Abstract
Sumoylation has emerged as a major post-translational modification of cellular proteins, affecting a variety of cellular processes. Viruses have exploited the sumoylation pathway to advance their own replication by evolving several ways to perturb the host sumoylation apparatus. However, there has been no report of virally encoded enzymes directly involved in catalyzing the sumoylation reaction. Here, we report that the K-bZIP protein encoded by Kaposi's sarcoma-associated herpesvirus (KSHV) is a SUMO E3 ligase with specificity toward SUMO2/3. K-bZIP is a nuclear factor that functions to modulate viral gene expression and to prolong the G1 phase, allowing viral transcription and translation to proceed at the early stage of infection. In addition to functioning as a transcriptional factor, we show that K-bZIP carries a SIM (SUMO-interacting motif), which specifically binds to SUMO-2/3 but not SUMO-1. K-bZIP catalyzes its own SUMO modification as well as that of its interacting partners such as the cellular tumor suppressor proteins p53 and Rb, both in vitro and in vivo. This reaction depends on an intact SIM. Sumoylation of p53 leads to its activation and K-bZIP is recruited to several p53 target chromatin sites in a SIM-dependent manner. In addition to the identification of a viral SUMO-2/3 E3 ligase, our results provide additional insights into the mechanisms whereby K-bZIP induces cell cycle arrest.
Keywords: p53, Post-translational Modification, Signal Transduction, Sumoylation, Viral Protein, KSHV K-bZIP, SUMO E3 ligase, SUMO-2/3, SUMO-interacting Motifs
Introduction
Increasing evidence indicates that sumoylation, i.e. post-translational modification of proteins by the small ubiquitin-like modifier (SUMO)2 plays a central role in cellular signal transduction. Like phosphorylation, sumoylation is rapid and reversible. In a manner similar to the binding of phosphorylated tyrosine by signal molecules carrying Src homology 2 (SH2) and phosphotyrosine binding (PTB) domains, sumoylated proteins are specifically engaged by proteins with a SUMO-interacting motif (SIM). Modulation of sumoylation has a profound effect on protein-protein interactions and the propagation of cellular signals. Viruses have evolved different mechanisms to exploit the host sumoylation pathway to create a cellular environment that is favorable for viral replication by modulating the functions of viral and cellular proteins (reviewed in Ref. 1). Many viral proteins are themselves sumoylated, and this post-translational modification affects specific functions of these targets. For DNA tumor viruses, the immediate-early and early gene products, which include transcriptional factors, are often sumoylated. Examples include immediate-early 1 (IE 1) and immediate-early 2 (IE 2) proteins of cytomegalovirus (CMV) (2, 3), E1 and E2 of human papillomavirus (HPV) (4, 5), BZLF1 of Epstein-Barr virus (EBV) (6), and K-bZIP of Kaposi's sarcoma-associated herpesvirus (KSHV) (7). Some viral proteins indirectly modulate the sumoylation status of specific cellular proteins. For example, the HPV E7 protein and adenovirus E1A protein block sumoylation of the cellular tumor suppressor Rb (8). Additionally, the KSHV viral protein kinase (ORF36) inhibits the sumoylation of KAP-1 (9). Viruses can also affect global sumoylation of cellular proteins by directly modulating the cellular SUMO machinery. Avian adenovirus Gam1 protein inhibits the SUMO E1-activating enzyme by targeting the SAE1/SAE2 (AOS1/UBA2) heterodimer to cullin RING ligases (CRLs) and promoting SAE1 ubiquitylation and degradation (10). Here, we describe a unique mechanism by which a virus directly modulates sumoylation via the expression of a virally encoded SUMO E3 ligase.
Analogous to ubiquitylation, conjugation of SUMO to target proteins is a multistep process involving an E1-activating enzyme heterodimer SAE1/SAE2, an E2-conjugating enzyme Ubc9 (ubiquitin-like protein SUMO-1-conjugating enzyme 9), and an E3 ligase, which is believed to provide specificity for the sumoylation pathway (11). In stark contrast to ubiquitin E3 ligases, which number in the hundreds to thousands, there are very few cellular SUMO E3 ligases. These include the PIAS (protein inhibitor of activated STATS) family (12, 13), the RanBP2 (Ran-binding protein 2) family (14), and the Pc2 (polycomb 2) family (15). These ligases share the common properties of binding to 1) the SUMO moiety via their SIM domains, 2) Ubc9, the E2 conjugating enzyme, and 3) substrates to which SUMO is subsequently covalently attached. The ability of these ligases to catalyze the sumoylation reaction is demonstrated by reconstituted in vitro sumoylation reactions using purified proteins, including the enzymes, E1, E2, and E3, substrate target proteins, and SUMO proteins. PIAS family members contain a RING domain commonly found in ubiquitin ligases, whereas RanBP2 and Pc2 do not share this feature. Although expression of certain viral proteins has been associated with sumoylation of cellular proteins in vivo, a clear demonstration of a viral protein with SUMO E3 ligase activity has not previously been reported.
KSHV, a gamma herpesvirus, is the causative agent for Kaposi's sarcoma as well as primary effusion lymphoma and multicentric Castleman's disease (16, 17). This virus encodes two major transcriptional factors, K-Rta and K-bZIP (KSHV basic-leucine-zipper). K-Rta is a strong transactivator and a trigger of lytic replication (18, 19), whereas K-bZIP is a strong transcriptional repressor that attenuates the activity of K-Rta (20). We previously reported that K-bZIP is highly sumoylated at a single lysine residue (Lys-158) and that its repressive activity depends on sumoylation (7, 21). Upon phosphorylation of K-bZIP at Thr-111 by either the cell cycle protein CDK2 or the viral protein kinase (ORF36), the sumoylation of K-bZIP is inhibited; thus, phosphorylation converts K-bZIP from functioning as a transcriptional repressor to a transactivator (21). K-bZIP is a nuclear protein distributed in the nucleoplasm as well as in PML bodies (7). This viral protein has multiple interacting partners, including the cellular tumor suppressor p53 (22, 23), and is a modulator of p53 and its recruitment to PML bodies (24). The latter process is likely to be SUMO-related because PML bodies are storage sites for sumoylated proteins (25). Sumoylation of most transcriptional factors results in a reduction of their transactivation activities. However, p53 is a rare exception, because sumoylation increases its transcriptional activity on target promoters (26, 27). In addition to its activity as a transcriptional factor, K-bZIP plays a role in viral genome replication (28), and G1 cell cycle arrest (29–31). We hypothesize that these activities of K-bZIP, taken together, provide a cellular environment conducive for the expression of viral early genes (reviewed in Ref. 32).
