Abstract
The regulation of gene expression at the translational level not only allows for rapid changes in specific protein levels but also provides an opportunity to alter codon specificity. For the incorporation of selenocysteine (Sec) into protein, the UGA codon is transformed from one that signals translation termination to one specific for Sec. This review provides a look at Sec incorporation from the perspective of the individual steps involved in protein synthesis: initiation, elongation and termination. The roles of the factors known to be required for Sec incorporation are considered in the context of each step in translation including structural modeling of the differences between the standard elongation factor eEF1A and the Sec-specific counterpart, eEFSec.
Keywords: Translation, Initiation, Elongation, Termination, Selenocysteine insertion sequence (SECIS), SECIS binding protein
1. Introduction
The regulation of the machinery involved in protein synthesis is as complex and varied as the constituents of the machinery itself. Of course, most of the regulation of translation occurs at its most rate-limiting step: initiation. The potential points of regulation during this sequence of events is staggering, as is becoming the body of literature on the subject (reviewed in, Dever, 2002). Considerably less attention is paid to another rate-limiting step during translation: termination. One surprise for new students in the field is that tRNAs do not mediate translation termination. The naïve and arguably preferable way to efficiently terminate translation would be for the cell to possess unacylated tRNAs whose anticodons recognize the three stop codons UAA, UGA and UAG. This is, of course, not the case, but the question is rarely asked: why not? At least part of the answer lies in the fact that the universal genetic code is not quite universal; that is, stop codons do not always encode ‘stop’. For example, UAA and UGA encode glutamine in some ciliates (Preer et al., 1985), UGA encodes cysteine in Euplotes octocarinatus (Meyer et al., 1991), and UGA encodes tryptophan in mitochondria (Macino et al., 1979), and selenocysteine (Sec) in metazoans, archaebacteria and eubacteria. The overall process of Sec incorporation has been recently reviewed from several perspectives (Berry et al., 2001; Copeland and Driscoll, 2001; Hatfield and Gladyshev, 2002; Driscoll and Copeland, 2003). This review should provide a unique perspective by considering the mechanism of Sec incorporation in the context of the three main steps in protein synthesis: initiation, elongation and termination.
2. Overview of Sec incorporation
The dietary requirement for selenium is manifested in its incorporation into an essential set of proteins that utilize selenium as a functional group. There are 19 reported human selenoproteins that are required for a variety of functions ranging from cellular protection against oxidative stress to growth hormone synthesis (reviewed in Gladyshev, 2001; Hatfield and Gladyshev, 2002). The elements of Sec incorporation in eubacteria, archeae and eukaryotes are identical with regard to the requirement for an in-frame UGA codon to specify Sec, a unique Sec-tRNASec, a unique elongation factor to deliver the tRNA and a cis-acting element in the selenoprotein mRNA that forms a stem-loop structure. Beyond these general specifications, the mechanisms for Sec incorporation differ dramatically. The stem loop is universally referred to as a selenocysteine insertion sequence (SECIS) element, but the structure and conserved sequences are not shared among the three lines of descent. In Escherichia coli, the SECIS element is immediately downstream of the Sec codon within the coding region. In archaebacteria and eukarya, the SECIS element is found in the 3′ untranslated region (UTR). The elongation factor required in each case is distinct, but they do appear to be distantly related. The most significant difference between bacterial and eukaryal Sec incorporation is that in the former the insertion event requires only a single elongation factor (SelB) that binds to the SECIS element and the Sec-tRNASec thus delivering Sec to the polypeptide chain in a SECIS element and codon-specific manner. In eukaryotes, Sec incorporation requires both an elongation factor (eEFSec) and a separate SECIS binding protein (SBP2). The simple model for the function of these two factors dictates that the SBP2/SECIS complex interacts with the eEFSec/Sec-tRNASec complex to deliver Sec to SECIS containing mRNAs. The mechanistic details concerning the activity of these two factors is discussed in detail below.
3. The players
3.1. Selenoprotein mRNAs
Eukaryotic selenoprotein mRNAs share two invariant features: one SECIS element in the 3′ UTR and one in-frame UGA codon within the coding region. There is, of course, one exception to this rule. Mammalian selenoprotein P mRNA contains ten in-frame UGA codons and two functional SECIS elements. Most SECIS elements contain a conserved AUGA motif 9–12 nt upstream of a conserved AAA/G (AAR) motif found in the terminal bulge or loop. The 200 nucleotide SECIS element found in the phospholipid hydroperoxide glutahione peroxidase (PHGPx) 3′ UTR is shown in Fig. 1 as an example. Only those nucleotides in red are conserved among all known SECIS elements. Opposite the AUGA motif lies a conserved dinucleotide, GA, which via a non-Watson/Crick base pair with the AUGA forms what is termed the SECIS core. There are several exceptions to these rules which possess variations at the adenosine in the AUGA motif and several variations in the AAR motif including the mammalian selenoprotein M (Sel M) SECIS, which possess a CCC in place of AAR. All of these variants have been described in detail very recently (Krol, 2002). Interestingly, mutations in this CCC motif showed that either AAC or CCC promoted Sec incorporation equally well, but changes to UUC or GGC completely abolished Sec incorporation (Korotkov et al., 2002). Since no specific function has been assigned to the AAR motif, it is difficult to speculate how this single SECIS element evolved to relax the sequence requirements at this position. Although it is not immediately apparent, it is tempting to suggest that a compensatory mechanism lies within the Sel M SECIS itself.
