Abstract
Chemical examination of the secondary metabolites of a marine Saccharomonospora sp., isolated from marine sediments collected at the mouth of the La Jolla Submarine Canyon, yielded the unprecedented alkaloid lodopyridone (1). The low proton-to-carbon ratio of 1 precluded structure elucidation by NMR spectroscopic methods, thus the structure was defined by X-ray crystallography. Lodopyridone is cytotoxic to HCT-116 human colon cancer cells with IC50 = 3.6 μM.
As a group, the actinomycete bacteria are a prolific source of structurally interesting and biologically active compounds, responsible for over 45% of all microbial natural products,1 including the clinicially important antibiotics tetracycline, erythromycin, and vancomycin.2 Over the past 15 years, we have focused our efforts on an underexplored subgroup of these bacteria—the actinomycetes dwelling in marine sediments—and this effort has yielded a cornucopia of novel bioactive molecules,3 including salinisporamide A,4 which recently completed phase I clinical trials for the expected treatment of multiple myeloma.
The usually calm waters off the coast of La Jolla belie a dramatic underwater topography. The La Jolla Submarine Canyon is a major subsurface canyon with a depth in excess of 3000 ft that nearly reaches the pier at Scripps Institution of Oceanography. This canyon, and the millions of years of fault activity that created it, led to a very unusual pattern of sediment deposition on the ocean floor.5 As part of our ongoing efforts to discover novel secondary metabolites from marine sediment actinomycetes, we decided to explore the sediments of this geologically unique area.6 We report here the isolation of a unique actinomycete, strain CNQ490, and the production by this strain of lodopyridone (1), a modified alkaloid with an unprecedented carbon skeleton. The new strain was collected near the mouth of the La Jolla Canyon.
Marine bacterial strain CNQ490 was isolated from a sediment sample collected approximately 2 km west of the Scripps pier at a depth of 45 m. Sequence analysis of its 16S rRNA gene showed only modest identity with Saccharomonospora caesia, while subsequent phylogenetic analysis indicated this strain was a new operational taxonomic unit (OTU) within the genus Saccharomonospora.6 Strain CNQ490 requires saltwater for growth, indicating a high level of adaptation to the marine environment.7
LCMS analysis of a crude extract of a CNQ490 shake flask culture revealed trace amounts of several high molecular weight, chlorinated molecules with an unusual UV chromophore. A large-scale fermentation was undertaken to obtain enough material for structure elucidation. Two rounds of flash chromatography followed by HPLC gave pure 1 (4.0 mg), along with other lodopyridone isomers too minor (<0.02 mg) to define.
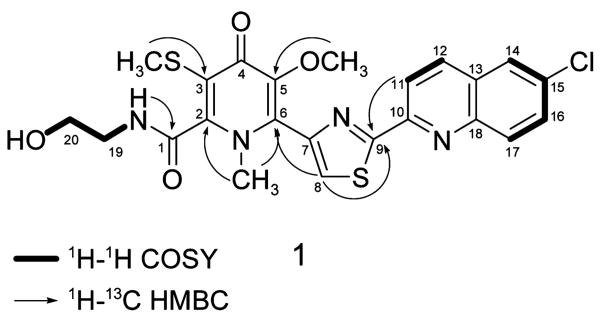
Lodopyridone (1) was isolated as a colorless glass.8 High-resolution ESI-MS (HRESIMS) analysis of 1 gave an [M + H]+ ion at m/z 517.0760 consistent with a molecular formula of C23H21ClN4O4S2 (calcd for C23H2235ClN4O4S2, 517.0765) and 15 degrees of unsaturation. Analysis of the 13C and DEPT NMR spectra of 1 revealed three methyl carbons, two methylene carbons, six methine carbons, and an intimidating 12 quaternary carbons. The 1H–1H COSY spectrum revealed only three spin systems (Figure 1): a pair of ortho-coupled aromatic protons at δH 8.33 and 8.55 (H-11 and H-12, J = 8.5 Hz); a proton at δH 8.25 (H-14) displaying weak meta-coupling to a proton at 7.87 (H-16, J = 2.0 Hz), which in turn was ortho-coupled to an aromatic proton at 8.14 (H-17, J = 9.0 Hz); and last, a spin system encompassing an exchangeable proton at δH 4.74 (OH), two pairs of adjacent methylene protons at 3.35 and 3.55 (H-19 and H-20), and a second exchangeable proton at 8.96 (NH), corresponding to ethanolamine.
