Abstract
The YidC/Oxa1/Alb3 family of membrane proteins controls the insertion and assembly of membrane proteins in bacteria, mitochondria, and chloroplasts. Here we describe the molecular mechanisms underlying the interaction of Alb3 with the chloroplast signal recognition particle (cpSRP). The Alb3 C-terminal domain (A3CT) is intrinsically disordered and recruits cpSRP to the thylakoid membrane by a coupled binding and folding mechanism. Two conserved, positively charged motifs reminiscent of chromodomain interaction motifs in histone tails are identified in A3CT that are essential for the Alb3-cpSRP43 interaction. They are absent in the C-terminal domain of Alb4, which therefore does not interact with cpSRP43. Chromodomain 2 in cpSRP43 appears as a central binding platform that can interact simultaneously with A3CT and cpSRP54. The observed negative cooperativity of the two binding events provides the first insights into cargo release at the thylakoid membrane. Taken together, our data show how Alb3 participates in cpSRP-dependent membrane targeting, and our data provide a molecular explanation why Alb4 cannot compensate for the loss of Alb3. Oxa1 and YidC utilize their positively charged, C-terminal domains for ribosome interaction in co-translational targeting. Alb3 is adapted for the chloroplast-specific Alb3-cpSRP43 interaction in post-translational targeting by extending the spectrum of chromodomain interactions.
Keywords: Protein Domains, Protein Folding, Protein Motifs, Protein Targeting, Protein Translocation, Protein-Protein Interactions, Oxa1/YidC/Alb3 Membrane Insertases, Chromodomains, Intrinsic Disorder, Signal Recognition Particle
Introduction
Integral membrane proteins comprise about 20–30% of the proteome. They preferentially use the co-translational targeting and insertion machinery of the universally conserved signal recognition particle (SRP)3 in conjunction with the SecYEG translocation channel (1–3). However, membrane proteins that play important roles in respiration and energy transduction mechanisms are the main substrates for the YidC/Oxa1/Alb3 family of membrane insertases (4, 5). Members of the YidC/Oxa1/Alb3 family are involved in the insertion and folding of membrane proteins as well as their assembly into larger protein complexes and quality control (4). In Escherichia coli, YidC is essential and catalyzes membrane protein insertion into the inner membrane (6). YidC is homologous to Oxa1 and Alb3, which are involved in the integration of proteins into the inner membrane of mitochondria and the thylakoid membrane of chloroplasts, respectively (7). Alb3 cooperates with the chloroplast SRP (cpSRP) in the post-translational insertion of light-harvesting chlorophyll a,b-binding proteins (LHCPs) (8, 9). LHCPs serve as antenna systems in photosynthesis and belong to the most abundant membrane proteins on earth. They are post-translationally imported into the chloroplast where they are sequestered into a soluble transit complex (cpSRP43-cpSRP54-LHCP) of yet unknown stoichiometry (10). The transit complex is directed to the thylakoid membrane by the interaction of cpSRP54 with the membrane bound SRP receptor cpFtsY (11). cpSRP54 acts both co-translationally and post-translationally in the insertion of thylakoid membrane proteins (1, 12). In post-translational targeting, cpSRP54 together with cpSRP43 forms the heterodimeric cpSRP (13). cpSRP43 has a unique, modular structure (14) with three chromodomains (CD1–3) and four ankyrin repeats (Ank1–4) that create a platform for multiple interactions. CD2 interacts with a positively charged extension of the M-domain of cpSRP54 (15, 16). The ankyrin repeats of cpSRP43 bind to an extended, internal signal sequence in LHCPs, the L18 region (14, 17). Alb3 was previously shown to co-localize with cpSRP43 in double-knock-out plants lacking cpSRP54 and cpFtsY and to interact with cpSRP43 (18). However, how Alb3 interacts with cpSRP43 and how it participates in LHCP targeting and membrane insertion is not known.
Despite the low sequence conservation in the YidC/Oxa1/Alb3 family, all members share a hydrophobic core region of about 200 residues representing five transmembrane helices (TMs) (19). A cryo-electronmicroscopy projection structure of E. coli YidC shows the overall arrangement of the TM regions (20), which harbor the membrane insertase function. Hydrophilic domains of various lengths can be present at the N or the C terminus and are thought to be adaptors for additional, specialized functions. E. coli YidC contains an additional N-terminal TM followed by a large periplasmic domain (21, 22), which is absent in the organellar proteins or homologs in Gram-positive bacteria. Mitochondria and most Gram-positive bacteria contain two YidC paralogs, whereas Gram-negative bacteria contain only one (23). They are distinguished mainly by their C-terminal domains (24). Chloroplasts of green algae and plants also contain two paralogs of the YidC/Oxa1/Alb3 family (23), which in Arabidopsis thaliana are named Alb3 and Alb4. Although Alb3 and Alb4 share a high sequence similarity, Alb4 cannot compensate for the loss of Alb3 (25).
In this study, we present a detailed biochemical analysis of the C-terminal domain of Alb3. We demonstrate that it interacts with cpSRP43, whereas the paralog Alb4 does not. Two conserved motifs in Alb3 are essential for cpSRP43 interaction. Alb3 and cpSRP54 can bind simultaneously to CD2 of cpSRP43. The observed negative cooperativity of the two binding events allows a first glimpse on transit complex delivery to the membrane.
EXPERIMENTAL PROCEDURES
Immobilized Peptide Libraries
Peptide arrays were synthesized by Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry at activated polyethylene glycol spacers on cellulose membranes using a semiautomated spot robot (ASP222; Intavis, Germany) as described (13, 26). A. thaliana Alb3 was synthesized as overlapping peptides (15-mer off-set by one amino acid) covering stromal domains of Alb3 as follows: stromal loop S1 including amino acids 152–204, and stromal loop S2 including amino acids 254–277, and the C terminus 339–462. After 2 h of blocking with 4% (w/v) skim milk (Sigma) in the incubation buffer (30 mm Tris-HCl, pH 7.6, 100 mm KCl, 5 mm MgCl2, 5% sucrose, and 0.05% Tween 20), the membranes were incubated in the incubation buffer overnight at 4 °C with 0.25 μm purified His-tagged cpSRP43. Bound cpSRP43 was detected with an anti-His antibody and visualized using an ECL anti-mouse IgG secondary antibody (GE Healthcare). The incubations with the antibodies were performed in the incubation buffer with 0.5% (w/v) skim milk. All incubation steps were followed by three consecutive washes of 10 min with the incubation buffer. Binding signals were detected by enhanced chemiluminescence (ECL, GE Healthcare).
