Abstract
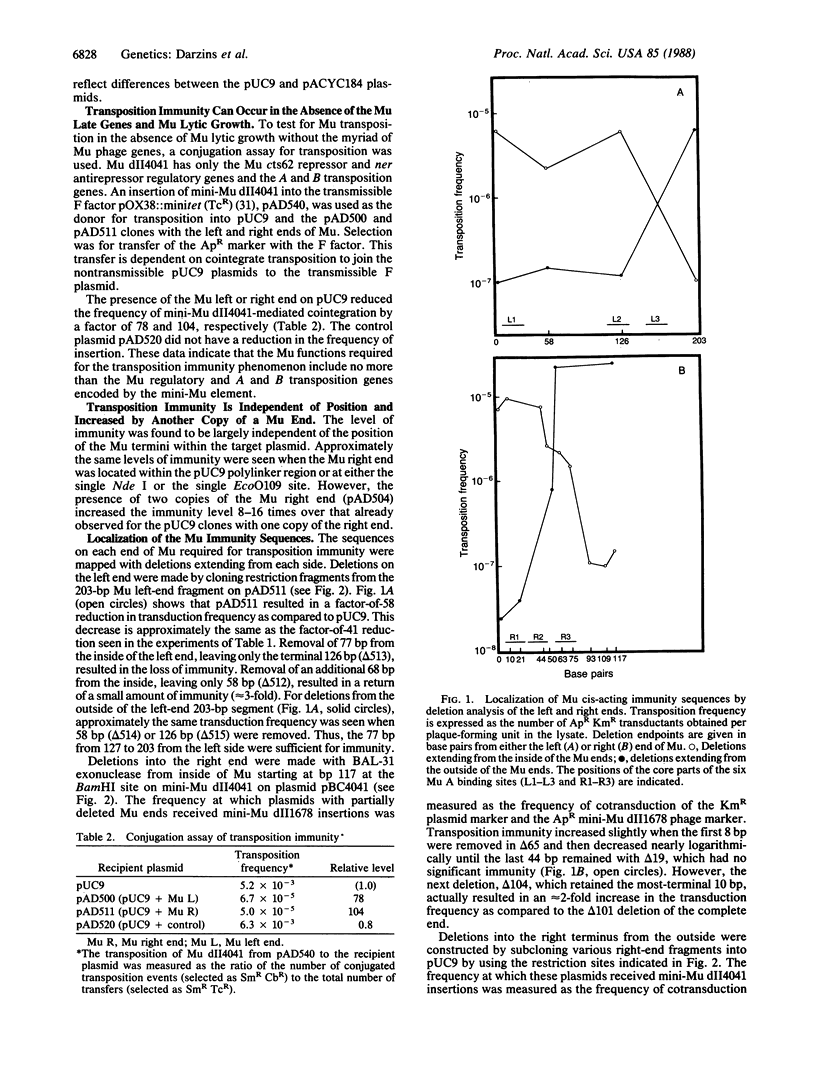
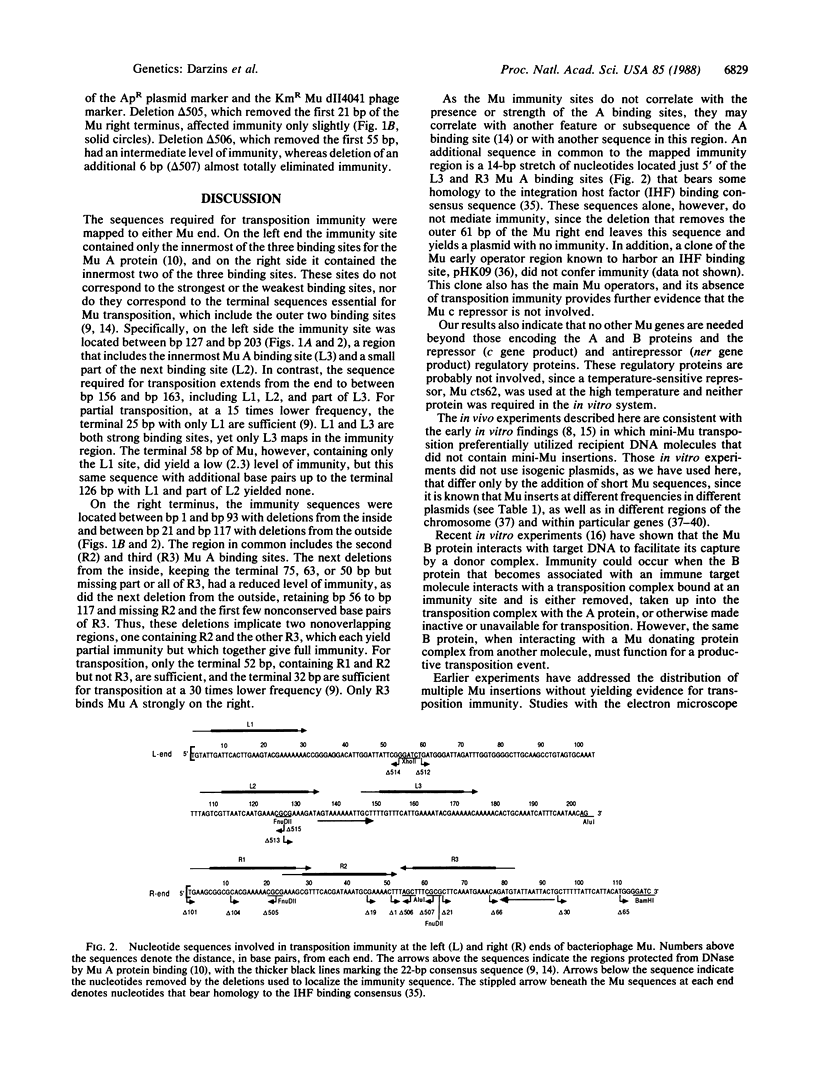
Plasmids with bacteriophage Mu sequences receive additional Mu insertions 20-700 times less frequently than plasmids without Mu sequences. The Mu sites required for this transposition immunity were mapped near each end, either of which was sufficient. The left site was between 127 and 203 base pairs from the left end, and the right site was between 22 and 93 base pairs from the right end. These sequences include the innermost but not the outermost of the three binding sites for the Mu A transposition protein at each end of Mu. Transposition immunity was cis-acting and independent of its location on a target plasmid. An additional copy of an immunity site reduced transposition a factor of 10 further. Transposition immunity was seen both during full phage lytic growth, with all the bacteriophage Mu genes, and during normal cellular growth, with a mini-Mu element containing only the Mu c and ner regulatory and A and B transposition genes.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adzuma K., Mizuuchi K. Target immunity of Mu transposition reflects a differential distribution of Mu B protein. Cell. 1988 Apr 22;53(2):257–266. doi: 10.1016/0092-8674(88)90387-x. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I. Bacteriophage mu as a transposition element. Annu Rev Genet. 1976;10:389–412. doi: 10.1146/annurev.ge.10.120176.002133. [DOI] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G., Harshey R. M., Sarvetnick N., Bukhari A. I. Predominant end-products of prophage Mu DNA transposition during the lytic cycle are replicon fusions. J Mol Biol. 1981 Aug 15;150(3):341–359. doi: 10.1016/0022-2836(81)90551-9. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Transposition of Mu DNA: joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell. 1987 Nov 6;51(3):493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Daniell E., Roberts R., Abelson J. Mutations in the lactose operon caused by bacteriophage Mu. J Mol Biol. 1972 Aug 14;69(1):1–8. doi: 10.1016/0022-2836(72)90019-8. [DOI] [PubMed] [Google Scholar]