Figure 1.
Selected 2D NMR correlations of 1.
The 1H–13C HMBC spectrum of 1 permitted assembly of larger molecular fragments (Figure 1). The quinoline moiety was identified using correlations from various of the five aromatic protons to four quaternary carbons at δC 150.5, 129.1, 131.9, and 145.5 (C-10, C-13, C-15, and C-18, respectively; see Table 1 for specific correlations). A correlation from the exchangeable proton at δ 8.96 (NH of the ethanolamine) to a carbonyl at δC 161.6 (C-1) indicated attachment via an amide bond. The methyl singlets at δH 2.30 and 3.67 (SCH3, OCH3) each showed a single, strong correlation to an olefinic carbon (δC 119.7 (C-3), 146.8 (C-5)), while the methyl singlet at δH 3.36 showed two strong three-bond correlations (to δC 148.9 (C-2) and 136.8 (C-6)) exposing it as an N-methyl group. The proton at δH 8.27 (H-8) also showed a correlation to C-6, indicating that this proton was near the N-methyl group. H-8 showed additional correlations to carbons at δC 145.0, 168.3, and 150.5 (C-7, C-9, and a four-bond correlation to C-10). Correlations from H-11 and H-12 (four-bond) to C-9 suggested that C-9 is the point of attachment to the quinoline moiety, but it was not possible to clarify the nature of that connection using NMR data alone.
Table 1.
1H (500 MHz) and 13C (125 MHz) NMR Data for 1 in DMSO (δ in ppm)
| δC | δH (mult, J (Hz)) | COSY | HMBC | |
|---|---|---|---|---|
| 1 | 161.6 | |||
| 2 | 148.9 | |||
| 3 | 119.7 | |||
| 4 | 170.9 | |||
| 5 | 146.8 | |||
| 6 | 136.8 | |||
| 7 | 145.0 | |||
| 8 | 126.3 | 8.27 (1H, s) | 6,7,9,10 | |
| 9 | 168.3 | |||
| 10 | 150.5 | |||
| 11 | 118.4 | 8.33 (1H, d, 8.5) | 12 | 9,10,12,13 |
| 12 | 137.3 | 8.55 (1H, d, 8.5) | 11 | 9,10,11,13,14,18 |
| 13 | 129.1 | |||
| 14 | 126.7 | 8.25 (1H, d, 2.0) | 16 | 12,15,18 |
| 15 | 131.9 | |||
| 16 | 131.6 | 7.87 (1H, dd, 9.0, 2.0) | 14,17 | 14,15,18 |
| 17 | 130.8 | 8.14 (1H, d, 9.0) | 16 | 13,15,18 |
| 18 | 145.5 | |||
| 19 | 41.6 | 3.35a | 20, NH | |
| 20 | 59.2 | 3.55 (2H, m) | 19, OH | |
| SCH3 | 16.1 | 2.30 (3H, s) | 3 | |
| OCH3 | 59.4 | 3.67 (3H, s) | 5 | |
| NCH3 | 39.1 | 3.36 (3H, s) | 2,6 | |
| OH | 4.74 (1H, m) | 20 | ||
| NH | 8.96 (1H, t, 6.0) | 19 | 1,19 |
Multiplicity and integral obscured by water peak at δH 3.32.
NMR data ultimately proved to be insufficient to solve the structure of lodopyridone. The pyridone carbonyl C-4 was completely invisible by both HMBC and HSQC, and connections between C-1 and C-2, C2 and C-3, C-5 and C-6, and C-7 and C-9 could not be confirmed by NMR. To unambiguously assign the structure, we undertook a single X-ray diffraction study.