Cloning
The different constructs of the Alb3 C-terminal domain (A3CT) encoding the amino acids 339–462, 363–462, and 363–450 were cloned into pET21d using the NcoI and XhoI restriction sites leading to a C-terminal His tag. The A3CT constructs encoding amino acids 363–405 and 406–462 were cloned into pETtrx_1a (Gunter Stier, Heidelberg) using NcoI and XhoI leading to C-terminal fusion to His-tagged thioredoxin with a tobacco etch virus cleavage site.
Recombinant Protein Production and Purification
C-terminally His-tagged A3CT was produced in BL21 (DE3) cells. Protein production was induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside at an A600 of 0.6. After 3 h at 37 °C, the cells were harvested and stored at −80 °C. For the purification of His-tagged A3CT, cells were resuspended in lysis buffer (100 mm Hepes/NaOH, 300 mm NaCl, 5 mm MgCl2, 10% (w/v) glycerol, 0.02% 1-thioglycerol, pH 7.5). The cells were lysed by an M1-10L Microfluidizer (Microfluidics). The lysate was clarified, and the supernatant was applied onto a 1-ml HisTrap HP column (GE Healthcare). The column was washed with washing buffer (50 mm Hepes/NaOH, 300 mm NaCl, 5 mm MgCl2, 5% (w/v) glycerol, 0,02% 1-thioglycerol, pH 7.5) containing 0, 10, and 50 mm imidazole. The proteins were eluted in washing buffer containing 300 mm imidazole. The eluted protein was diluted into SP buffer (50 mm Hepes/NaOH, 150 mm NaCl, 5% (w/v) glycerol, 0.02% 1-thioglycerol, pH 7.5), applied onto SP Sepharose Fast Flow (GE Healthcare), and eluted with a gradient from 150 mm to 1 m NaCl. A3CT was further purified on an S75 26/60 size-exclusion column equilibrated in 20 mm Hepes/NaOH, 150 mm NaCl, 2 mm MgCl2, 1 mm EDTA, 1 mm DTT, pH 7.5.
Purification of Proteins Fused to Thioredoxin
Cell lysis and purification via HisTrap HP column were essentially the same as described above. 200 μg of recombinant tobacco etch virus protease was added, and cleavage occurred overnight during dialysis against 20 mm Hepes/NaOH, 150 mm NaCl, 2 mm MgCl2, 1 mm DTT. After the cleavage, the sample was reloaded on a HisTrap HP column, and the flow-through was collected. The proteins were further purified by size-exclusion chromatography using an S30 16/60 column equilibrated in 20 mm Hepes/NaOH, 150 mm NaCl, 2 mm MgCl2, 1 mm EDTA, 1 mm DTT, pH 7.5.
Coproduction and Copurification of the cpSRP43-A3CT Complex
cpSRP43-A3CTII–IV-His6 and cpSRP43Δ3-A3CTII–IV-His6 were coproduced in Rosetta2 (DE3) pLysS. Expression was induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside at A600 of 0.6. The copurification of the cpSRP43-A3CT complex was essentially the same as that for A3CT described above with the exception that the SP-Sepharose column was omitted.
Circular Dichroism
Circular dichroism spectra were recorded on a Jasco J-600 spectropolarimeter in a 0.1-cm path length cuvette at 15 °C. Proteins were exchanged into 10 mm potassium phosphate, pH 7.5, and diluted to 6 μm. To measure the spectrum of the complex, both proteins were mixed (6 μm each) and incubated for 1 h at 15 °C. For each experiment, 16 scans were taken from 250 to 190 nm in 0.2-nm increments with a 1-s averaging time and a bandwidth of 1 nm. The scans were averaged, and the buffer spectrum was subtracted.
Pulldown Assays
20 μm of the cpSRP43-A3CT-His6 or cpSRP43Δ3-A3CT-His6 complex was mixed with increasing amounts of cpSRP54 M-domain (0–250 μm) in 350 μl of 20 mm Hepes/NaOH, 150 mm NaCl, 2 mm MgCl2, 1 mm EDTA, 1 mm DTT, pH 7.5, and incubated for 1 h at 20 °C. 20 μl of nickel-nitrilotriacetic acid agarose was added and then incubated for 1 h at 4 °C. The beads were washed three times with 500 μl of the same buffer, and bound proteins were eluted with buffer including 300 mm imidazole. Proteins were detected by SDS-PAGE on NuPAGE 4–12% polyacrylamide gels (Invitrogen) with subsequent Coomassie Brilliant Blue R (Sigma) staining.
Analytical Size-exclusion Chromatography (SEC)
The hydrodynamic properties of A3CT, cpSRP43, dimeric cpSRP43-A3CT or cpSRP43-cpSRP54 M-domain complexes, or the trimeric complex were measured by size-exclusion chromatography (Superdex 200 10/300). Purified A3CT, cpSRP43, or the different complexes were loaded on an S200 10/300 column equilibrated in 20 mm Hepes/NaOH, 150 mm NaCl, 2 mm MgCl2, 1 mm EDTA, 1 mm DTT, pH 7.5. The elution volume (Ve) was monitored by absorbance at 280 nm. Fractions were collected and analyzed by SDS-PAGE. We estimated the Stokes radius of A3CT according to the method of Laurent and Killander (as modified by Siegel and Monty in Ref. 59) (27). The absolute molecular weight was determined by static light scattering (miniDAWN Tristar; Wyatt Technologies) and refractive index analysis (WGE Dr. Bures, Δn-1000 differential refractive index detector, Dallgow, Germany).
PONDR Prediction of Unstructured Regions
We submitted the sequences of Alb3 and Alb4 to the Predictors of Natural Disordered Region (PONDR) server using the default integrated predictor VL-XT (28). Access to PONDR was provided by Molecular Kinetics (Pullman, WA) under license from the Washington State University (WSU) Research Foundation. PONDR was copyrighted in 1999 by the WSU Research Foundation, all rights reserved.
Isothermal Titration Calorimetry
All isothermal titration calorimetry (ITC) experiments were carried out in a buffer containing 20 mm Hepes/NaOH, 200 mm NaCl, 2 mm MgCl2, 1 mm EDTA, 5% (w/v) glycerol, 0.25 mm tris(2-carboxyethyl)phosphine, pH 7.5. The samples were extensively dialyzed against the same buffer. Solutions were degassed immediately before measuring, and all binding experiments were performed using a VP-ITC microcalorimeter (MicroCal, Northampton, MA) equilibrated at 20 °C. Typically, protein concentrations between 50 and 100 μm in the cell and 0.5–1 mm in the syringe were used in an ITC experiment. A titration consisted of injections of 12-μl aliquots of the titrant into the solution in the cell under constant stirring at 307 rpm at time intervals of 5 min to ensure that the titration peak returned to the baseline. For data analysis, the Origin 7.0 software was used.