In this study, using purified components required for SUMO modification, we provide evidence that K-bZIP is a SIM-containing poly-SUMO-specific E3 ligase. Overexpression of K- bZIP increased global sumoylation of cellular proteins in a SUMO-2/3- and SIM-dependent manner. Enhancement of the sumoylation of p53 by K-bZIP was accompanied by the recruitment of K-bZIP to p53 target promoters, and an increase in SUMO-2/3 content at these promoters. Cumulatively, these events lead to an increase of p21, providing an additional mechanism whereby K-bZIP expression prolongs the G1 phase of the cell cycle. Perhaps the most significant finding of this study is the first demonstration of a viral protein, which functions as a SUMO-2/3 E3 ligase.
EXPERIMENTAL PROCEDURES
Cell Culture and Plasmid DNA
293 and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). SaOS-2 cells were cultured in McCoy's 5a Medium containing 10% FBS. Inducible TREx-F-K-bZIP wt, L75A, and dDL 293 cells were generated by Flp-In TREx system and cultured in DMEM, 10% FBS, 15 μg/ml of blasticidin, and 100 μg/ml of hygromycin (Invitrogen). The pcDNA3-F-K-bZIP, pcDNA3-T7-Ubc9, pcDNA3-F-K-Rta, pGL3-Orf57-Luc, pGEX-SUMO-1GG, pGEX-SUMO-2GG, pGEX-SUMO-3GG, and pFastBacTM1-F-KbZIP were described previously (7, 21). SIM mutants of K-bZIP, L75A and dDL, were generated by site-directed mutagenesis using mutagenesis oligonucleotide primers (K-bZIP-L75A-F: 5′-GAAACGGTCATTGACGCTACAGCGCCTTCCC-3′; K-bZIP-L75A-R: 5′-GGGAAGGCGCTGTAGCGTCAATGACCGTTTC-3′; K- bZIP-dDL-F: 5′-GTGAAACGGTCATTACAGCGCCTTCCC-3′; K-bZIP-dDL-R: 5′-GGGAAGGCGCTGTAATGACCGTTTCAC-3′). To generate inducible TREx-F-K-bZIP wt, L75A, and dDL 293 cells, wild-type K-bZIP and its SIM mutants were subcloned into pcDNA5/FRT/TO. To generate recombinant proteins of F-K-bZIP mutants, L75A and dDL in the baculovirus protein expression system, K-bZIP, L75A, and dDL were subcloned into pFastBacTM1-F-KbZIP. pEGFP-C1 was used to generate GFP-SUMO-1, -2, and -3. p53-responsive luciferase reporter vector, 4XBS2WT-Luc, containing two copies of wild-type p53 binding site, was purchased from Addgene Inc. (Cambridge, MA).
Protein Expression and GST Pull-down Assay
GST-tagged proteins were expressed in BL21(DE3) cells and purified with glutathione-SepharoseTM-High Performance beads (GE Healthcare). Equal amounts of GST and GST-tagged proteins were mixed with 0.1 μg of FLAG-tagged K-bZIP (F-K-bZIP) protein in 0.5 ml of GST pull-down buffer (20 mm HEPES, pH 7.9, 100 mm NaCl, 1 mm EDTA, 4 mm MgCl2, 1 mm dithiothreitol, 0.02% Nonidet P-40, 10% glycerol) and rotated at 4 °C for overnight. The beads were washed four times with GST binding buffer, eluted with SDS sample buffer, and analyzed by immunoblotting using anti-K-bZIP antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunoprecipitation and Western Blot Analysis
Transfected 293T cells were collected in modified radioimmune precipitation assay buffer. TCLs were incubated with anti-FLAG M2-agarose (Sigma) overnight at 4 °C. Beads were washed, and the bound proteins were analyzed by immunoblotting using anti-K-bZIP antibody. For detection of SUMO-conjugated proteins, cells were lysed in SUMO lysis buffer (1× PBS, 2% SDS, 10% glycerol, 20 mm N-ethylmaleimide (NEM)) followed by immunoblotting with anti-SUMO-1 or anti-SUMO-2/3 antibody (Abcam, Cambridge, MA). To immunoprecipitate SUMO-conjugated proteins, cells were lysed in SUMO IP buffer (1× PBS, 0.15 m NaCl, 5 mm EDTA, 0.5% Triton X-100, 0.5% Nonidet P-40, 0.1% sodium deoxycholate, 20 mm NEM). Lysates were diluted 1:10 with PBS containing 0.5% Nonidet P-40, and incubated overnight at 4 °C with anti-p53 mouse antibody (Cell Signaling Technologies, Beverly, MA).
Surface Plasmon Resonance (SPR) Analysis Using a Biacore 3000 Instrument (Biacore, Pittsburgh, PA)
FLAG-K-bZIP in 10 mm acetate buffer (pH 4.0) was immobilized on the CM5 sensor chip using a standard amine coupling procedure (33). The GST, GST-SUMO-1, and GST-SUMO-2 were individually 2-fold serially diluted from 2 μm to 125 nm, and GST-SUMO-3 was 2-fold serially diluted from 1 μm to 62.5 nm in HBS-EP buffer (pH 7.4). Samples were injected at a rate of 30 μl/min for 2 min. After the injection, HBS-EP buffer was continuously pumped into the flow channels at the same flow rate for 4 min to allow the bound GST fusion protein to dissociate from immobilized K-bZIP. The KD was calculated with BIAevaluation software by fitting the obtained binding curves with 1:1 Langmuir binding model.
ChIP Assay
ChIP assays were performed following the protocol provided in Ref. 50. The antibodies were anti-FLAG (Sigma) and rabbit IgG (Alpha Diagnostic International, San Antonio, TX). Immunoprecipitated chromatin DNA was analyzed by SYBR Green-based quantitative PCR (qPCR) (Bio-Rad) (34).