Fig. 1.
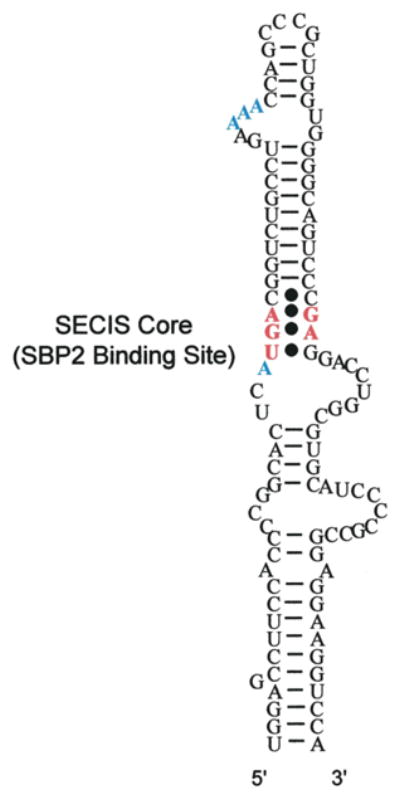
SECIS element of phospholipid hydroperoxide glutathione peroxidase. Residues that are found in all SECIS elements are in red boldface. Residues that are conserved but not universal are shown in blue boldface.
3.2. Selenocysteyl tRNA
The Sec codon is deciphered by a unique tRNA possessing a UCA anticodon (Lee et al., 1989). The Sec-tRNA is first serylated, then the Ser-tRNASec is converted to Sec-tRNASec by the action of selenocysteine synthase. Elimination of the Sec-tRNA gene in the mouse is early embryonic lethal, but heterozygotes are normal despite enduring a 50–80% reduction in Sec-tRNA levels, suggesting that the Sec-tRNASec is not limiting in vivo (Bosl et al., 1997). The Sec-tRNASec is unusual in that it is expressed as two equally abundant isoforms: with or without a methylated U in the anticodon (position 34). The level of the methylated isoform is reduced when a mutant tRNA that lacks the isopentenyladenosine at position 37 is overexpressed in the mouse. Interestingly, the relative levels of selenoproteins in a variety of tissues is also changed under these conditions, suggesting a relationship between isoform abundance and efficiency of Sec incorporation (Moustafa et al., 2001). This effect does not appear to be at the level of elongation factor binding affinity as each isoform binds eEFSec with equal affinity (Tujebajeva et al., 2000).
3.3. eEFSec
The effector of Sec-tRNASec delivery, eEFSec (also known as mSelB), specifically interacts with aminoacylated Sec-tRNASec but not Ser-tRNASer or Ser-tRNASec (Fagegaltier et al., 2000; Tujebajeva et al., 2000). eEFSec shares minimal sequence identity with eEF1A (20%), and it is postulated that like SelB, eEFSec does not require a guanine nucleotide exchange factor (GEF). This is experimentally supported by the fact that the affinity for GDP is two to three times lower than for GTP (Fagegaltier et al., 2000; Tujebajeva et al., 2000). Although eEFSec has not been demonstrated to interact directly with the SECIS element, it does appear to either directly or indirectly interact with SBP2 or the SBP2/SECIS complex (Tujebajeva et al., 2000; Zavacki et al., 2003).
3.4. SBP2
SBP2 is an 846 amino acid protein whose C-terminal 450 amino acids are sufficient for the execution of all known SBP2 functions (Copeland et al., 2000, 2001). These functions include SECIS binding, Sec incorporation and ribosome binding. SBP2 binds specifically to the SECIS core as demonstrated by both mutagenesis of the SECIS element and RNA footprinting (Lesoon et al., 1997; Fletcher et al., 2001). SBP2 binding is not affected by mutations in the conserved AAR motif, leaving this part of the SECIS with no known function or binding partner. The introduction of SBP2 into an in vitro translation reaction increases selenoprotein synthesis by ~20-fold, and immunodepletion eliminates Sec incorporation in vitro (Copeland et al., 2000). The precise role of SBP2 in Sec incorporation remains unclear, but recent evidence indicates that it stably interacts with ribosomes and it appears to do so by binding 28S rRNA. This suggests that SBP2 may be modifying a subset of ribosomes, thereby ‘marking’ those that are competent for Sec incorporation (Copeland et al., 2001). A structure/function analysis utilizing truncated and mutagenized forms of SBP2 described distinct SECIS binding and Sec incorporation domains (Copeland et al., 2001), the latter of which is a likely candidate for non-SECIS interactions with either the ribosome and/or eEFSec as discussed below.