RESULTS
The Alb3 C-terminal Domain Interacts with cpSRP43
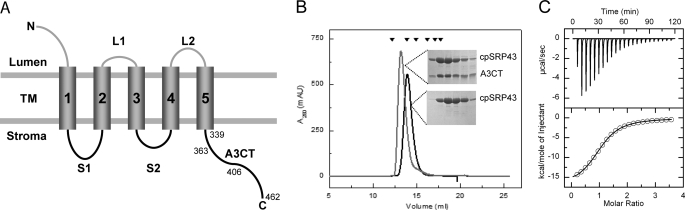
To characterize the interaction of Alb3 with cpSRP43, we first probed the stromal regions of Alb3 with purified cpSRP43 using immobilized peptide libraries (26) (Fig. 1A; supplemental Fig. S1A). Strong signals were observed with the C terminus of Alb3 (A3CT), whereas the region corresponding to the first stromal loop (S1) of Alb3 gave only weak signals. The peptides representing the second stromal loop of Alb3 (S2) gave no signals. These data support our previously reported interaction between cpSRP43 and Alb3 (18) and suggest that the binding site is located in the S1 loop and/or at the C terminus of Alb3. To validate these interactions, recombinantly produced Alb3 S1 loop and A3CT were tested for cpSRP43 binding by SEC and ITC. Binding of the S1 loop to cpSRP43 could not be confirmed using ITC or SEC. A3CT (residues Leu-339–Val-462) formed a stable complex with cpSRP43 as shown by SEC (Fig. 1B, gray line). A dissociation constant of 9.7 μm was determined by ITC with a stoichiometry of 1:1 (Fig. 1C; Table 1). These data show that Alb3 interacts with cpSRP43. To test whether the C-terminal domain of Alb4 (A4CT) can also interact with cpSRP43, recombinantly produced A4CT was analyzed for cpSRP43 interaction by ITC. No interaction could be detected (Table 1; supplemental Fig. S2D). Therefore, cpSRP43 selectively interacts with Alb3.
FIGURE 1.
The Alb3 C-terminal domain interacts with cpSRP43. A, membrane topology of Alb3. Mature Alb3 is a polytopic membrane protein comprising five TMs. The topology prediction was generated by the use of the TopPred II software (57). The luminal, stromal, and C-terminal (C) domains are labeled. N, N terminus; B, analysis of cpSRP43-A3CT complex formation. cpSRP43 alone (black line) or a mixture of cpSRP43 and A3CT in a 1:2 ratio (gray line) was subjected to SEC. Peak fractions were analyzed by SDS-PAGE (inset). The standard proteins used for calibration are indicated by black triangles: γ-globulin (158 kDa), bovine serum albumin (66 kDa), ovalbumin (44 kDa), carboanhydrase (29 kDa), myoglobin (17 kDa), and cytochrome c (12.4 kDa). C, determination of binding affinities for cpSRP43-A3CT interaction ITC. Analysis of the titration isotherms resulted in a Kd of 9.7 μm for A3CT and a stoichiometry of 1:1. Data analysis was performed using the Origin 7.0 software. See also Table 1.
TABLE 1.
Analysis of Alb3-cpSRP interactions by ITC
| Cell | Syringe | KD | n |
|---|---|---|---|
| μm | |||
| Alb3-cpSRP43 interaction: contribution of four conserved motifs in A3CT | |||
| A3CT | cpSRP43 | 9.7 (±0.3) | 1.09 (±0.01) |
| A3CTII–IV | cpSRP43 | 11.4 (±0.2) | 1.07 (±0.02) |
| A3CTIII | cpSRP43 | No interaction observed | |
| A3CTII+III long | cpSRP43 | 194.2 (±22.3) | 1.35 (±0.25) |
| A3CTII+III short | cpSRP43 | 152.4 (±28.8) | 1.67 (±0.36) |
| A3CTIV | cpSRP43 | 207.9 (±24.0) | 1.63 (±0.24) |
| Alb4-cpSRP43 interaction | |||
| A4CTFL | cpSRP43 | No interaction observed | |
| Alb3-cpSRP43 interaction: contribution of the cpSRP43 chromodomains | |||
| A3CT | cpSRP43Δ3 | 53.2 (±6.8) | 0.89 (±0.14) |
| A3CT | cpSRP43Δ2Δ3 | No interaction observed | |
| A3CT | cpSRP43ΔCD1-Ank4 | 25.3 (±0.8) | 1.10 (±0.01) |
| Influence of mutations in motifs II and IV of A3CT and mutations of the modified aromatic cage of CD2 | |||
| A3CTII–IV/K367A/R377A | cpSRP43 | 91.7 (±8.5) | 1.16 (±0.08) |
| A3CTII–IV/K454A/R455A | cpSRP43 | 95.2 (±3.9) | 0.8 (±0.07) |
| A3CTII–IV K367A/R377A/K454A/R455A | cpSRP43 | No interaction observed | |
| A3CTII–IV | cpSRP43 (Y269A) | 30.4 (±3.1) | 0.92 (±0.05) |
| A3CTII–IV | cpSRP43 (W291A) | 100.6 (±3.4) | 0.95 (±0.02) |
| A3CTII–IV | cpSRP43 (D293A) | 20.8 (±1.3) | 0.97 (±0.02) |
| Formation of the trimeric complex: cpSRP43-cpSRP54 M-domain (RRKR-peptide)-A3CT | |||
| cpSRP43-RRKR | A3CTII–IV | 50.8 (±4.7) | 1.11 (±0.06) |
| cpSRP43 | RRKR | 0.5 (±0.04) | 1.01 (±0.00) |
| cpSRP43-A3CTII–IV | RRKR | 2.2 (±0.1) | 1.03 (±0.01) |
The Alb3 C-terminal Domain Is Intrinsically Disordered and Folds upon Interaction with cpSRP43
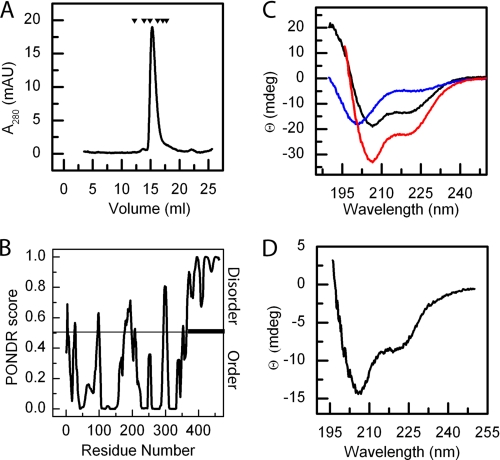
A3CT showed an aberrant migration behavior in SEC and eluted in a fraction indicating a significantly larger molecular mass (Fig. 2A). Although for a 124-residue globular protein a stokes radius of about 19 Å would be expected (29), A3CT eluted in SEC in fractions corresponding to a Stokes radius of 33 Å. This suggests an apparent molecular mass of 38 kDa. The apparent larger hydrodynamic radius observed for A3CT could be due to an extended conformation as observed for intrinsically disordered proteins. Static light scattering refractive index measurements confirmed A3CT to be a monomer. Potential intrinsic disorder of A3CT was analyzed by predictors suited to identify unstructured regions in proteins (Fig. 2B). The region containing the five TMs (between residues Phe-56 and Asp-363) is predicted to be folded, whereas the C terminus including residues Glu-364–Val-462 is clearly predicted as an intrinsically disordered region. The intrinsic disorder of purified A3CT was analyzed by far-UV