Reverse Transcription and Quantitative PCR
Total RNA was extracted from non-induced and doxycycline-induced TREx-F-K-bZIP wt and L75A 293 cells using TRIzol reagent (Invitrogen, Carlsbad, CA). Total RNA (5 μg) was used for reverse transcription using SuperScript II reverse transcriptase (Invitrogen) and Oligo-dT. Relative mRNA levels were determined by qPCR and normalized to GAPDH (35).
Reporter Assay
293 cells were cultured in 12-well plates, and plasmids were transfected using TransFectin (Bio-Rad). At 48 h after transfection, luciferase activity was determined by the Luciferase Assay System (Promega Corp., Madison, WI).
In Vitro Sumoylation Assay
Reactions were performed using the SUMOlinkTM SUMO-1 and SUMO-2/3 kit (Active Motif, Carlsbad, CA), with recombinant F-K-bZIP wt, F-K-bZIP L75A, F-K-bZIP dDL, F-Rb, and p53 proteins.
RESULTS
K-bZIP Contains a SUMO-interacting Motif and Binds to SUMO-2/3 and Ubc9
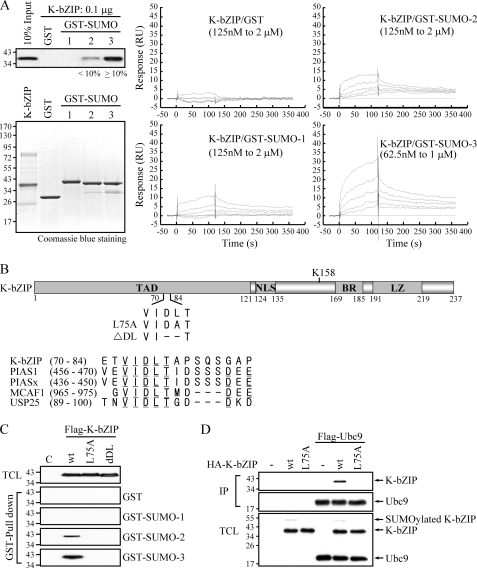
The common properties of SUMO ligases include their ability to bind Ubc9 and contain a SIM, which recognizes and interacts with specific SUMO paralogs. We asked whether K-bZIP directly interacts with SUMO isoforms. The GST pull-down assay was used to analyze these interactions in a cell-free system. When glutathione-Sepharose beads conjugated with GST-SUMO-1, -2, -3 or GST alone were incubated with purified FLAG-tagged K-bZIP protein (F-K-bZIP), K-bZIP was pulled-down by GST-SUMO-2 and -3, but not by GST-SUMO-1 or control beads (Fig. 1A, left panel). These binding data suggest that K-bZIP may be a ligase with potential selectivity toward poly-SUMO chains. We consistently observed a higher level of K-bZIP binding to SUMO-3 than SUMO-2. The differential binding properties toward SUMO-1 and SUMO-2/3 were confirmed by surface plasmon resonance (Biacore) analysis (Fig. 1A, right panel). F-K-bZIP was immobilized on a sensor chip, and a solution of either GST, GST-SUMO-1, -SUMO-2, or -SUMO-3 was passed over the chip. The results showed that GST-SUMO-2 and -SUMO-3 bind K-bZIP with an apparent KD of ∼ 0.2 μm, compared with an apparent KD of ∼2 μm of SUMO-1. The KD was non-analyzable for GST. Consistent with our pull-down assay, the resonance responses elicited by 1 μm of SUMO-3 (30 resonance units) were higher than the response elicited by the same amount of SUMO-2 (∼10 resonance units).
FIGURE 1.
K-bZIP is a SUMO-2/3- and Ubc9-binding protein. A, left panel, direct binding of K-bZIP to SUMO-2 and -3 with a preference for SUMO-3. Baculovirus-expressed FLAG-tagged KSHV K-bZIP protein (F-K-bZIP) was incubated with glutathione-Sepharose beads (GST beads) bound to GST-SUMO-1, GST-SUMO-2, GST-SUMO-3, or GST. Equal amounts of GST proteins were confirmed by gel electrophoresis and Coomassie Blue staining (lower panel). The purified K-bZIP protein (10% of the input) was loaded in lane 1 (upper panel). Proteins were visualized by immunoblotting using anti-FLAG antibody. Right panel, Biacore analysis of interaction between K-bZIP and SUMO proteins. F-K-bZIP was immobilized to a CM5 sensor chip. GST, GST-SUMO-1, and GST-SUMO-2 were 2-fold serially diluted from 2 μm to 125 nm. GST-SUMO-3 was serially diluted from 1 μm to 65 nm. B, schematic representation of KSHV K-bZIP wt and its SIM domain mutant L75A (Leu-75 to Ala) or dDL (deletion of Asp-74 and Leu-75). Sequence comparison of K-bZIP SIM with previously identified SIM domains. TAD, transactivation domain; NLS, nuclear localization signal; BR, basic region; LZ, leucine zipper domain. C, wild-type K-bZIP, but not its SIM domain mutants, interacts with SUMO-2 and -3. TCLs of 293T cells transiently transfected with an expression vector encoding F-K-bZIP wt, or its SIM domain mutant L75A or dDL, were incubated with GST beads bound to GST-SUMO-1, GST-SUMO-2, GST-SUMO-3, or GST. Following incubation, the proteins associated with the GST fusion proteins were analyzed by immunoblotting using anti-FLAG antibody. Immunoblotting of TCLs confirmed equal transfection efficiency. D, wild-type K-bZIP, but not its SIM domain mutants, interacts with Ubc9. TCLs of 293T cells transiently transfected with an expression vector encoding HA-K-bZIP wt, or its SIM domain mutant L75A alone or co-transfected with FLAG-Ubc9 were incubated with anti-FLAG M2 beads. Following incubation, the proteins associated with F-Ubc9 were analyzed by immunoblotting using anti-K-bZIP antibody. Immunoblotting of TCLs using anti-FLAG and anti-HA antibodies confirmed equal levels of transfection efficiency.