4. Initiation
The link between Sec incorporation and translation initiation may seem tenuous, but the well established communication between the 3′ end of mRNA and the initiation apparatus suggests that the SBP2/SECIS complex may participate in the initiation process. There appear to be two major pathways for 3′–5′ communication during translation initiation. First, the poly(A) tail positively influences translation by delivering the poly(A) binding protein (PABP) to the cap binding complex, specifically eIF4G (Sachs, 2000). The interaction between PABP and eIF4G stimulates translation synergistically, perhaps by promoting rapid reinitiation on a ‘circular’ mRNA. Second, the 3′ UTRs of many mRNAs also regulate translation, albeit usually in a negative fashion. One of the best-studied examples is that of 15-lipoxygenase (LOX) being regulated by hnRNP K and E1/E2 by means of a differentiation control element (DICE) in the 3′ UTR. This protein/RNA complex interferes with translation initiation by preventing 60S ribosomal subunit joining (Ostareck et al., 2001). Other well-defined examples of translational regulation by 3′ UTRs include tra-2 in Caenorhabditis elegans (Thompson et al., 2000), Vg1 in Xenopus (Otero et al., 2001), ceruloplasmin in human monocytes (Mazumder et al., 2001) and the multiple targets of the PUF proteins which appear to function similarly in all eukaryotes (reviewed in Wickens et al., 2002).
The above is presented as support for the argument that the SECIS element/SBP2 complex may regulate translation initiation in a SECIS-dependent fashion. Interestingly, a SECIS-like element was recently found to modulate translation initiation of the Sel AB transcript in E. coli, apparently as a negative feedback translational repressor that is both Sel B and tRNA-dependent (Thanbichler and Bock, 2002). Although experimental evidence for a specific role for initiation in eukaryotic Sec incorporation is lacking, it is logical that the set of ribosomes decoding the 10 Sec codons in Sel P, for instance, might be specifically and rapidly recruited during initiation to allow the processive incorporation of Sec. One caveat to this model is that SBP2 does not appear to behave in a fashion similar to PABP or hnRNP K/E1/E2 proteins which are brought to the initiation reaction by the mRNA. Since SBP2 stably binds ribosomes, and does so in a SECIS-independent fashion (Copeland et al., 2001), it is likely that SBP2 does not directly recruit SECIS elements to the ribosome, but that this may require one or more translation factors. Whether these factors are involved in initiation or more specifically for delivering selenoprotein mRNAs to an SBP2-bound ribosome requires further investigation. Of course, one of the key questions regarding SBP2 is the functional relevance of ribosome binding, and this may be partly addressed by an analysis of exactly where SBP2 interacts with the ribosome. Interestingly, the SECIS element can be considered a member of a recently identified RNA motif, called the kink-turn, six of which are found in the ribosomal large subunit (Klein et al., 2001), and preliminary evidence suggests that one or more of these does, indeed, allow SBP2 binding (P.R. Copeland, unpublished observation). The function of these elements during translation is not known, so determining which kink-turn may be linking SBP2 to the ribosome will only begin to answer questions about the functional significance of the SBP2/ribosome interaction.
5. Elongation
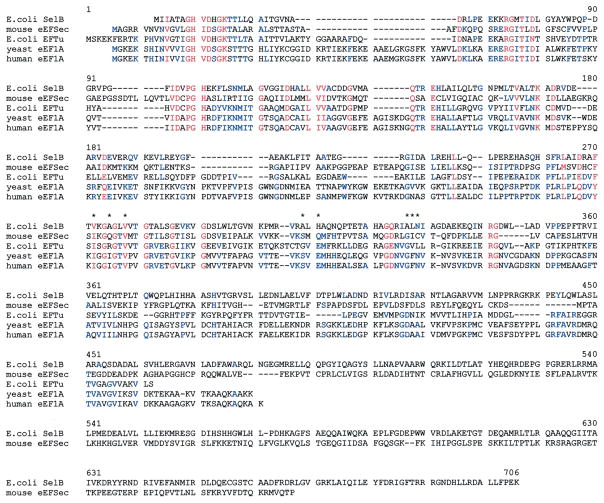
The translation elongation cycle revolves around delivery of a cognate aminoacyl-tRNA to the ribosomal A-site by GTP-bound eEF1A (reviewed in Anand et al., 2001). GTP is hydrolyzed during the codon recognition step and eEF1A-GDP leaves the A-site. eEF1A is then bound by eEF1B which acts as a GEF thus returning eEF1A to the GTP bound state. To investigate the potential significance of the major differences between eEFSec and eEF1A, we analyzed a sequence alignment of the relevant elongation factors shown in Fig. 2. All amino acid positions noted below refer to the human eEF1A sequence. The most striking differences between the two proteins are as follows: a deletion of D35 through W58, an insertion between T88 and I89, a deletion from A125 through G131, and a deletion of N182 through W194. Fig. 3 shows two views of the structural data from the eEF1A-eEF1Bα-GDPNP co-crystal (Andersen et al., 2001) modeled using the Protein Explorer software developed by Eric Martz (http://www.proteinexplorer.org). The D35–W58 deletion (Fig. 3A) significantly shortens the switch I region. The A125–G131 deletion seems minor, but a mutation in this region is known to cause a defect in translational fidelity (increased +1 frameshifting) in yeast (Kinzy and Woolford, 1995). Interestingly, the N182–W194 deletion removes residues that make contacts with GTP/GDP in eEF1A. This significant change in the binding site for nucleotide may be part of the explanation for the observed higher affinity for GTP than GDP. The insertion between T88 and I89 is also in an interesting location as it may well project into the cleft where both eEF1Bα and aminoacyl-tRNA bind. In such a position, it may provide part of the specificity for Sec-tRNASec.
