ resin coupled to fast performance liquid chromatography (FPLC). The purified protein was washed three times with 50 mM HEPES-Na+ buffer and concentrated a final time. Concentrated protein was stored in 20% glycerol at −80 °C.
FrbH Activity
Wild-type and mutant FrbH activity was assayed by HPLC and LC-MS. The assay mixture (100 μL) contained 2 mM (±)-2-amino-4-phosphonobutyric acid, 1 mM CTP, 10 mM MgCl2 and His6-FrbH in 50 mM HEPES-Na+ buffer at pH 7.25. The reaction was initiated with the addition of 25 μg of purified wild-type or mutant FrbH and incubated at 30 °C for 2 hours. Reaction mixtures were transferred into Microcon® centrifugal filter device (MWCO 10,000) and centrifuged for 15 minutes at maximum speed. The reactions were then analyzed by LC-MS.
FrbG and FrbF Activity
The activity of FrbG and FrbF was demonstrated in a coupled assay with FrbH. The assay mixture (100 μL) contained 2 mM (±)-2-amino-4-phosphonobutyric acid, 1 mM CTP, 10 mM MgCl2, 1 mM AcCoA, 1 mM NADPH, His6-FrbH, His6-FrbF, and His6-FrbG in 50 mM HEPES-Na+ buffer at pH 7.25. The reaction was initiated with the addition of 25 μg of purified FrbF and 35 μg of purified FrbG. Reaction mixtures were transferred into Microcon® centrifugal filter device (MWCO 10,000) and centrifuged for 15 minutes at maximum speed. The reactions were then analyzed by LC-MS.
FrbI Activity
The activity of FrbI was demonstrated in a coupled assay with FrbH, FrbF, and FrbG. The assay mixture (100 μL) contained 2 mM (±)-2-amino-4-phosphonobutyric acid, 1 mM CTP, 10 mM MgCl2, 1 mM AcCoA, 1 mM NADPH, His6-FrbH, His6-FrbF, His6-FrbG, and His6-FrbI in 50 mM HEPES-Na+ buffer at pH 7.25. The reaction was initiated with the addition of 25 μg of purified FrbI and incubated at 30 °C for 2 hours. Reaction mixtures were transferred into Microcon® centrifugal filter device (MWCO 10,000) and centrifuged for 15 minutes at maximum speed. The reactions were then analyzed by LC-MS.
FrbJ Activity
FrbJ activity was assayed by HPLC and LC-MS. The assay mixture (100 μL) contained 1 mM FR-900098, 1 mM α-ketoglutarate, 1 mM FeSO4, 4 mM ascorbic acid, and His6-FrbJ in 50 mM HEPES-Na+ buffer at pH 7.25. The reaction was initiated with the addition of 25 μg of purified FrbJ and incubated at 25 °C for 4 hours. Reaction mixtures were transferred into Microcon® centrifugal filter device (MWCO 10,000) and centrifuged for 15 minutes at maximum speed. The reactions were then analyzed by LC-MS.
LC-MS Analysis of CMP-containing Compounds
Products of the enzymatic reactions were analyzed by an Agilent 1100 series LC/MSD XCT plus ion trap mass spectrometer (Agilent, Palo Alto, CA) at the Roy J. Carver Metabolomics Center at the University of Illinois (Urbana, IL). The liquid chromatography (LC) was performed using an Agilent 1100 series HPLC equipped with an online degasser, binary pump, and autosampler. Analytes were resolved on a 150 × 4.6 mm Agilent C18 reverse-phase HPLC column. HPLC parameters were as follows: 25 °C; solvent A: 15 mM ammonium formate; solvent B: 20 mM ammonium acetate; gradient: 25% B at 0 minutes; then to 100% B in 10 minutes, and finally maintain 100% B for 3 minutes; flow rate: 0.3 mL min−1; detection by UV absorbance at 254 nm. The mass spectrometry (MS) was performed using an Agilent XCT ion trap MSD mass spectrometer. Mass spectra were acquired in ultra scan mode using electrospray ionization (ESI) with negative polarity. The MS system was operated using a drying temperature of 350 °C, a nebulizer pressure of 35 psi, a drying gas flow of 8.5 L min−1, and a capillary voltage of 4500 V.