CD spectroscopy (Fig. 2C, blue line). The far-UV CD spectrum of A3CT exhibited a pronounced minimum around 200 nm and only weak ellipticity above 210 nm, characteristic for unfolded proteins. The formation of an α-helical conformation could be induced by the addition of trifluoroethanol (supplemental Fig. S1B), and secondary structure predictions confirmed the presence of regions in A3CT that could form α-helices (supplemental Fig. S1C). Because A3CT bound to cpSRP43, we tested whether A3CT folds upon binding. CD spectra of the individual components were compared with a CD spectrum of the complex. The observed spectrum of the equimolar mixture (Fig. 2C, red line) differed from the theoretically calculated spectrum of a 1:1 mixture of the single components. The difference spectrum showed two minima around 207 and 222 nm typical for α-helices (Fig. 2D). Therefore, upon binding to cpSRP43, A3CT folds into a conformation that contains α-helical regions. In addition, cpSRP43 was stabilized by about 8 °C upon interaction with A3CT (supplemental Fig. S1D). Taken together, our data show that A3CT is intrinsically disordered. Upon binding to cpSRP43, A3CT adopts a more compact structure and α-helices are formed, supporting a coupled folding and binding model.
FIGURE 2.
A3CT is intrinsically disordered and folds upon binding to cpSRP43. A, analysis of A3CT using SEC. A3CT (molecular mass 15 kDa) shows an aberrant migration behavior corresponding to an apparent molecular mass of 38 kDa. The protein standards (black triangles) are the same as in Fig. 1B. mAU, milliabsorbance units; B, sequence analysis predicts A3CT as intrinsically disordered. PONDR (28) analysis of Alb3 (1–462) from A. thaliana. A large segment of predicted disorder is indicated in the C-terminal region. mdeg, millidegrees. C, analysis of A3CT secondary structure using CD spectroscopy indicates that A3CT is unfolded in solution. Far-UV CD spectra of A3CT (blue line), cpSRP43 (black line), and A3CTFL cpSRP43 in an equimolar mixture (red line) are shown. D, A3CT folds and adopts an α-helical conformation upon binding to cpSRP43. The difference spectrum obtained by subtracting the CD spectrum of cpSRP43 (black line in C) from the spectrum of cpSRP43 and A3CT mixture (red line in C) shows two minima at around 207 and 222 nm typical for α-helices.
The Alb3-cpSRP43 Interaction Requires Two Positively Charged Motifs in Alb3
To analyze the cpSRP43-A3CT interaction in more detail, we performed multiple sequence alignments of Alb3. Although A3CT is more divergent than the transmembrane part, four conserved regions enriched in positively charged residues could be identified (Fig. 3A, motifs I–IV). A number of A3CT variants containing different combinations of these motifs were expressed and purified (Fig. 3B). The A3CT variants were tested for their ability to form a complex with cpSRP43 using SEC, ITC (Table 1; supplemental Fig. S3A), and pulldown experiments (supplemental Fig. S3B). The relative affinities are given in Fig. 3B. ITC experiments showed that A3CT and A3CTII–IV bind to cpSRP43 in a 1:1 stoichiometry with dissociation constants of 9.7 and 11.4 μm, respectively. Removal of motif I therefore did not affect the binding affinity. Further truncations of A3CT from either the C terminus or the N terminus resulted in a drastically reduced binding affinity, suggesting an important role for both motifs II and IV (Table 1; supplemental Fig. S3A). The A3CT variants containing only motif IV or motifs II and III showed a 20-fold reduction in binding affinity. A synthetic peptide (A3CTIII) comprising only motif III was not able to bind to cpSRP43. The deviation from a 1:1 stoichiometry observed for A3CT variants that contain only motifs II and III or IV may indicate the presence of two interaction sites for A3CT in cpSRP43. Our results show that motifs I and III in A3CT are dispensable for binding to cpSRP43, but motifs II and IV are essential for an efficient interaction.
FIGURE 3.
Alb3-cpSRP43 interaction requires two positively charged motifs absent in Alb4. A, multiple sequence alignment of Alb3 C-terminal domains from different plants. Four conserved positively charged motifs (motif I–IV) are present. Sequence numbers for A. thaliana are given above the sequence. Species names are given on the left. Highly conserved residues are shown in dark blue, less conserved residues are in light blue. P. sativum, Pisum sativum; V. vinifera, Vitus vinifera; P. trifoliata, Ptelea trifoliata; O. sativa, Oryza sativa. B, schematic representation of the A3CT constructs used in this study. Dark gray areas indicate the conserved motifs I–IV that have been identified in A. The relative affinities of the truncated A3CT constructs for cpSRP43 as compared with A3CT (containing motifs I–IV) derived from ITC are indicated on the right (see Table 1). For details of the A3CT variants, see the supplemental material. C, sequence alignment of Alb3 and Alb4 C-terminal domains from A. thaliana. The conserved motifs I and III identified in Alb3 proteins in A are present in Alb4 but modified (indicated by an asterisk); motifs II and IV are absent.