Fig. 2.
Amino acid alignment of canonical and Sec-specific elongation factors. Similar amino acids are in blue, highly conserved and identical residues in red. Asterisks denote the contacts between yeast eEF1A and F163 in eEF1Bα as described in Section 3 in the text. The alignment was generated by Multalin (Corpet, 1988).
Fig. 3.
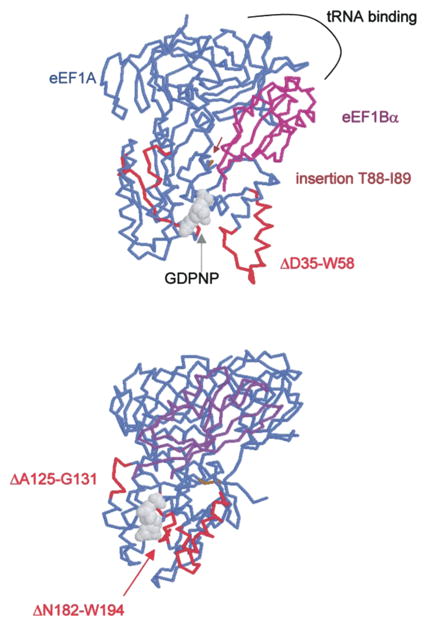
Crystal structure of the eEF1A/eEF1Bα/GDPNP complex as reported in Andersen et al. (2000). Modeling was performed with Protein Explorer (Eric Martz) using PDB coordinates 1G7C. eEF1A is in blue, eEF1Ba is in magenta, and regions of eEF1A that are deleted in eEFSec are in red. The two amino acids flanking an insertion in eEFSec are in brown.
As mentioned above, eEFSec is likely not to require GEF activity, but it may still interact with eEF1B which has been hypothesized to play a non-GEF function in yeast (Anand et al., 2001). Indeed, the co-crystal structure shown in Fig. 3 illustrates that that the majority of eEF1A-eEF1Bα interactions occur not at the eEF1A GTP-binding domain, but in the tRNA binding domain (domain II). Phenylalanine 163 of eEF1Bα projects into a hydrophobic pocket making specific contacts with eEF1A, and this site significantly overlaps with the proposed tRNA binding site in eEF1A. This data is supported by the observation that an eEF1Bα mutation that eliminates GEF activity has no affect on eEF1Bα binding to eEF1A (Andersen et al., 2000). These results led the authors to speculate that eEF1Bα may physically interact with eEF1A until the ternary complex is formed, perhaps channeling the molecules necessary for the next round of peptide bond formation. In the context of Sec incorporation, eEF1Bα could play a similar role in allowing processive incorporation of Sec into Sel P. In Fig. 3, the amino acid positions marked with an asterisk represent those known to make contacts with eEF1Bα at F163 (L. Valente and T.G. Kinzy, pers. comm; Andersen et al., 2000). While the eEFSec and eEF1A sequences are not identical at these positions, they may be conserved enough to support eEF1Bα binding, and the differences may help to explain the specificity for eEFSec binding to Sec-tRNASec. It is, of course, also possible that the other two members of the eEF1B complex (β and γ) interact with eEFSec in a larger complex as well. These hypotheses remain to be tested experimentally.
The role of SBP2 during elongation is less clear. The principal issue is whether eEFSec and SBP2 interact, when they interact and whether the interaction is essential for Sec incorporation. The first point has been addressed in two reports. First, Tujebajeva et al. (2000) showed that SBP2 and eEFSec were co-immunoprecipitated from cells over-expressing the two factors. Interestingly, this interaction appeared to be RNA-dependent as RNAse treatment prior to immunoprecipitation eliminated or significantly reduced the interaction. Subsequently, this interaction was shown to be strongly tRNA dependent and directed by the C-terminal extension of eEFSec (amino acids 448–520; see Fig. 2; Zavacki et al., 2003). SBP2 does not appear to interact with full-length eEFSec in the absence of excess Sec-tRNASec, but it does directly interact with the C-terminus in isolation, perhaps suggesting that Sec-tRNASec binding causes a conformational change allowing SBP2 binding (Zavacki et al., 2003). Whether this interaction is required is still formally not proven, but it seems likely that the SBP2/SECIS complex is somehow informing Sec-tRNASec-bound eEFSec which UGA codons should be decoded as Sec, perhaps by providing a high-affinity binding site on the ribosome. The tRNA requirement assures the formation of active complex and allows free eEFSec to bind another charged Sec-tRNASec.