LC-MS Analysis of FR-900098 and FR-33289
The LC-MS was performed using an Agilent 1100 series LC/MSD XCT plus ion trap mass spectrometer (Agilent, Palo Alto, CA). The liquid chromatography (LC) was performed using an Agilent 1100 series HPLC equipped with an online degasser, binary pump, and autosampler. The mobile phase contained 0.1% formic acid in ddH2O (solvent A) and 100% acetonitrile (solvent B). Analytes were resolved on a 100 × 4.6 mm Synergi 4u Fusion-RP 80A analytical column (Phenomenex, Torrance, CA) at a flow rate of 200 μL min−1 with a gradient of 0 to 40% solvent B for 10 minutes, a gradient of 40 to 100% solvent B for 3 minutes, and 100% solvent B for 5 minutes. The MS was performed using an Agilent XCT ion trap MSD mass spectrometer. Mass spectra were acquired in ultra scan mode using ESI with positive polarity. The system was operated using a drying temperature of 350 °C, a nebuliser pressure of 35 psi, a drying gas flow of 8.5 L min−1, and a capillary voltage of 4500 V. For determination of titers, extracted ion chromatograms were generated from suitable ions of each analyte, and the integrated areas corresponding to each compound were compared to standards. Fosmidomycin was used as internal standard to measure FR-900098 concentrations.
Acknowledgements
We thank Lucas Li at the Metabolomics Center at the University of Illinois for help in LC-MS analysis. This work was supported by grants from the National Institute of General Medical Sciences GM077596 (H.Z., W.W.M., and N.L.K.) and GM59334 (W.W.M.). M.A.D. acknowledges support from the US National Institutes of Health under Ruth L. Kirschstein National Research Award 5 T32 GM070421 from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barry RJ, Bowman E, McQueney M, Dunaway-Mariano D. Elucidation of the 2-aminoethylphosphonate biosynthetic pathway in Tetrahymena pyriformis. Biochem. Biophys. Res. Commun. 1988;153:177–182. doi: 10.1016/s0006-291x(88)81205-1. [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O'Handley SF. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- Beytia ED, Porter JW. Biochemistry of polyisoprenoid biosynthesis. Annu. Rev. Biochem. 1976;45:113–142. doi: 10.1146/annurev.bi.45.070176.000553. [DOI] [PubMed] [Google Scholar]
- Blodgett JA, Thomas PM, Li G, Velasquez JE, van der Donk WA, Kelleher NL, Metcalf WW. Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nat. Chem. Biol. 2007;3:480–485. doi: 10.1038/nchembio.2007.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blodgett JA, Zhang JK, Metcalf WW. Molecular cloning, sequence analysis, and heterologous expression of the phosphinothricin tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob. Agents Chemother. 2005;49:230–240. doi: 10.1128/AAC.49.1.230-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrmann S, Adegnika AA, Matsiegui PB, Issifou S, Schindler A, Mawili-Mboumba DP, Baranek T, Wiesner J, Jomaa H, Kremsner PG. Fosmidomycin-clindamycin for Plasmodium falciparum infections in African children. J. Infect. Dis. 2004;189:901–908. doi: 10.1086/381785. [DOI] [PubMed] [Google Scholar]
- Borrmann S, Lundgren I, Oyakhirome S, Impouma B, Matsiegui PB, Adegnika AA, Issifou S, Kun JF, Hutchinson D, Wiesner J, et al. Fosmidomycin plus clindamycin for treatment of pediatric patients aged 1 to 14 years with Plasmodium falciparum malaria. Antimicrob. Agents Chemother. 2006;50:2713–2718. doi: 10.1128/AAC.00392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher Y, Doolittle WF. The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol. Microbiol. 2000;37:703–716. doi: 10.1046/j.1365-2958.2000.02004.x. [DOI] [PubMed] [Google Scholar]