Because A4CT did not interact with cpSRP43 (Table 1), we compared the sequences of A4CT and A3CT (Fig. 3C). A4CT is 53 residues longer and is also predicted as intrinsically disordered (supplemental Fig. S2, A and C). As observed for A3CT, A4CT showed an aberrant migration behavior in SEC (supplemental Fig. S2B). Two regions correspond to motifs I and III in A3CT; however, motifs II and IV are absent in A4CT (Fig. 3C). This explains how cpSRP43 discriminates between Alb3 and Alb4. In contrast to Alb3, Alb4 seems not to play a role in post-translational cpSRP-dependent targeting to the thylakoid membrane.
The cpSRP43 Chromodomains 2 and 3 Are Required for the Interaction with Alb3
We next investigated which domains of cpSRP43 are involved in the interaction with Alb3. We produced and purified truncated variants of cpSRP43 (Fig. 4A) and analyzed their ability to interact with A3CT using SEC and ITC (supplemental Fig. S4; Table 1). Deletion of both chromodomains CD2 and CD3 (cpSRP43Δ2Δ3) totally abolished the binding to A3CT. The interaction therefore requires both CD2 and CD3, whereas CD1 at the N terminus of cpSRP43 does not seem to be involved. The CD2CD3 construct bound to A3CT with a dissociation constant of 25 μm, which is about half the affinity measured for the full-length protein. Deletion of only CD3 led to a 5-fold decrease in affinity, but cpSRP43Δ3 still formed a SEC stable complex with A3CT. These data strongly suggest that the interaction between cpSRP43 and Alb3 is mediated mainly by the two chromodomains CD2 and CD3 with CD2 playing an essential role. The higher affinities observed with full-length cpSRP43 and also with the CD2CD3 construct as compared with cpSRP43Δ3 might be due to a stabilizing effect of CD3 on CD2, or CD3 may contribute an additional interaction site.
FIGURE 4.
Alb3 interaction requires cpSRP43 chromodomains 2 and 3. A, schematic representation of cpSRP43 constructs used in this study. The domain organization of cpSRP43 with chromodomains (CD1–3) and ankyrin repeats (Ank1–4) is shown. The relative affinities (rel. aff.) of the truncated cpSRP43 constructs for A3CT as compared with cpSRP43 (derived from ITC, Table 1 and supplemental Fig. S4) are indicated on the right. B, sequence alignment of the three chromodomains of cpSRP43 (A. thaliana) (CD1–3) as compared with canonical chromodomains from HP1 and polycomb (Drosophila melanogaster). The positions of the three aromatic residues forming the aromatic cage in HP1 and PC are indicated by black dots. The positions of the three point mutations (Y269A/W291A/D293A) in cpSRP43 CD2 used in interaction studies are indicated by black stars. Highly conserved residues are shown in dark blue, and less conserved residues are in light blue. C, ribbon representation of the structure of the HP1 in complex with the histone H3 tail (PDB (Protein Data Bank) code: 1kne). HP1 is shown in blue, and the peptide of the histone H3 tail is represented in orange. Canonical chromodomains bind methylated lysines (M3K) in a conserved aromatic cage. The adjacent peptide extends the β-sheet of the chromodomain. Panels C and D were created using PyMOL (58). D, ribbon representation of cpSRP43 CD2 (model based on the crystal structure of CD1, PDB code 3deo; in green). The third aromatic residue forming the aromatic cage is absent in cpSRP43 chromodomains, and the aromatic cage is not assembled in the absence of an interacting peptide. The arrow indicates a possible movement of the N-terminal region upon binding of a histone-like peptide derived from A3CT.
The cpSRP43 Chromodomains Interact with an AKRS Motif in Alb3
cpSRP43 interacts with the motifs II and IV in the C-terminal region of Alb3, and CD2 plays a central role in this interaction. The interaction of chromodomains with their substrates has been characterized in detail for their roles e.g. in chromatin remodeling (30, 31). There, chromodomains interact with histone tails by β-augmentation and accommodate methylated lysines in a so-called “aromatic cage” formed by three conserved aromatic residues (Trp, Tyr, or Phe) (32) (Fig. 4, B and C). Sequence comparison of polycomb (PC) and heterochromatin protein 1 (HP1) with CD1–3 of cpSRP43, however, showed that they are different (Fig. 4B). In CD1–3 of cpSRP43, the third aromatic residue forming the cage is missing. The crystal structure of cpSRP43 revealed an aspartate residue adjacent to the (missing) third aromatic residue that points toward the cage in CD1 (Fig. 4D) (14). This aspartate is conserved in CD1–3 of cpSRP43 and might contribute to a modified aromatic cage. It could form a salt bridge with a cation (e.g. a lysine or arginine residue) bound to the modified aromatic cage. We hypothesized that if motifs II and IV in Alb3 are recognized by CD2CD3 of cpSRP43 in a similar manner as described for HP1 or PC, the removal of the aromatic cage residues should have an impact on the binding affinity. The D293A, Y269A, and W291A mutants showed indeed a drop in A3CT binding affinity by a factor of 2, 3, and 10, respectively (Table 1; supplemental Fig. S5B). CD2 might therefore interact with A3CT employing a modified aromatic cage. However, the effects of the point mutations in CD2 on A3CT interaction were lower than what was observed for corresponding mutations in HP1 and PC. Therefore, the precise mode of interaction may yet be different. The chromodomains of polycomb and HP1 were shown to specifically recognize conserved ARK(S/T) motifs in the tail of histone H3 (supplemental Fig. S5A) (31–33). A closer inspection of motifs II and IV in A3CT showed that they contain AKRS and SKRS sequences reminiscent of ARK(S/T) motifs in the histone H3 tail (Fig. 3A; supplemental Fig. S5A). To validate this observation, we mutated both the lysine and the arginine residues in motifs II and IV to alanines (A3CTII–IVK376A/R377A, A3CTII–IVK454A/R455A). These mutations reduced the affinity for cpSRP43 by about 10-fold as shown by ITC (Table 1; supplemental Fig. S5B). When both motifs were mutated at the same time (A3CTII–IVK376A/R377A/K454A/R455A), binding was not observed by ITC. This suggests that the AKRS/SKRS sequences in Alb3 motifs II and IV are directly involved in the interaction with cpSRP43.