- Faelen M., Huisman O., Toussaint A. Involvement of phage Mu-1 early functions in Mu-mediated chromosomal rearrangements. Nature. 1978 Feb 9;271(5645):580–582. doi: 10.1038/271580a0. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A., Resibois A. Mini-muduction: a new mode of gene transfer mediated by mini-mu. Mol Gen Genet. 1979 Oct 3;176(2):191–197. doi: 10.1007/BF00273213. [DOI] [PubMed] [Google Scholar]
- Groenen M. A., Timmers E., van de Putte P. DNA sequences at the ends of the genome of bacteriophage Mu essential for transposition. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2087–2091. doi: 10.1073/pnas.82.7.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M. A., van de Putte P. Analysis of the ends of bacteriophage Mu using site-directed mutagenesis. J Mol Biol. 1986 Jun 20;189(4):597–602. doi: 10.1016/0022-2836(86)90490-0. [DOI] [PubMed] [Google Scholar]
- Groisman E. A., Casadaban M. J. Mini-mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusing. J Bacteriol. 1986 Oct;168(1):357–364. doi: 10.1128/jb.168.1.357-364.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A., Castilho B. A., Casadaban M. J. In vivo DNA cloning and adjacent gene fusing with a mini-Mu-lac bacteriophage containing a plasmid replicon. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1480–1483. doi: 10.1073/pnas.81.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Harshey R. M., McKay R., Bukhari A. I. DNA intermediates in transposition of phage Mu. Cell. 1982 Jun;29(2):561–571. doi: 10.1016/0092-8674(82)90172-6. [DOI] [PubMed] [Google Scholar]
- Howe M. M., Bade E. G. Molecular biology of bacteriophage mu. Science. 1975 Nov 14;190(4215):624–632. doi: 10.1126/science.1103291. [DOI] [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986 Mar 15;261(8):3744–3752. [PubMed] [Google Scholar]
- Kupersztoch Y. M., Helinski D. R. A catenated DNA molecule as an intermediate in the replication of the resistance transfer factor R6K in Escherichia coli. Biochem Biophys Res Commun. 1973 Oct 15;54(4):1451–1459. doi: 10.1016/0006-291x(73)91149-2. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Bhagwat A., Heffron F. Identification of a transposon Tn3 sequence required for transposition immunity. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6765–6769. doi: 10.1073/pnas.80.22.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K. In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell. 1983 Dec;35(3 Pt 2):785–794. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- Mogutov M. A., Velikodvorskaia G. A., Kobets N. S., Piruzian E. S. Immunnost' transpozitsii bakteriofaga Mu. Vliianie mutatsii po genu kil na ustanovlenie immunnosti. Genetika. 1985 Jun;21(6):927–935. [PubMed] [Google Scholar]