The question of when the SBP2/eEFSec interaction occurs is only important in the context of selenoprotein mRNA recruitment. Since SBP2 is known to bind the SECIS element in the absence of Sec-tRNASec and eEFSec binding, it is likely that the recruitment step occurs in conjunction with or just subsequent to initiation. Resolving this issue depends largely on whether SBP2 forms a homo-multimeric complex, and if so, the timing of SBP2 multimerization (if it is dynamic) may be the most relevant. One could envision a model where one molecule of SBP2 is bound to a SECIS element and another on the ribosome and self-association would not occur until the Sec codon is reached. Alternatively, SBP2 could form a complex in the absence of a SECIS or ribosome. There are several pieces of evidence that support SBP2 multimerization: SBP2 was shown to sediment in a ≥500 kDa complex during gel filtration of a partially purified SBP2 preparation (Copeland and Driscoll, 1999), glycerol gradient sedimentation of purified, recombinant SBP2 showed the formation of a salt-sensitive complex (Copeland et al., 2001), and in mobility shift assays, increasing concentrations of SBP2 yielded both high and low-mobility complexes (Fletcher et al., 2001; Lescure et al., 2002). An attractive model, therefore, would be that SBP2 is bound to the ribosome as a dimer with one RNA binding domain interacting with 28S rRNA and the other with an incoming SECIS element (see Fig. 4). This would suggest that SBP2 is modifying a subset of ribosomes which would be primed for processive Sec incorporation. This model is supported by data regarding the sequences within SBP2 that are required for ribosome binding. Interestingly, ribosome binding was negated or severely diminished as a result of deletions in either the SECIS binding or Sec incorporation domain. However, a point mutation in the SECIS binding domain (G669R) eliminated SECIS binding and Sec incorporation, but had no effect on ribosome binding (Copeland et al., 2001). So, although ribosome binding may utilize the SECIS binding domain, the protein/RNA interactions are not identical.
Fig. 4.
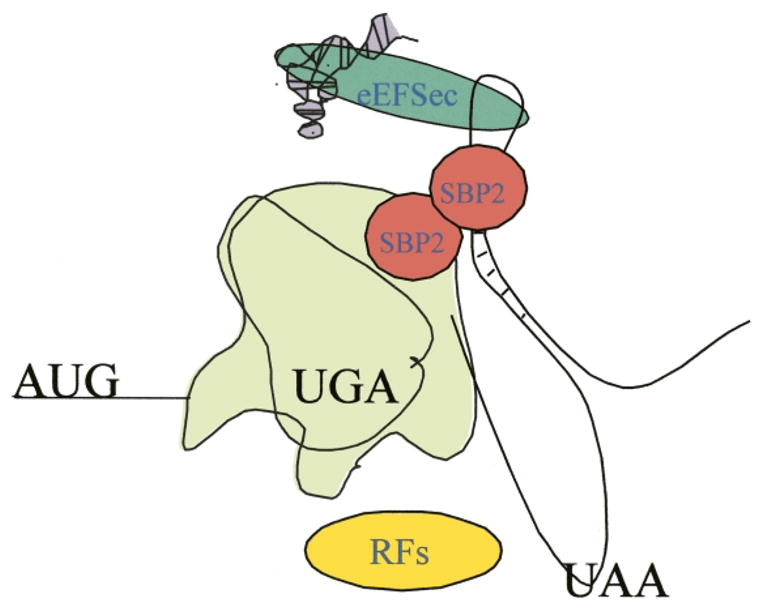
A model of Sec incorporation. We propose that SBP2 binds as at least a dimer simultaneously to the ribosome and the SECIS element and thus may prevent release factor access. In addition, SBP2 may play an active role in delivering the eEFSec/Sec-tRNA complex to the ribosomal A site.
The model in Fig. 4 is unproven, but it seems to be the only model that incorporates all of the currently available data. If SBP2 does not play a role in recruiting SECIS-containing mRNAs then, depending on the concentration of SBP2, the amount of termination at Sec codons would be significant. The combination of low levels of SBP2 and relatively rare selenoprtotein mRNAs might lead to too great a dilution of necessary factors among the large pool of ribosomes. Therefore, it may be more likely that SBP2 does recruit SECIS-containing RNAs and the reason that they were not observed in the SBP2/ribosome complex is that the recruitment requires active translation (see above).