The cpSRP43 Chromodomains Can Bind Alb3 and cpSRP54 Simultaneously
It was previously shown that cpSRP43 CD2 is necessary and sufficient to bind the cpSRP54 M-domain (15, 34, 35). A stretch of 10 amino acids (RRKR peptide, supplemental Fig. S5A) in the C-terminal region of the M-domain was identified as the interacting region (16). Because we identified two motifs in Alb3 that interact with the CD2CD3 domains of cpSRP43, we asked whether cpSRP43 can simultaneously bind both the cpSRP54 M-domain and A3CT. We performed a competition pulldown assay using cpSRP43, A3CT-His6, and cpSRP54 M-domain (Fig. 5A). cpSRP43, the M-domain, and A3CT were all retained by the nickel-agarose beads. Neither cpSRP43 nor the M-domain bound nonspecifically to the beads, and the M-domain did not bind to A3CT (supplemental Fig. S6C). Therefore, this experiment indicates that the M-domain and A3CT bind concomitantly to cpSRP43. The trimeric complex was, however, not stable using SEC, and the dimeric cpSRP43-M-domain complex and A3CT eluted separately (supplemental Fig. S6, A and B). This indicates a decrease in A3CT-cpSRP43 binding affinity in the trimeric complex. Because full-length cpSRP43 bound A3CT more tightly than cpSRP43Δ3 (Table 1), we analyzed the contribution of CD3 to the trimeric complex and performed the competition pulldown assay with cpSRP43Δ3 (supplemental Fig. S6C). Deletion of CD3 did not affect the formation of the trimeric complex. cpSRP43Δ3 can bind both A3CT and the M-domain at the same time. Hence, CD2 appears to be the central binding platform that interacts with both cpSRP54 and Alb3. To analyze the formation of the trimeric complex quantitatively, we used ITC. There, the cpSRP54 M-domain was replaced by the RRKR peptide (16) (Fig. 5B; supplemental Fig. S6E). We first formed the cpSRP43-RRKR peptide complex. Subsequently, the trimeric complex was formed by adding A3CT. The binding affinities for the cpSRP43-RRKR peptide complex were similar as described before (35, 36) (Fig. 5B; Table 1). A 5-fold decrease in A3CT binding affinity was observed when the cpSRP43-RRKR complex was preassembled (53 μm) as compared with free cpSRP43 (11 μm). The same effect was observed when the trimeric complex was formed in reverse order. This confirms the results from SEC and shows that the cpSRP54 M-domain and Alb3 can interact simultaneously with CD2 of cpSRP43. The 5-fold decrease in affinity shows that the two binding events are coupled by negative cooperativity with a coupling energy ΔΔGint of −0.8 kcal/mol.
FIGURE 5.
cpSRP43 can interact with Alb3 and cpSRP54 simultaneously. A, analysis of complex formation using a competition pulldown assay. cpSRP43-A3CT-His6 (labeled 1 and 3) was incubated with increasing amounts of cpSRP54 M-domain (labeled 2) up to 12.5 molar excess (for details, see the supplemental material). A3CT, cpSRP43, and the cpSRP54 M-domain form a trimeric complex. B, thermodynamic cycle of trimeric complex formation by cpSRP43 (labeled 43), A3CT, and the RRKR peptide (RRKR, representing the M-domain of cpSRP54). The two binding events, A3CT and RRKR peptide binding to cpSRP43, are coupled and show negative cooperativity. The affinities were derived from the ITC data (Table 1).
DISCUSSION
Alb3 is a member of the YidC/Oxa1/Alb3 family that controls the membrane insertion of a large and diverse group of membrane proteins. cpSRP and Alb3 cooperate in the post-translational targeting and membrane insertion of members of the LHCP family in chloroplasts of higher plants and algae (8, 37). LHCP insertion into the thylakoid membrane strictly depends on both cpSRP and Alb3 (9, 38, 39). However, the interactions that link the post-translational targeting machinery (cpSRP) to the membrane-embedded insertase Alb3 were not known.
The Alb3-cpSRP43 Interaction Follows a Coupled Folding and Binding Mechanism
In this study, we have demonstrated that the C-terminal region of Alb3 is intrinsically disordered, binds to cpSRP43, and folds upon interaction. Alb3 thereby recruits the transit complex (cpSRP43-cpSRP54-LHCP) to the thylakoid membrane, which is a prerequisite for efficient insertion of LHCPs. The intrinsic disorder of the C-terminal domain of Alb3 might be advantageous to provide orientational freedom in searching for and binding to the transit complex in the stroma. Such a mechanism was recently proposed for the intrinsically disordered C-terminal domain of the Shaker potassium channel where it might modulate the kinetics of channel activation (40). Intrinsically disordered regions are associated with a broad repertoire of biological functions including cellular control mechanisms and signaling. Folding of intrinsically disordered regions upon binding to their partners has been described for a number of physiologically relevant protein-protein interactions, and general mechanisms are arising (41). The moderate affinities measured in our study for the Alb3-cpSRP43 interaction would support relatively short interaction times typical for dynamic processes where transient binding and unbinding reactions are important.
The C-terminal Domains of YidC/Oxa1/Alb3 Membrane Insertases Determine Their Roles in Co- and Post-translational Targeting
The C-terminal domain of Alb3 contains two motifs with net charges of +3 and +8 that are essential for cpSRP43 interaction in post-translational targeting. Oxa1 also has a positively charged C terminus, which is required for ribosome binding. This interaction is crucial for its function in co-translational membrane insertion (42, 43). Cox18, the paralog of Oxa1 in Saccharomyces cerevisiae, lacks the C-terminal extension and is restricted to post-translational activity only (44). Positively charged C-terminal extensions similar to Oxa1 are found in YidC of some bacteria (45, 46). Although the C-terminal region of E. coli YidC is shorter and contains less positive charges, it also mediates YidC-ribosome interaction (47). Therefore, the members of the YidC/Oxa1/Alb3 family utilize positively charged C-terminal regions for specific interactions. In the case of a co-translational function, the interaction partner is the ribosome. For the post-translational function of Alb3, the interaction partner is cpSRP43. This suggests conserved mechanisms of co-and post-translational membrane targeting in chloroplasts, mitochondria, and bacteria.
Alb3 and Alb4 mainly differ in their C-terminal domains, and Alb4 lacks motifs II and IV. Because A4CT does not bind to cpSRP43, it should therefore not be able to recruit the transit complex to the thylakoid membrane. This may explain why Alb4 cannot compensate for the loss of Alb3 (25). A4CT might function as an interaction module but for a different yet unidentified binding partner. Similar to Cox18, the Oxa1 paralog in mitochondria (48), Alb4, may have a function in post-translational targeting, independent of cpSRP. Despite the low conservation of Alb3 and Alb4 C-terminal domains, they share two motifs (motif I and III) that might interact with partners that can bind to both homologs. Obvious candidates include cpSecY and the ribosome. Although an interaction of Alb3 with ribosomes has not been described, evidence for an Alb3-cpSecY interaction was provided from co-localization studies and using a yeast split-ubiquitin system (49–51). It is tempting to speculate that motif I or III might be the binding sites for cpSecY. In this case, Alb4 should also interact with cpSecY.