To map the SUMO-2/3-interacting domain of K-bZIP, we initially performed a sequence alignment analysis and noticed that amino acids 73–77 matched the core sequence of the SIM found in the PIAS family of E3 ligases and in MCAF1, a chromatin-remodeling protein that binds SUMOs (36) (Fig. 1B). To test the functional importance of this putative SIM, two K-bZIP mutants were constructed. The first, K-bZIP L75A, is a point mutant with the central leucine of the SIM changed to an alanine. The second, K-bZIP dDL, deletes two amino acids within the SIM (Fig. 1B). These mutant K-bZIP plasmids and a plasmid expressing wild-type K-bZIP were individually transfected into 293T cells. Total cell lysates (TCLs) were tested in the pull-down assay for binding to GST or GST-SUMOs. The results revealed that alterations of the VIDLT sequence of K-bZIP abrogated the ability of GST-SUMO-2 and -3 to interact with K-bZIP, establishing the role of this sequence as a SIM (Fig. 1C). No binding was detected with any of these K-bZIP proteins to GST-SUMO-1 or the GST control.
We previously demonstrated that K-bZIP associates with Ubc9 (7). This result was reproduced here in co-transfection and co-immunoprecipitation experiments (Fig. 1D). Furthermore, we showed that this association depends on the SIM domain, because K-bZIP L75A fails to bind Ubc9 (Fig. 1D), suggesting that SUMO-Ubc9 is likely to be the active binding component.
K-bZIP Mediates Its Own Sumoylation via SIM Binding to SUMO
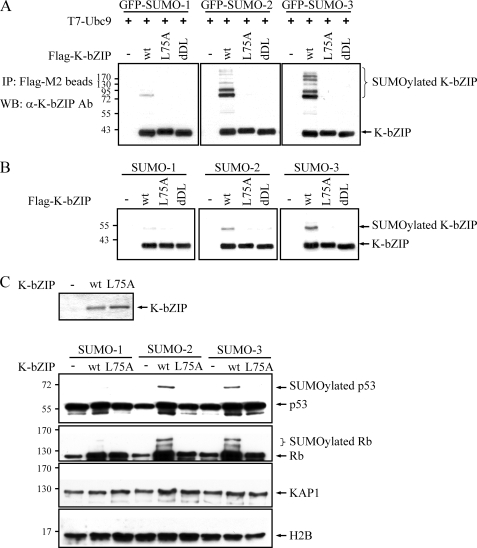
Our previous report showed that K-bZIP is sumoylated at lysine 158, which resides outside the SIM (7). We set out to determine whether K-bZIP was responsible for its own sumoylation, and whether binding of SUMO via its SIM was required as well. Cultures of 293T cells were transiently transfected with vectors expressing GFP-SUMO isoforms together with F-K-bZIP wild-type or its SIM mutants F-K-bZIP L75A or dDL, followed by immunoprecipitation using FLAG-M2 beads. The use of GFP-SUMO is based on the work of others (37) and permits a better resolution of the multisumoylated K-bZIP. This analysis showed that K-bZIP is efficiently conjugated to SUMO-2/3 in vivo, but to a much lower extent, to SUMO-1, reflecting the binding capacity of the SIM. The SIM mutants K-bZIP L75A and dDL failed to conjugate any of the SUMO moieties, indicating that binding to SUMO is a prerequisite for sumoylation of K-bZIP (Fig. 2A). To test whether K-bZIP is a SUMO E3 ligase mediating its own sumoylation, a cell-free in vitro sumoylation reaction was reconstituted with purified SUMO isoforms, SUMO E1, E2, and K-bZIP or its SIM mutants. The experiments showed that K-bZIP catalyzed its own conjugation to SUMO-2/3, but not to SUMO-1, and this reaction depended on an intact SIM (Fig. 2B). These results provide the first suggestive evidence that K-bZIP possesses E3 ligase activity.
FIGURE 2.
K-bZIP is a SIM-containing SUMO E3 ligase. A, 293T cells were co-transfected with the indicated plasmids. Forty-eight hours after transfection, TCLs were prepared for co-immunoprecipitation assay with anti-FLAG M2 beads followed by immunoblotting with anti-K-bZIP antibody. B, purified F-K-bZIP was incubated in a sumoylation reaction containing purified E1-activating and E2-conjugating enzyme, in the presence of either SUMO-1, -2, or -3. Immunoblotting with anti-K-bZIP antibody was then used to reveal the presence of SUMO-conjugated K-bZIP. C, in vitro sumoylation of p53 and Rb but not KAP-1 and histone H2B by K-bZIP. Purified p53, Rb, KAP-1, and histone H2B were incubated in a sumoylation mix containing purified E1 activation and E2-conjugating enzyme together with either SUMO-1, -2, or -3 in absence or presence of recombinant wild-type K-bZIP or its L75A mutant. Immunoblotting with anti-p53, anti-Rb, anti-KAP-1, or anti-histone 2B antibody was then used to reveal the SUMO-conjugated proteins. Equal amounts of wild-type F-K-bZIP and its L75A mutant were confirmed by gel electrophoresis and Coomassie Blue staining (upper panel).
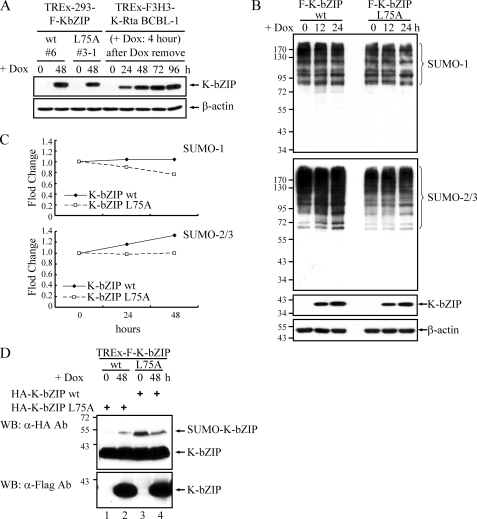
Because K-bZIP can dimerize (38), we asked whether SUMO-Ubc9-bound by the wild-type K-bZIP is able to act in trans to sumoylate the SIM mutant in the context of a heterodimer. The HA-tagged K-bZIP wild-type or L75A mutant was transfected into inducible cell lines expressing the FLAG-tagged-L75A mutant or wild-type-K-bZIP, respectively. Doxycycline was added 6 h after transfection. Forty-eight hours after doxycycline induction, the extent of sumoylation of HA-K-bZIP was analyzed (Fig. 3D). The results in Fig. 3D showed that while HA-L75A was unable to induce sumoylation on its own (lane 1), it could be sumoylated by the wild-type F-K-bZIP, in trans (lane 2), although at a lower efficiency than the wild-type HA-K-bZIP self-sumoylation (lane 3). In fact, the inactive F-K-bZIP L75A mutant seemed to have a dominant-negative effect on the self-sumoylation activity of the wild-type HA-K-bZIP (lane 4), consistent with a trans-sumoylation model. These data further support the contention that K-bZIP is a SUMO E3 ligase.