6. Termination
The most overt relationship between Sec incorporation and canonical translation is the inherent conflict in interpreting UGA as Sec or stop. There are two models that describe this interaction. In the simple model, there is no reason to assume that the two processes are even in direct conflict. It is possible that the Sec incorporation ‘switch’ is either on or off. When on, all of the required factors are in place, and if any one of these factors is missing, then the switch is off and the Sec-tRNASec will not be able to act as an authentic suppressor tRNA. In the complex model, the two processes are in dynamic competition. What is of primary interest is whether a ribosomal pause in the absence of termination is required for Sec incorporation. One of the intriguing hypotheses is that the sequence context surrounding the Sec codon plays a critical role determining the existence or length of the pause by providing an inefficient termination codon context. There is some evidence for this hypothesis. First, the context around UGA has been studied in transfected cells, and those contexts that support termination, particularly at the ‘fourth’ base position and the immediately upstream codon, generally are less well suited for Sec incorporation (McCaughan et al., 1995; Nasim et al., 2000; Grundner-Culemann et al., 2001). Selenoprotein P, with its ten Sec codons, is also a good example of the apparent importance of context as it allows both termination and Sec incorporation within the same coding region. Termination apparently ‘outcompetes’ Sec incorporation at codons 2, 3 and 7, allowing the synthesis of distinct isoforms of this selenium-rich protein (Ma et al., 2002). It is worth noting, however, that in nature Sec codons are not always found in poor termination contexts (Driscoll and Copeland, 2003). Thus it is likely that if context is important, it is working in tandem with another factor or cis-element. This brings us back to the central question: does Sec incorporation interfere with translation termination, and if so, at what point? There are at least two factors involved in translation termination in eukaryotes: release factors 1 and 3 (eRF1 and eRF3). eRF1 has been proposed to be a tRNA mimetic by binding to the A-site thus terminating chain elongation. This hypothesis is corroborated by the recently determined crystal structure (Song et al., 2000) and direct crosslinking to the stop codon (Chavatte et al., 2001). The role of eRF3 is unclear, but it does form a complex with eRF1 in vivo and in vitro (Zhouravleva et al., 1995), and is essential in yeast (Ter-Avanesyan et al., 1993). Interestingly, eRF3 has recently been proposed to be yet another link between the 3′ and 5′ end of translating mRNAs. Hoshino et al. (1999) demonstrated that eRF3 also interacts with PABP, and more recent work has suggested that eRF3 may play a role in ribosome re-initiation (Uchida et al., 2002). The lack of knowledge regarding SBP2 function precludes a guess about where it might interfere with termination, but it is not likely to be direct as SBP2 does not physically interact with either eRF1 or eRF3 in vitro (P.R. Copeland, unpublished observation). If SBP2 does interfere with termination, then there might be a noticeable pause in translation even when Sec incorporation is efficient and SBP2 is in excess, perhaps even at non-Sec codons. Two recent analyses of the polyribosomes that translate selenoprotein mRNAs have, indeed, indicated that there is considerable pausing and termination at the Sec codon (Fletcher et al., 2000; Martin and Berry, 2001). When SBP2 was added in vitro, ribosome loading increased, but these results are difficult to interpret because the vast majority of the mRNA is not found in the polyribosome fraction. It is worth noting that the polyribosome analysis performed for an endogenous selenoprotein mRNA (PHGPx) was also found to be underloaded in cells and mouse liver, but it is likely that SBP2 was limiting in these experiments (Fletcher et al., 2000). The polyribosome profile of selenoprotein mRNAs in testis, where SBP2 is apparently abundant (Copeland et al., 2000), has not been reported.
Translation termination is also linked to another process that has bearing on the success or failure of Sec incorporation: nonsense mediated decay (NMD). This is a process by which most mRNAs that possess premature stop codons are rapidly degraded. Since most, if not all, seleno-protein mRNAs fit the rules for being subject to NMD (Sun et al., 2000), it is not surprising that mRNA decay is a factor in the regulation of selenoprotein expression. In fact, under conditions of limiting selenium, differential mRNA stability appears to be the primary means of regulation for seleno-protein expression. However, some selenoprotein mRNAs are resistant to NMD even under conditions of limited selenium (Lei et al., 1995). It is possible that SBP2 might play a direct role in mRNA stability by functioning like the iron responsive protein (IRP-1) which binds to and stabilizes the transferrin mRNA (Erlitzki et al., 2002). In this scenario, SECIS elements with the highest affinity for SBP2 would be more stable than those with lower affinity. Currently, there is no data to support this model. Since the SECIS element alone is not sufficient to alter mRNA stability (Weiss and Sunde, 1998), it is more likely that variable mRNA stability is a direct result of the efficiency of translation termination which is regulated both by the codon context and the efficiency of Sec incorporation. This hypothesis is clearly supported by analyses of NMD in yeast where components of the termination complex are known to interact with factors required for NMD (Wang et al., 2001). One would therefore predict that when Sec incorporation is successful, selenoprotein mRNAs are stable and when translation results in termination, they are unstable. However, this does not appear to be the case, as it has been shown that the steady-state level of GI-GPx mRNA is actually increased (in the absence of increased transcription) during selenium deficiency even though the level of protein decreases to undetectable levels (Wingler et al., 1999). Something about the GI-GPx mRNA and its relationship to the Sec incorporation machinery keeps it from being degraded even in the absence of complete translation. One as-yet untested possibility is that GI-GPx does not allow termination and thus remains polyribosome associated but stalled at the Sec codon while other selenoprotein mRNAs are efficiently terminated. The resolution of this seeming paradox will undoubtedly shed light on both the intricacies of Sec incorporation and translation termination.