Chromodomain 2 Emerges as a Central Binding Platform in cpSRP-dependent Targeting
Using a number of cpSRP43 truncations, the binding site for Alb3 was identified in the two terminal chromodomains CD2 and CD3. Although CD2 is essential for the interaction, CD3 is not strictly required but contributes to the interaction. Chromodomains are well characterized for their pivotal roles in chromatin remodeling and regulation of gene expression (31). They typically act as adaptors, bringing together different proteins in multiprotein complexes and locating them in heterochromatin by protein-protein interactions (52). They are found in eukaryotes, plants, and algae, but cpSRP43 is so far the only example for the use of chromodomains in protein targeting. Previously, cpSRP54 has also been reported to bind to CD2 by a positively charged region located at the C terminus of the M-domain (RRKR peptide) (15, 16). We show that cpSRP54 and the membrane insertase Alb3 both interact with CD2 in cpSRP43 and that a trimeric complex can be formed. We noticed that the binding of Alb3 to cpSRP43 weakens the binding of the M-domain (and the RRKR peptide) by a factor of five and vice versa. The two binding events show negative cooperativity. Two nonexclusive models could explain this behavior. (i) A3CT and the M-domain have a partly overlapping binding interface on cpSRP43. (ii) cpSRP43 can adopt two conformational states that differ in their affinities for the two interaction partners. Evidence that CD2 adopts different conformations was provided recently by an NMR study (35). A conformational change in CD2 occurs upon binding of the RRKR peptide, which interacts in an extended conformation with CD2. The binding mode of the RRKR peptide differs significantly from other chromodomain-peptide complexes (32, 53).
Integrated Model for LHCP Targeting to Alb3
Our data show that cpSRP43 and Alb3 play a central role in post-translational targeting (Fig. 6). In the transit complex, cpSRP43 acts as a LHCP-specific chaperone that, together with cpSRP54, keeps the hydrophobic LHCPs in a conformation competent for membrane insertion and assembly (14) (step 1). Formation of cpSRP allows cpSRP43 to utilize the components of the SRP system for post-translational targeting. These include the membrane-bound SRP receptor cpFtsY (54–56) and the membrane insertase Alb3 (this study). cpSRP43 interacts with the C terminus of Alb3 (step 2) by a coupled binding and folding mechanism. The interaction depends on two positively charged motifs in Alb3 reminiscent of interaction motifs in histone tails recognized by chromodomains in HP1 and PC. The interactions of cpSRP43 with cpSRP54 and Alb3 extend the spectrum of canonical chromodomain interactions, and the cpSRP43 chromodomains are employed in a novel functional context. The negative cooperativity of the two binding events that both require CD2 offers an additional level of regulation that might be necessary for the controlled release of LHCPs from the transit complex into the membrane (step 3). Docking the transit complex to Alb3 could act as a conformational switch that triggers the release of LHCPs. It will therefore be interesting to study the influence of LHCPs on the interactions between cpSRP43 and cpSRP54 as well as with Alb3. Alb4 does not interact with cpSRP43 due to the absence of motifs II and IV and should therefore not participate in cpSRP-dependent post-translational targeting (step 4).
FIGURE 6.
Schematic view on Alb3 function in post-translational targeting. cpSRP54 and cpSRP43 form cpSRP, which sequesters LHCPs into a soluble transit complex (LHCP-cpSRP43-cpSRP54) (step 1). The Alb3 C-terminal domain contains four motifs enriched in positively charged residues (motifs I–IV) (step 2) and recruits the transit complex to the membrane utilizing motifs II and IV. A3CT folds upon binding to cpSRP43 (step 3). cpSRP54 and A3CT can bind simultaneously to CD2. Thereby the transit complex is recruited to the membrane, and cpSRP54 can interact with the membrane-bound receptor cpFtsY. The C-terminal domain of Alb4 lacks motifs II and IV and therefore does not bind cpSRP43 (step 4).
Conclusions
Our results provide the basis for a detailed analysis of YidC/Oxa1/Alb3-mediated membrane insertion. All members of the family that interact with ribosomes in co-translational targeting use positively charged C-terminal regions for binding. Alb3 utilizes positively charged C-terminal motifs in post-translational targeting. Common principles seem to apply even when cpSRP is employed in a most SRP-unlike post-translational targeting complex that involves neither SRP RNA nor ribosomes. Chromodomains once again proved their versatility as adaptors for interactions in multiprotein complexes.
Acknowledgments
We thank Gert Bange, Gunes Bozkurt, Iris Holdermann, Goran Stjepanovic, Klemens Wild, Vladimir Rybin (EMBL-Heidelberg), and Franz Xaver Schmid (Bayreuth) for stimulating discussions and helpful comments on the manuscript, Gabriela Müller for excellent technical assistance, and Valerie Panneels for the schematic.