FIGURE 3.
K-bZIP enhances global level of SUMO-2/3 modification. A, Tet-inducible TREx-F-K-bZIP wt and L75A 293 cells were treated with 1 μg/ml doxycycline for 48 h. Tet-inducible TREx-F3H3-K-Rta BCBL-1 cells were treated with 0.2 μg/ml doxycycline for 4 h, and TCLs were collected at the indicated time points. Expression of K-bZIP from both cell lines was examined by immunoblotting. B, Tet-inducible TREx-F-K-bZIP wt and L75A 293 cells were treated with 1 μg/ml doxycycline for 12 and 24 h. TCLs were prepared and immunoblotted with anti-SUMO-1, SUMO-2/3, K-bZIP, or β-actin antibody. C, relative expression levels of total sumoylated proteins to β-actin were quantitated using a Molecular Imager Fx with Quantity One® software (Bio-Rad) and are shown in the plot. D, Tet-inducible TREx-F-K-bZIP wt and L75A 293 cells were transfected with HA-tagged K-bZIP-L75A or its wild-type, respectively. Four hours after transfection, cells were treated with 1 μg/ml doxycycline for 48 h. TCLs were prepared and immunoblotted with anti-FLAG or anti-HA antibody.
K-bZIP Is a SUMO E3 Ligase
To provide additional evidence that K-bZIP is a SUMO E3 ligase, we examined K-bZIP-interacting partners, including p53 and Rb proteins, known to be sumoylated in certain contexts (8, 13). For these experiments, F-K-bZIP wild-type and F-K-bZIP L75A were employed. The in vitro sumoylation reaction was performed on each of these cellular proteins in the presence or absence of K-bZIP. This sumoylation analysis, using limited amounts of Ubc9 enzyme, showed that K-bZIP catalyzed the conjugation of SUMO-2 and -3, but not SUMO-1, to p53. In addition, this reaction depends on the binding of SUMO-2/3 to K-bZIP, because the L75A mutant does not conjugate SUMO to p53 (Fig. 2C). Identical results were obtained with the Rb protein (Fig. 2C). By contrast, the sumoylation of proteins not known to directly associate with K-bZIP, such as the cellular co-repressor KAP-1 and histone H2B, were not enhanced by K-bZIP (Fig. 2C). These findings show that K-bZIP is a SIM containing SUMO E3 ligase that provides a platform for Ubc9, SUMO, and substrate to engage in the sumoylation reaction.
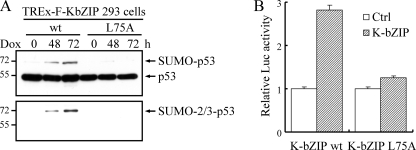
To study the function of K-bZIP as a SUMO ligase in vivo, we established inducible cell lines for K-bZIP wild-type and the L75A mutant (TREx-F-K-bZIP wt and L75A). 293T cells are permissive for KSHV and widely used for studying infection and replication with this virus. The expression level of K-bZIP wild-type and the L75A mutant after induction in the respective cell lines was comparable to that during KSHV lytic replication after K-Rta induction in BCBL-1 cells (Fig. 3A). Immunoblot analysis of total cell lysates from non-induced and doxycycline-induced TREx-F-K-bZIP wt and L75A 293 cells demonstrated that the profiles of SUMO-1-modified proteins did not differ noticeably. In contrast, an increase in SUMO-2/3-modified proteins was detected in doxycycline-induced wild-type K-bZIP but not its L75A mutant, consistent with a role of K-bZIP in the conjugation of a selected set of cellular proteins (Fig. 3, B and C). Additionally, the increase of cellular sumoylation was accompanied by an increase of the SUMO-2/3 modification of p53 by wild-type K-bZIP but not in its L75A mutant (Fig. 4A). Thus, the presence of K-bZIP induces the sumoylation of p53 both in vitro and in vivo.
FIGURE 4.
Sumoylation and modulation of p53 by K-bZIP. A, after 48 or 72 h of incubation with 1 μg/ml doxycycline, TCLs from TREx-F-K-bZIP wt and L75A mutant 293 cells were immunoprecipitated using anti-p53 mouse antibody. Western blot analysis with anti-p53 rabbit antibody detected a major 55kDa p53 band in all immunoprecipitates. An additional ∼72-kDa band, representing the SUMO-modified form of p53, was detected by anti-p53 and anti-SUMO-2/3 antibodies only in immunoprecipitates prepared from wild-type K-bZIP overexpressed TREx-F-K-bZIP 293 cells. B, SaOS-2 cells were co-transfected with p4XBS2WT-Luc (0.1 μg/well), a p53-response reporter plasmid, pcDNA3- F-K-bZIP wt or L75A mutant (0.4 μg/well), pcDNA3-HA-p53 (0.001 μg/well), and an internal control plasmid, pTK-RL (0.01 μg/well) in a 12-well plate. Forty-eight hours after transfection, luciferase reporter activity was assayed and normalized to internal Renilla luciferase activity. Values from three independent data points are reported as the mean + S.D.