7. Conclusions
The incorporation of selenocysteine into protein provides a unique vantage point for the observation of protein synthesis. Each phase of canonical translation is likely modified specifically for this process, and the elucidation of its mechanics will be an essential part of the complete body of knowledge regarding eukaryotic translation.
Acknowledgments
The author thanks Louis Valente, Terri Goss Kinzy, Pedro A. Ortiz and Cheryl Rebsch for helpful comments and discussions.
Abbreviations
- SECIS
selenocysteine insertion sequence
- UTR
untranslated region
- Sec
selenocysteine
- GEF
guanine nucleotide exchange factor
- NMD
nonsense mediated decay
References
- Anand M, Valente L, Carr-Schmid A, Munshi R, Olarewaju O, Ortiz PA, Kinzy TG. Translation elongation factor 1 functions in the yeast Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 2001;66:439–448. doi: 10.1101/sqb.2001.66.439. [DOI] [PubMed] [Google Scholar]
- Andersen GR, Pedersen L, Valente L, Chatterjee I, Kinzy TG, Kjeldgaard M, Nyborg J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Bα. Mol Cell. 2000;6:1261–1266. doi: 10.1016/s1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- Andersen GR, Valente L, Pedersen L, Kinzy TG, Nyborg J. Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Bα complex. Nat Struct Biol. 2001;8:531–534. doi: 10.1038/88598. [DOI] [PubMed] [Google Scholar]
- Berry MJ, Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Martin IG, Low SC, Mansell JB, Grundner-Culemann E, Harney JW, Driscoll DM, Hatfield DL. Selenocysteine incorporation directed from the 3′UTR: characterization of eukaryotic EFsec and mechanistic implications. Biofactors. 2001;14:17–24. doi: 10.1002/biof.5520140104. [DOI] [PubMed] [Google Scholar]
- Bosl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Natl Acad Sci USA. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Frolova L, Kisselev L, Favre A. The polypeptide chain release factor eRF1 specifically contacts the sUGA stop codon located in the A site of eukaryotic ribosomes. Eur J Biochem. 2001;268:2896–2904. doi: 10.1046/j.1432-1327.2001.02177.x. [DOI] [PubMed] [Google Scholar]
- Copeland PR, Driscoll DM. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J Biol Chem. 1999;274:25447–25454. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- Copeland PR, Driscoll DM. RNA binding proteins and selenocysteine. Biofactors. 2001;14:11–16. doi: 10.1002/biof.5520140103. [DOI] [PubMed] [Google Scholar]
- Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland PR, Stepanik VA, Driscoll DM. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol Cell Biol. 2001;21:1491–1498. doi: 10.1128/MCB.21.5.1491-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- Erlitzki R, Long JC, Theil EC. Multiple, conserved iron responsive elements in the 3′ untranslated region of transferrin receptor mRNA enhance binding of iron regulatory protein 2. J Biol Chem. 2002;27:27. doi: 10.1074/jbc.M207918200. [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JE, Copeland PR, Driscoll DM. Polysome distribution of phospholipid hydroperoxide glutathione peroxidase mRNA: evidence for a block in elongation at the UGA/selenocysteine codon. RNA. 2000;6:1573–1584. doi: 10.1017/s1355838200000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JE, Copeland PR, Driscoll DM, Krol A. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA. 2001;7:1442–1453. [PMC free article] [PubMed] [Google Scholar]
- Gladyshev VN. Identity, evolution and function of selenoproteins and selenoprotein genes. In: Hatfield DL, editor. Selenium: Its Molecular Biology and Role in Human Health. Kluwer Academic Publishers; Boston: 2001. [Google Scholar]
- Grundner-Culemann E, Martin GW, 3rd, Tujebajeva R, Harney JW, Berry MJ. Interplay between termination and translation machinery in eukaryotic selenoprotein synthesis. J Mol Biol. 2001;310:699–707. doi: 10.1006/jmbi.2001.4809. [DOI] [PubMed] [Google Scholar]
- Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- Kinzy TG, Woolford JL., Jr Increased expression of Saccharomyces cerevisiae translation elongation factor 1 α bypasses the lethality of a TEF5 null allele encoding elongation factor 1 beta. Genetics. 1995;141:481–489. doi: 10.1093/genetics/141.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Novoselov SV, Hatfield DL, Gladyshev VN. Mammalian selenoprotein in which selenocysteine (sec) incorporation is supported by a new form of sec insertion sequence element. Mol Cell Biol. 2002;22:1402–1411. doi: 10.1128/mcb.22.5.1402-1411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A. Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis. Biochimie. 2002;84:765–774. doi: 10.1016/s0300-9084(02)01405-0. [DOI] [PubMed] [Google Scholar]
- Lee BJ, Worland PJ, Davis JN, Stadtman TC, Hatfield DL. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon. UGA J Biol Chem. 1989;264:9724–9727. [PubMed] [Google Scholar]