- New J. H., Eggleston A. K., Fennewald M. Binding of the Tn3 transposase to the inverted repeats of Tn3. J Mol Biol. 1988 Jun 5;201(3):589–599. doi: 10.1016/0022-2836(88)90640-7. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Roa M., Braun-Breton C., Schwartz M. Structure of the malB region in Escherichia coli K12. I. Genetic map of the malK-lamB operon. Mol Gen Genet. 1979 Jul 24;174(3):241–248. doi: 10.1007/BF00267796. [DOI] [PubMed] [Google Scholar]
- Reyes O., Beyou A., Mignotte-Vieux C., Richaud F. Mini-Mu transduction: cis-inhibition of the insertion of Mud transposons. Plasmid. 1987 Nov;18(3):183–192. doi: 10.1016/0147-619x(87)90061-8. [DOI] [PubMed] [Google Scholar]
- Robinson M. K., Bennett P. M., Richmond M. H. Inhibition of TnA translocation by TnA. J Bacteriol. 1977 Jan;129(1):407–414. doi: 10.1128/jb.129.1.407-414.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumm J. W., Howe M. M. Mu-specific properties of lambda phages containing both ends of Mu depend on the relative orientation of Mu end DNA fragments. Virology. 1981 Oct 30;114(2):429–450. doi: 10.1016/0042-6822(81)90224-5. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Brickman E., Bassford P. J., Jr, Casadaban M. J., Shuman H. A., Schwartz V., Guarente L., Schwartz M., Beckwith J. R. Structure of the malB region in Escherichia coli K12. II. Genetic map of the malE,F,G operon. Mol Gen Genet. 1979 Jul 24;174(3):249–259. doi: 10.1007/BF00267797. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tu C. P., Cohen S. N. Translocation specificity of the Tn3 element: characterization of sites of multiple insertions. Cell. 1980 Jan;19(1):151–160. doi: 10.1016/0092-8674(80)90396-7. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Waggoner B., Pato M., Toussaint A., Faelen M. Replication of mini-Mu prophage DNA. Virology. 1981 Aug;113(1):379–387. doi: 10.1016/0042-6822(81)90163-x. [DOI] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]