Modulation of p53 Activity by K-bZIP Is SIM-dependent
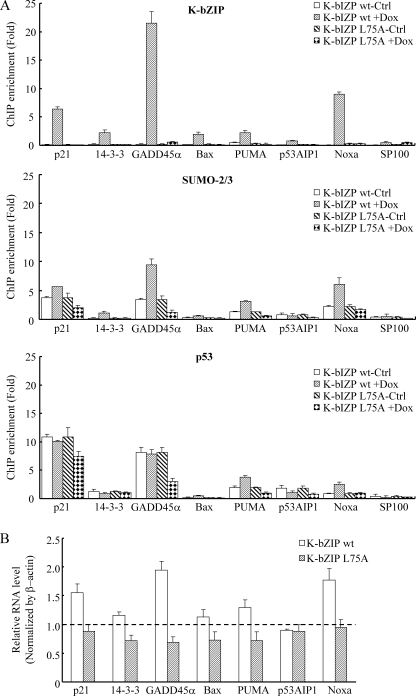
p53 is one of the few transcriptional factors that are activated by sumoylation (39). To explore whether K-bZIP sumoylation of p53 impacts transcriptional activation of this tumor suppressor protein, we transfected SaOS-2 cells with either K-bZIP wild-type or L75A mutant plasmids, together with p53 and a p53-responsive luciferase reporter vector, 4XBS2WT-Luc. Wild-type K-bZIP, but not the L75A mutant, activated the reporter ∼3-fold (Fig. 4B). These data corroborate the activation of p53 by SUMO modification reported previously (39) and suggest that sumoylation might contribute to K-bZIP-mediated activation of p53. If K-bZIP plays a role in p53 activation, we further surmise that K-bZIP and SUMO-2/3 should be part of the p53 complex assembled near the p53 target sites in the induced cells. Seven p53-responsive promoters (34), including those of three growth arrest- and four apoptosis-related genes, were selected for ChIP assay analysis in both the TREx-FLAG-K-bZIP wt and L75A inducible cell lines. After K-bZIP induction, K-bZIP was recruited to all seven p53 responsive promoters (>2× enrichment). The enrichment of K-bZIP near the GADD45α promoter is particularly striking, reaching a level of 22-fold above the background (Fig. 5A). This recruitment of K-bZIP to p53 responsive promoters is accompanied by a concomitant recruitment of SUMO-2/3 protein (Fig. 5A). Importantly, the K-bZIP L75A mutant protein is not recruited to these promoters, despite the same level of expression as its wild-type counterpart (Fig. 3A). The recruitment of p53 itself to these target sites however is largely unaffected by the expression of K-bZIP, as expected (Fig. 5A). These data suggest that K-bZIP brings SUMO-2/3 moieties to p53, enhancing this post-translational modification, which is known to stimulate the transactivation properties of p53 (Fig. 5A). RT-PCR data generally confirmed the increased expression of these genes (Fig. 5B), consistent with the published account of SUMO activation of p53 transcriptional activity (39).
FIGURE 5.
SIM of K-bZIP is crucial for K-bZIP occupancy to p53-responsive promoters and transactivation of p53 response genes. A, ChIP assay was performed on non-induced and F-K-bZIP wt or L75A-induced 293 cells using anti-FLAG (upper panel), anti-SUMO-2/3 (middle panel), or anti-p53 (lower panel) antibody or rabbit IgG. K-bZIP, SUMO-2/3, and p53 binding was analyzed by real-time qPCR using primers specific for p53 target gene p21, 14-3-3, GADD45α, Bax, PUMA, p53AIP1, and Noxa. Sp100 was used as a negative control. The level of 10% input DNA was set at 1. Values from three independent data points are reported as mean + S.D. B, RNA isolated from non-induced and F-K-bZIP wt or L75A-induced 293 cells was subjected to RT-qPCR to examine mRNA levels of the p53 target genes p21, 14-3-3, GADD45α, Bax, PUMA, p53AIP1, and Noxa. Values from three independent data points are reported as mean + S.D.
DISCUSSION
A large body of evidence supports a role for sumoylation in replication of many viruses. This post-translational modification modulates subcellular localization, transcriptional activity, stability and ability to interact with other proteins (40, 41). A handful of viruses have evolved means to directly regulate the host SUMO machinery. For example, avian adenovirus Gam1 interacts with and inhibits SUMO E1 activity (42), Ebola Zaire virus VP35 interacts with SUMO E2 Ubc9 and E3 PIAS to stimulate their activity (43), and herpes simplex virus ICP0 co-localizes SENP1 de-sumoylase with PML (44). Thus these viral proteins are likely to produce a global effect on the host SUMO status during viral replication. In this report, we show for the first time that a virus encodes a SUMO E3 ligase.
A SUMO ligase is defined by its ability to 1) bind Ubc9, SUMO, and a substrate, and 2) catalyze the sumoylation of the substrate both in vivo and in vitro. Previously, we showed that K-bZIP associates with Ubc9 (7). Here, we have demonstrated that K-bZIP contains a SIM, which binds SUMO-2 and SUMO-3, but not SUMO-1, both in vitro and in vivo. SUMO-2 and SUMO-3 are highly homologous (97% identity in the mature form), whereas SUMO-1 is more distantly related (48% identity to SUMO-2, and 46% to SUMO-3). SUMO-2 and -3 can form poly-chain structures, whereas SUMO-1 is usually added in monomeric form. SUMO-1 is found in nucleoli, nuclear envelopes, and cytoplasmic foci, whereas SUMO-2/3 are associated with chromatin. As such, these proteins are likely to convey different signals and exert distinct functions. All previously characterized cellular SUMO E3 ligases, such as RanBP2, PIAS, and Pc2, bind all three SUMO paralogues non-discriminatorily. K-bZIP, on the other hand, appears to be highly selective toward SUMO-2/3. MCAF-1, which has an identical core SIM motif (VIDLT) as K-bZIP, also shows selectivity toward SUMO-2/3 (36, 45). The KD for K-bZIP SIM/SUMO-2/3 (∼0.2 μm) is 10-fold less than that of K-bZIP SIM/SUMO-1 (∼2 μm). This measure is comparable to the 1.3 μm KD value reported for MCAF-1 SIM/SUMO-2/3 and 14 μm for MCAF-1 SIM/SUMO-1 interaction (36). Likewise, USP25, a ubiquitin-specific protease which selectively binds SUMO-2/3 possesses a TNVIDLT motif (Fig. 1B) (46), and these seven amino acids are sufficient to confer the selectivity toward SUMO-2/3. These data are in good agreement with our finding. On the other hand, PIASxa which carries the core VIDLT motif is known to display E3 ligase activity for all three paralogues. Thus, the preference toward SUMO-2/3 is also influenced by the surrounding amino acid sequences. Further structural studies are required to resolve these issues.