- Lei XG, Evenson JK, Thompson KM, Sunde RA. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J Nutr. 1995;125:1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- Lescure A, Allmang C, Yamada K, Carbon P, Krol A. cDNA cloning, expression pattern and RNA binding analysis of human selenocysteine insertion sequence (SECIS) binding protein 2. Gene. 2002;291:279–285. doi: 10.1016/s0378-1119(02)00629-7. [DOI] [PubMed] [Google Scholar]
- Lesoon A, Mehta A, Singh R, Chisolm GM, Driscoll DM. An RNA-binding protein recognizes a mammalian selenocysteine insertion sequence element required for cotranslational incorporation of selenocysteine. Mol Cell Biol. 1997;17:1977–1985. doi: 10.1128/mcb.17.4.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Hill KE, Caprioli RM, Burk RF. Mass spectrometric characterization of full-length rat selenoprotein P and three isoforms shortened at the C terminus. Evidence that 3 UGA codons in the mRNA open reading frame have alternative functions of specifying selenocysteine insertion or translation termination. J Biol Chem. 2002;30:30. doi: 10.1074/jbc.M111462200. [DOI] [PubMed] [Google Scholar]
- Macino G, Coruzzi G, Nobrega FG, Li M, Tzagoloff A. Use of the UGA terminator as a tryptophan codon in yeast mitochondria. Proc Natl Acad Sci USA. 1979;76:3784–3785. doi: 10.1073/pnas.76.8.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin IG, Berry MJ. Selenocysteine codons decrease polysome association on endogenous selenoprotein mRNAs. Genes Cells. 2001;6:121–129. doi: 10.1046/j.1365-2443.2001.00402.x. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Imataka H, Sonenberg N, Fox PL. Translational silencing of ceruloplasmin requires the essential elements of mRNA circularization: poly(A) tail, poly(A)-binding protein, and eukaryotic translation initiation factor 4G. Mol Cell Biol. 2001;21:6440–6449. doi: 10.1128/MCB.21.19.6440-6449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci USA. 1995;92:5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Schmidt HJ, Plumper E, Hasilik A, Mersmann G, Meyer HE, Engstrom A, Heckmann K. UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. Proc Natl Acad Sci USA. 1991;88:3758–3761. doi: 10.1073/pnas.88.9.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa ME, Carlson BA, El-Saadani MA, Kryukov GV, Sun QA, Harney JW, Hill KE, Combs GF, Feigenbaum L, Mansur DB, Burk RF, Berry MJ, Diamond AM, Lee BJ, Gladyshev VN, Hatfield DL. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol Cell Biol. 2001;21:3840–3852. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim MT, Jaenecke S, Belduz A, Kollmus H, Flohe L, McCarthy JE. Eukaryotic selenocysteine incorporation follows a non-processive mechanism that competes with translational termination. J Biol Chem. 2000;275:14846–14852. doi: 10.1074/jbc.275.20.14846. [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation. The 3′ UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Otero LJ, Devaux A, Standart N. A 250-nucleotide UA-rich element in the 3′ untranslated region of Xenopus laevis Vg1 mRNA represses translation both in vivo and in vitro. RNA. 2001;7:1753–1767. [PMC free article] [PubMed] [Google Scholar]
- Preer JR, Jr, Preer LB, Rudman BM, Barnett AJ. Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature. 1985;314:188–190. doi: 10.1038/314188a0. [DOI] [PubMed] [Google Scholar]
- Sachs A. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. pp. 447–456. [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1 – mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Sun X, Moriarty PM, Maquat LE. Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position. EMBO J. 2000;19:4734–4744. doi: 10.1093/emboj/19.17.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Bock A. The function of SECIS RNA in translational control of gene expression in Escherichia coli. EMBO J. 2002;21:6925–6934. doi: 10.1093/emboj/cdf673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR, Goodwin EB, Wickens M. Rapid deadenylation and poly(A)-dependent translational repression mediated by the Caenorhabditis elegans tra-2 3′ untranslated region in Xenopus embryos. Mol Cell Biol. 2000;20:2129–2137. doi: 10.1128/mcb.20.6.2129-2137.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:1–6. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J Biol Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, Peltz SW. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001;20:880–890. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SL, Sunde RA. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA. 1998;4:816–827. doi: 10.1017/s1355838298971990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wingler K, Bocher M, Flohe L, Kollmus H, Brigelius-Flohe R. mRNA stability and selenocysteine insertion sequence efficiency rank gastrointestinal glutathione peroxidase high in the hierarchy of selenoproteins. Eur J Biochem. 1999;259:149–157. doi: 10.1046/j.1432-1327.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- Zavacki AM, Mansell JB, Chung M, Klimovitsky B, Harney JW, Berry MJ. Coupled tRNASec-dependent assembly of the selenocysteine decoding apparatus. Mol Cell. 2003;11:773–781. doi: 10.1016/s1097-2765(03)00064-9. [DOI] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]