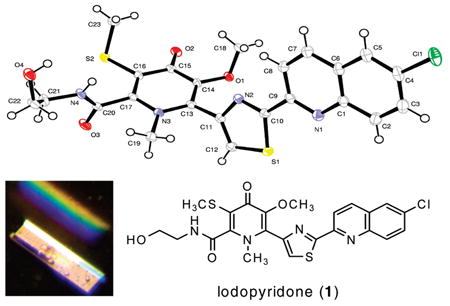
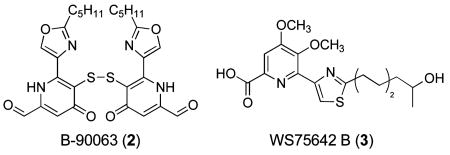
Slow evaporation of a solution of 1 in a mixture of methanol, water, and ethyl acetate gave X-ray quality crystals after six months. The X-ray structure of 1 revealed the unique carbon skeleton shown (Figure 2), including ethanolamine, thiomethyl-substituted 4-pyridone, thiazole, and chloroquinoline moieties.9 The 4-pyridone moiety is highly unusual among natural products. Searches in the AntiMarin Database and Crossfire Database Suite yielded fewer than 20 examples, all but one of which were isolated from plants or fungi.10 The only bacterial 4-pyridone we could find is B-90063 (2) isolated from a marine α-proteobacterium.11
Figure 2.
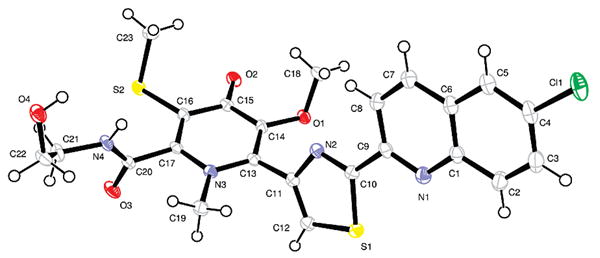
ORTEP representation of 1.
The closest molecule we could find from an actinomycete is WS75642 B (3) produced by Saccharothrix sp. 75624.12
In an attempt to increase the low production yields of the lodopyridones, we grew CNQ490 in media supplemented with presumed biosynthetic substrates. We guessed that lodopyridone is formed from the amino acids Gly, Cys (2 equiv), and Trp,13 with an additional two carbons coming from pyruvate. Hence we cultivated strain CNQ490 in media supplemented with Gly (100 mg/mL), Cys (50 mg/mL), Trp (50 mg/mL), or pyruvic acid (250 mg/mL). We also grew CNQ490 in the media used by Tsurumi et al. to obtain 3 in high yield.14 The reported yield of 3 was 2.9 mg/L, nearly 30× our yield of lodopyridone. The producing organism, Saccharothrix sp. 75624, like strain CNQ490, is in the suborder Pseudonocardineae, suggesting that the same medium might work. Unfortunately, although CNQ490 grew in all the media conditions we tried, we did not observe any increase in lodopyridone production.
Lodopyridone (1) showed no significant activity against methicillin-resistant Staphylococcus aureus. However, 1 showed modest activity against the human colon adenocar-cinoma cell line HCT-116 with IC50 = 3.6 μM.15
In summary, we have described the isolation and structure elucidation of a novel alkaloid produced by a marine actinomycete, Saccharomonospora sp. Lodopyridone (1) joins a growing list of structurally unprecedented, bioactive compounds produced by marine actinomycetes.
Supplementary Material
Acknowledgments
The authors thank Alejandra Prieto-Davó for providing taxonomic information regarding strain CNQ490, Sara M. Kelly for performing the MRSA and HCT-116 cytotoxicity assays, and Dan Udwary and Marcus Nett for discussions about the possible biosynthesis of 1. This work was supported by NIH R37 CA44848 (to WF) and by the UCSD Cancer Therapeutics Training Program, funded under NIH grant T32CA121938 (to KNM, S. Howell UCSD PI). The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Cancer Institute or the National Institutes of Health.