- Brushaber KR, O'Toole GA, Escalante-Semerena JC. CobD, a novel enzyme with L-threonine-O-3-phosphate decarboxylase activity, is responsible for the synthesis of (R)-1-amino-2-propanol O-2-phosphate, a proposed new intermediate in cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem. 1998;273:2684–2691. doi: 10.1074/jbc.273.5.2684. [DOI] [PubMed] [Google Scholar]
- Eliot AC, Griffin BM, Thomas PM, Johannes TW, Kelleher NL, Zhao H, Metcalf WW. Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chem. Biol. 2008;15:765–770. doi: 10.1016/j.chembiol.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokin AA, Yurchenko AG, Rodionov VN, Gunchenko PA, Yurchenko RI, Reichenberg A, Wiesner J, Hintz M, Jomaa H, Schreiner PR. Synthesis of the antimalarial drug FR900098 utilizing the nitroso-ene reaction. Org. Lett. 2007;9:4379–4382. doi: 10.1021/ol702082k. [DOI] [PubMed] [Google Scholar]
- Hemmi K, Takeno H, Hashimoto M, Kamiya T. Studies on phosphonic acid antibiotics. IV. Synthesis and antibacterial activity of analogs of 3-(N-acetyl-N-hydroxyamino)-propylphosphonic acid (FR-900098) Chem. Pharm. Bull. (Tokyo) 1982;30:111–118. doi: 10.1248/cpb.30.111. [DOI] [PubMed] [Google Scholar]
- Hidaka T, Goda M, Kuzuyama T, Takei N, Hidaka M, Seto H. Cloning and nucleotide sequence of fosfomycin biosynthetic genes of Streptomyces wedmorensis. Mol. Gen. Genet. 1995;249:274–280. doi: 10.1007/BF00290527. [DOI] [PubMed] [Google Scholar]
- Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler HK, et al. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- Lange BM, Rujan T, Martin W, Croteau R. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac Sweeney A, Lange R, Fernandes RP, Schulz H, Dale GE, Douangamath A, Proteau PJ, Oefner C. The crystal structure of E. coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation. J. Mol. Biol. 2005;345:115–127. doi: 10.1016/j.jmb.2004.10.030. [DOI] [PubMed] [Google Scholar]
- McLennan AG. The Nudix hydrolase superfamily. Cell Mol. Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuhara M, Kuroda Y, Goto T, Okamoto M, Terano H, Kohsaka M, Aoki H, Imanaka H. Studies on new phosphonic acid antibiotics. I. FR-900098, isolation and characterization. J. Antibiot. (Tokyo) 1980a;33:13–17. doi: 10.7164/antibiotics.33.13. [DOI] [PubMed] [Google Scholar]
- Okuhara M, Kuroda Y, Goto T, Okamoto M, Terano H, Kohsaka M, Aoki H, Imanaka H. Studies on new phosphonic acid antibiotics. III. Isolation and characterization of FR-31564, FR-32863 and FR-33289. J. Antibiot. (Tokyo) 1980b;33:24–28. doi: 10.7164/antibiotics.33.24. [DOI] [PubMed] [Google Scholar]
- Richard SB, Lillo AM, Tetzlaff CN, Bowman ME, Noel JP, Cane DE. Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and mutants, reveals roles of active site amino acids. Biochemistry. 2004;43:12189–12197. doi: 10.1021/bi0487241. [DOI] [PubMed] [Google Scholar]
- Ridley RG. Medical need, scientific opportunity and the drive for antimalarial drugs. Nature. 2002;415:686–693. doi: 10.1038/415686a. [DOI] [PubMed] [Google Scholar]
- Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk MH. Cytidine 5'-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Berger S, Heinzelmann E, Muschko K, Welzel K, Wohlleben W. Biosynthetic gene cluster of the herbicide phosphinothricin tripeptide from Streptomyces viridochromogenes Tu494. Appl. Environ. Microbiol. 2004;70:7093–7102. doi: 10.1128/AEM.70.12.7093-7102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner J, Ortmann R, Jomaa H, Schlitzer M. New antimalarial drugs. Angew. Chem. Int. Ed. Engl. 2003;42:5274–5293. doi: 10.1002/anie.200200569. [DOI] [PubMed] [Google Scholar]
- Woodyer RD, Shao Z, Thomas PM, Kelleher NL, Blodgett JA, Metcalf WW, van der Donk WA, Zhao H. Heterologous production of fosfomycin and identification of the minimal biosynthetic gene cluster. Chem. Biol. 2006;13:1171–1182. doi: 10.1016/j.chembiol.2006.09.007. [DOI] [PubMed] [Google Scholar]