To demonstrate that K-bZIP functions as an E3 ligase, the ability of this viral protein to catalyze sumoylation was demonstrated in cell-free reconstituted sumoylation reactions. We showed that p53 and Rb, two K-bZIP-interacting proteins, are sumoylated in the presence of K-bZIP, and further, that the conjugation depended on an intact SIM domain of K-bZIP with a preference for SUMO-2/3 modification. The increased sumoylation of p53 was also shown in TREx-F-K-bZIP inducible cells in a SIM domain-dependent manner. This increased sumoylation is supported by heightened transactivation activity of p53 toward a reporter gene driven by p53-responsive element mediated by wild-type K-bZIP, but not the SIM mutant. Our data are consistent with earlier work showing that sumoylated p53 is more transcriptionally active (26, 27) and that K-bZIP is able to enhance the transactivation activity of p53 (23). In this regard, a recent report by Li et al. (39) showed that expression of SUMO-2/3, but not SUMO-1, induces p53 activity with consequent cellular senescence, and that treatment of cells with H2O2 induced SUMO-2/3, but not SUMO-1, resulting in p53-mediated growth arrest. Growth arrest is a common outcome associated with the early phase of herpesvirus infection (reviewed in Ref. 32) and provides an optimal environment for the virus to transcribe and translate viral RNA before the onset of viral DNA replication. At the same time, this viral-mediated growth arrest protects host cells from undergoing apoptosis. For KSHV, K-bZIP performs similar functions (29–31). Previously, we have shown that K-bZIP expression induced growth arrest via the inactivation of CDK2 (30). Here we provide an additional possible mechanism by which K-bZIP induces growth arrest, i.e. by activating p53 pathway via sumoylation of this cell regulatory protein. Many p53 target genes are induced by K-bZIP in a SUMO-dependent manner. Perhaps the most striking finding is that K-bZIP is recruited to p53 target sites in a SIM-dependent manner, suggesting that the SUMO-rich environment stabilizes the complex between p53 and K-bZIP, and the K-bZIP presence ensures the sumoylated status of p53.
Why does KSHV encode a SUMO-2/3 E3 ligase? Dynamic sumoylation of host and viral proteins is thought to play an important role in herpesvirus replication. The assembly and disassembly of PML bodies (or ND10) at herpesvirus replication complexes are SUMO-dependent (47). Condensation of viral chromatin to achieve a latent, heterochromatin state is likely to be facilitated by sumoylation (48), and the reverse is true for reactivation. Exploitation of the SUMO pathway for viral replication may be especially important for KSHV, because this virus encodes not only a SUMO ligase (K-bZIP, as reported here) but also a SUMO-dependent ubiquitin ligase, K-Rta, which degrades SUMO and sumoylated proteins.3 Overexpression of K-Rta leads to disassembly of PML bodies and reactivation of the latent genome, presumably by providing a SUMO-depleted environment. We showed previously that K-bZIP is an interacting partner and a modulator of K-Rta, thus counter-balancing the transactivation activity of K-Rta (20). Moreover, during viral replication, depending on the relative levels of K-Rta and K-bZIP, the abundance of SUMO-2/3 and SUMO-conjugated proteins might be modulated, thereby allowing the viral replication to proceed or in some cases, enabling viral latency to be established. Another intriguing role of K-bZIP, as a modulator of gene expression, might well be to facilitate the degradation functions of K-Rta, by “marking” the proteins to be targeted with SUMO-2/3 conjugation. The latter scenario is supported by a recent report showing that KSHV LANA2 increases PML sumoylation and facilitates its degradation by a cellular ligase (49). Additional studies are required to understand the critical regulatory interplay between these two major KSHV factors, the K-bZIP repressor and the K-Rta transactivator.
In summary, we report the discovery of K-bZIP as the first viral SUMO-2/3 E3 ligase, which critically depends on an intact SIM for its enzymatic function. We also demonstrate its ability to catalyze the sumoylation and modulate the transactivation of p53, which, in turn, modulates the assembly of a sumoylated complex near p53 transcriptional target sites, all in a SIM-dependent manner.
Acknowledgment
We thank Dr. Xinbin Chen for the pcDNA3-HA-p53 plasmid.
The work was supported, in whole or in part, by National Institutes of Health Grants R01 CA114575 and DE019085.
Y. Izumiya, C. Izumiya, M. Pochampalli, M. Campbell, P. A. Luciw, and H.-J. Kung, data to be published.
- SUMO
- small ubiquitin-like modifier
- KSHV
- Kaposi's sarcoma-associated herpesvirus
- IE
- immediate-early
- wt
- wild type
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin
- SIM
- SUMO-interacting motif
- TCL
- total cell lysate.