Footnotes
Supporting Information Available: Full experimental procedures, HRESIMS, and 1D and 2D NMR spectral data of lodopyridone (1). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bérdy J. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Walsh C. Antibiotics: Actions, origins, resistance. Chapter 11. ASM Press; Washington, D.C.: 2003. pp. 160–161. [Google Scholar]
- 3.Reviewed in: Fenical W, Jensen PR. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841.
- 4.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew Chem, Int Ed Engl. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 5.Covault JA, Normark WR, Romans BW, Graham SA. Geology. 2007;35:783–786. [Google Scholar]
- 6.Prieto-Davó A, Fenical W, Jensen PR. Aquat Microb Ecol. 2008;52:1–11. [Google Scholar]
- 7.MacLeod RA. Bacteriol Rev. 1965;29:9–23. [PMC free article] [PubMed] [Google Scholar]
- 8.The name Lodopyridone was derived from the Spanish Lodo (mud), as inspired by the muddy La Jolla marine sediments from which CNQ490 was isolated. Lodopyridone (1): colorless glass; UV (MeOH) λmax: 219, 257, 315, 331, 346 nm. IR (NaCl) νmax: 3378, 2926, 2855, 1722, 1660, 1591, 1460, 1243 cm−1. 1H and 13C NMR data: see Table 1. HRESIMS m/z 517.0760 [M + H]+ (C23H22ClN4O4S2 calcd 517.0765).
- 9.Crystal data for lodopyridone (1): monoclinic, space group P21/c; a = 8.4180(4) Å, b = 8.7144(4) Å, c = 31.3841(15) Å, α = 90°, β = 90.294(2)°, γ = 90°, V = 2302.24(19) Å3, Z = 4, d = 1.492 Mg/m3, crystal dimensions 0.10 × 0.04 × 0.04 mm. A total of 9700 reflections were collected covering the indices, −10 ≤ h ≤ 10, −10 ≤ k ≤ 10, −37 ≤ l ≤ 37. 3859 reflections were found to be symmetry independent, with an Rint of 0.0378. Final R(F2) = 0.0392. Crystallographic data for lodopyridone have been deposited with the Cambridge Crystallographic Data Centre (deposition number CCDC 725224). Copies of the data can be obtained, free of charge, via www.ccdc.cam.ac.uk/data_request/cif, or by emailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12, Union Road, Cambridge CB2 1EZ, UK; fax: + 44 1223 336033.
- 10.(a) Blunt JW, Munro MHG, Laatsch H, editors. AntiMarin Database. University of Canterbury; Christchurch, New Zealand: University of Göttingen; Göttingen, Germany: 2008. [Google Scholar]; (b) Elsevier; 2009. Crossfire Database Suite. http://www.info.crossfiredatabases.com/index.shtml. [Google Scholar]
- 11.Takaishi S, Tuchiya N, Sato A, Negishi T, Takamatsu Y, Matsushita Y, Watanabe T, Iijima Y, Haruyama H, Kinoshita T, Tanaka M, Kodama K. J Antibiot. 1998;51:805–815. doi: 10.7164/antibiotics.51.805. [DOI] [PubMed] [Google Scholar]
- 12.Tsurumi Y, Ueda H, Hayashi K, Takase S, Nishikawa M, Kiyoto S, Okuhara M. J Antibiot. 1995;48:1066–1072. doi: 10.7164/antibiotics.48.1066. [DOI] [PubMed] [Google Scholar]
- 13.Hallard D, Geerlings A, van der Heijden R, Lopes Cardozo MI, Hoge JHC, Verpoorte R. In: Hairy roots, culture and applications. Doran PM, editor. Harwood; 1997. pp. 43–49. [Google Scholar]
- 14.We prepared the production medium according to ref 12, substituting seawater in place of deionized water.
- 15.The HCT-116 cytotoxicity assay was performed using the MTT method, as described in: Mosmann T. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.