REFERENCES
- 1.Wilson V. G., Rangasamy D. (2001) Virus Res. 81, 17–27 [DOI] [PubMed] [Google Scholar]
- 2.Müller S., Dejean A. (1999) J. Virol. 73, 5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann H., Flöss S., Stamminger T. (2000) J. Virol. 74, 2510–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosas-Acosta G., Langereis M. A., Deyrieux A., Wilson V. G. (2005) Virology 331, 190–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y. C., Deyrieux A. F., Wilson V. G. (2007) Biochem. Soc. Trans. 35, 1433–1435 [DOI] [PubMed] [Google Scholar]
- 6.Adamson A. L., Kenney S. (2001) J. Virol. 75, 2388–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izumiya Y., Ellison T. J., Yeh E. T., Jung J. U., Luciw P. A., Kung H. J. (2005) J. Virol. 79, 9912–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ledl A., Schmidt D., Müller S. (2005) Oncogene 24, 3810–3818 [DOI] [PubMed] [Google Scholar]
- 9.Chang P. C., Fitzgerald L. D., Van Geelen A., Izumiya Y., Ellison T. J., Wang D. H., Ann D. K., Luciw P. A., Kung H. J. (2009) Cancer Res. 69, 5681–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boggio R., Passafaro A., Chiocca S. (2007) J. Biol. Chem. 282, 15376–15382 [DOI] [PubMed] [Google Scholar]
- 11.Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 12.Bischof O., Schwamborn K., Martin N., Werner A., Sustmann C., Grosschedl R., Dejean A. (2006) Mol. Cell 22, 783–794 [DOI] [PubMed] [Google Scholar]
- 13.Kahyo T., Nishida T., Yasuda H. (2001) Mol. Cell 8, 713–718 [DOI] [PubMed] [Google Scholar]
- 14.Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. (2002) Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
- 15.Kagey M. H., Melhuish T. A., Wotton D. (2003) Cell 113, 127–137 [DOI] [PubMed] [Google Scholar]
- 16.Chang Y., Cesarman E., Pessin M. S., Lee F., Culpepper J., Knowles D. M., Moore P. S. (1994) Science 266, 1865–1869 [DOI] [PubMed] [Google Scholar]
- 17.Dupin N., Fisher C., Kellam P., Ariad S., Tulliez M., Franck N., van Marck E., Salmon D., Gorin I., Escande J. P., Weiss R. A., Alitalo K., Boshoff C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4546–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukac D. M., Garibyan L., Kirshner J. R., Palmeri D., Ganem D. (2001) J. Virol. 75, 6786–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun R., Lin S. F., Gradoville L., Yuan Y., Zhu F., Miller G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10866–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izumiya Y., Lin S. F., Ellison T., Chen L. Y., Izumiya C., Luciw P., Kung H. J. (2003) J. Virol. 77, 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumiya Y., Izumiya C., Van Geelen A., Wang D. H., Lam K. S., Luciw P. A., Kung H. J. (2007) J. Virol. 81, 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J., Seo T., Hwang S., Lee D., Gwack Y., Choe J. (2000) J. Virol. 74, 11977–11982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamanegi K., Tang S., Zheng Z. M. (2005) J. Virol. 79, 14207–14221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katano H., Ogawa-Goto K., Hasegawa H., Kurata T., Sata T. (2001) Virology 286, 446–455 [DOI] [PubMed] [Google Scholar]
- 25.Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. (2006) Mol. Cell 24, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gostissa M., Hengstermann A., Fogal V., Sandy P., Schwarz S. E., Scheffner M., Del Sal G. (1999) EMBO J. 18, 6462–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez M. S., Desterro J. M., Lain S., Midgley C. A., Lane D. P., Hay R. T. (1999) EMBO J. 18, 6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.AuCoin D. P., Colletti K. S., Cei S. A., Papousková I., Tarrant M., Pari G. S. (2004) Virology 318, 542–555 [DOI] [PubMed] [Google Scholar]
- 29.Wu F. Y., Tang Q. Q., Chen H., ApRhys C., Farrell C., Chen J., Fujimuro M., Lane M. D., Hayward G. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10683–10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izumiya Y., Lin S. F., Ellison T. J., Levy A. M., Mayeur G. L., Izumiya C., Kung H. J. (2003) J. Virol. 77, 9652–9661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu F. Y., Wang S. E., Tang Q. Q., Fujimuro M., Chiou C. J., Zheng Q., Chen H., Hayward S. D., Lane M. D., Hayward G. S. (2003) J. Virol. 77, 8893–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flemington E. K. (2001) J. Virol. 75, 4475–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu P. L., Lee S. C., Chuang C. C., Mori S., Akakura N., Wu W. G., Takada Y. (2006) J. Biol. Chem. 281, 7937–7945 [DOI] [PubMed] [Google Scholar]
- 34.Jackson J. G., Pereira-Smith O. M. (2006) Cancer Res. 66, 8356–8360 [DOI] [PubMed] [Google Scholar]
- 35.Xie P., Tian C., An L., Nie J., Lu K., Xing G., Zhang L., He F. (2008) Cell Signal 20, 1671–1678 [DOI] [PubMed] [Google Scholar]
- 36.Sekiyama N., Ikegami T., Yamane T., Ikeguchi M., Uchimura Y., Baba D., Ariyoshi M., Tochio H., Saitoh H., Shirakawa M. (2008) J. Biol. Chem. 283, 35966–35975 [DOI] [PubMed] [Google Scholar]
- 37.Fu C., Ahmed K., Ding H., Ding X., Lan J., Yang Z., Miao Y., Zhu Y., Shi Y., Zhu J., Huang H., Yao X. (2005) Oncogene 24, 5401–5413 [DOI] [PubMed] [Google Scholar]
- 38.Lin S. F., Robinson D. R., Miller G., Kung H. J. (1999) J. Virol. 73, 1909–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T., Santockyte R., Shen R. F., Tekle E., Wang G., Yang D. C., Chock P. B. (2006) J. Biol. Chem. 281, 36221–36227 [DOI] [PubMed] [Google Scholar]
- 40.Huh Y. H., Kim Y. E., Kim E. T., Park J. J., Song M. J., Zhu H., Hayward G. S., Ahn J. H. (2008) J. Virol. 82, 10444–10454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y. C., Bian X. L., Heaton P. R., Deyrieux A. F., Wilson V. G. (2009) Virology 387, 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boggio R., Colombo R., Hay R. T., Draetta G. F., Chiocca S. (2004) Mol. Cell 16, 549–561 [DOI] [PubMed] [Google Scholar]
- 43.Chang T. H., Kubota T., Matsuoka M., Jones S., Bradfute S. B., Bray M., Ozato K. (2009) PLoS Pathog. 5, e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bailey D., O'Hare P. (2002) J. Gen. Virol 83, 2951–2964 [DOI] [PubMed] [Google Scholar]
- 45.Hecker C. M., Rabiller M., Haglund K., Bayer P., Dikic I. (2006) J. Biol. Chem. 281, 16117–16127 [DOI] [PubMed] [Google Scholar]
- 46.Meulmeester E., Kunze M., Hsiao H. H., Urlaub H., Melchior F. (2008) Mol. Cell 30, 610–619 [DOI] [PubMed] [Google Scholar]
- 47.Boggio R., Chiocca S. (2006) Curr. Opin. Microbiol. 9, 430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uchimura Y., Ichimura T., Uwada J., Tachibana T., Sugahara S., Nakao M., Saitoh H. (2006) J. Biol. Chem. 281, 23180–23190 [DOI] [PubMed] [Google Scholar]
- 49.Marcos-Villar L., Lopitz-Otsoa F., Gallego P., Muñoz-Fontela C., González-Santamaria J., Campagna M., Shou-Jiang G., Rodriguez M. S., Rivas C. (2009) J. Virol. 83, 8849–8858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Geen H., Squazzo S. L., Iyengar S., Blahnik K., Rinn J. L., Chang H. Y., Green R., Farnham P. J. (2007) PLoS Genet. 3, e89. [DOI] [PMC free article] [PubMed] [Google Scholar]