This work was supported by Collaborative Research Grants SFB 638 and FOR 967) and Grant GRK1188 from the Deutsche Forschungsgemeinschaft and by the German-Israeli-Foundation (to I. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
- SRP
- signal recognition particle
- cpSRP
- chloroplast SRP
- A3CT
- Alb3 C-terminal domain
- A4CT
- Alb4 C-terminal domain
- LHCP
- light-harvesting chlorophyll a,b-binding proteins
- CD
- chromodomain
- TM
- transmembrane helix
- HP1
- heterochromatin protein 1
- PC
- polycomb
- DTT
- dithiothreitol
- SEC
- size-exclusion chromatography
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1.Cross B. C., Sinning I., Luirink J., High S. (2009) Nat. Rev. Mol. Cell Biol. 10, 255–264 [DOI] [PubMed] [Google Scholar]
- 2.Grudnik P., Bange G., Sinning I. (2009) Biol. Chem. 390, 775–782 [DOI] [PubMed] [Google Scholar]
- 3.Luirink J., von Heijne G., Houben E., de Gier J. W. (2005) Annu. Rev. Microbiol. 59, 329–355 [DOI] [PubMed] [Google Scholar]
- 4.Driessen A. J., Nouwen N. (2008) Annu. Rev. Biochem. 77, 643–667 [DOI] [PubMed] [Google Scholar]
- 5.Yi L., Dalbey R. E. (2005) Mol. Membr. Biol. 22, 101–111 [DOI] [PubMed] [Google Scholar]
- 6.Samuelson J. C., Chen M., Jiang F., Möller I., Wiedmann M., Kuhn A., Phillips G. J., Dalbey R. E. (2000) Nature 406, 637–641 [DOI] [PubMed] [Google Scholar]
- 7.Luirink J., Samuelsson T., de Gier J. W. (2001) FEBS Lett. 501, 1–5 [DOI] [PubMed] [Google Scholar]
- 8.Moore M., Harrison M. S., Peterson E. C., Henry R. (2000) J. Biol. Chem. 275, 1529–1532 [DOI] [PubMed] [Google Scholar]
- 9.Sundberg E., Slagter J. G., Fridborg I., Cleary S. P., Robinson C., Coupland G. (1997) Plant Cell 9, 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuenemann D., Gupta S., Persello-Cartieaux F., Klimyuk V. I., Jones J. D., Nussaume L., Hoffman N. E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10312–10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaru-Ampornpan P., Nguyen T. X., Shan S. O. (2009) Mol. Biol. Cell 20, 3965–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schünemann D. (2007) Biol. Chem. 388, 907–915 [DOI] [PubMed] [Google Scholar]
- 13.Groves M. R., Mant A., Kuhn A., Koch J., Dübel S., Robinson C., Sinning I. (2001) J. Biol. Chem. 276, 27778–27786 [DOI] [PubMed] [Google Scholar]
- 14.Stengel K. F., Holdermann I., Cain P., Robinson C., Wild K., Sinning I. (2008) Science 321, 253–256 [DOI] [PubMed] [Google Scholar]
- 15.Goforth R. L., Peterson E. C., Yuan J., Moore M. J., Kight A. D., Lohse M. B., Sakon J., Henry R. L. (2004) J. Biol. Chem. 279, 43077–43084 [DOI] [PubMed] [Google Scholar]
- 16.Funke S., Knechten T., Ollesch J., Schünemann D. (2005) J. Biol. Chem. 280, 8912–8917 [DOI] [PubMed] [Google Scholar]
- 17.Tu C. J., Peterson E. C., Henry R., Hoffman N. E. (2000) J. Biol. Chem. 275, 13187–13190 [DOI] [PubMed] [Google Scholar]
- 18.Tzvetkova-Chevolleau T., Hutin C., Noël L. D., Goforth R., Carde J. P., Caffarri S., Sinning I., Groves M., Teulon J. M., Hoffman N. E., Henry R., Havaux M., Nussaume L. (2007) Plant Cell 19, 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yen M. R., Harley K. T., Tseng Y. H., Saier M. H., Jr. (2001) FEMS Microbiol. Lett. 204, 223–231 [DOI] [PubMed] [Google Scholar]
- 20.Lotz M., Haase W., Kühlbrandt W., Collinson I. (2008) J. Mol. Biol. 375, 901–907 [DOI] [PubMed] [Google Scholar]
- 21.Ravaud S., Stjepanovic G., Wild K., Sinning I. (2008) J. Biol. Chem. 283, 9350–9358 [DOI] [PubMed] [Google Scholar]
- 22.Oliver D. C., Paetzel M. (2008) J. Biol. Chem. 283, 5208–5216 [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y. J., Tian H. F., Wen J. F. (2009) BMC Evol. Biol. 9, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Funes S., Hasona A., Bauerschmitt H., Grubbauer C., Kauff F., Collins R., Crowley P. J., Palmer S. R., Brady L. J., Herrmann J. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6656–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdes L., Bals T., Klostermann E., Karl M., Philippar K., Hünken M., Soll J., Schünemann D. (2006) J. Biol. Chem. 281, 16632–16642 [DOI] [PubMed] [Google Scholar]
- 26.Groves M., Sinning I. (2002) in Peptide Arrays on Membrane Supports: Synthesis and Applications (Koch J., Mahler M. eds) pp. 83–96, Springer Verlag Heidelberg, Germany [Google Scholar]
- 27.Zeev-Ben-Mordehai T., Silman I., Sussman J. L. (2003) Biopolymers 68, 395–406 [DOI] [PubMed] [Google Scholar]
- 28.Li X., Romero P., Rani M., Dunker A. K., Obradovic Z. (1999) Genome Inform. Ser. Workshop Genome Inform 10, 30–40 [PubMed] [Google Scholar]
- 29.Uversky V. N. (2002) Eur. J. Biochem. 269, 2–12 [DOI] [PubMed] [Google Scholar]
- 30.Brehm A., Tufteland K. R., Aasland R., Becker P. B. (2004) Bioessays 26, 133–140 [DOI] [PubMed] [Google Scholar]
- 31.Grewal S. I., Jia S. (2007) Nat. Rev. Genet. 8, 35–46 [DOI] [PubMed] [Google Scholar]
- 32.Jacobs S. A., Khorasanizadeh S. (2002) Science 295, 2080–2083 [DOI] [PubMed] [Google Scholar]
- 33.Fischle W., Franz H., Jacobs S. A., Allis C. D., Khorasanizadeh S. (2008) J. Biol. Chem. 283, 19626–19635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hermkes R., Funke S., Richter C., Kuhlmann J., Schünemann D. (2006) FEBS Lett. 580, 3107–3111 [DOI] [PubMed] [Google Scholar]
- 35.Kathir K. M., Rajalingam D., Sivaraja V., Kight A., Goforth R. L., Yu C., Henry R., Kumar T. K. (2008) J. Mol. Biol. 381, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivaraja V., Kumar T. K., Leena P. S., Chang A. N., Vidya C., Goforth R. L., Rajalingam D., Arvind K., Ye J. L., Chou J., Henry R., Yu C. (2005) J. Biol. Chem. 280, 41465–41471 [DOI] [PubMed] [Google Scholar]
- 37.Schünemann D. (2004) Curr. Genet. 44, 295–304 [DOI] [PubMed] [Google Scholar]
- 38.Amin P., Sy D. A., Pilgrim M. L., Parry D. H., Nussaume L., Hoffman N. E. (1999) Plant Physiol. 121, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klimyuk V. I., Persello-Cartieaux F., Havaux M., Contard-David P., Schuenemann D., Meiherhoff K., Gouet P., Jones J. D., Hoffman N. E., Nussaume L. (1999) Plant Cell 11, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magidovich E., Orr I., Fass D., Abdu U., Yifrach O. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13022–13027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright P. E., Dyson H. J. (2009) Curr. Opin. Struct. Biol. 19, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia L., Dienhart M., Schramp M., McCauley M., Hell K., Stuart R. A. (2003) EMBO J. 22, 6438–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szyrach G., Ott M., Bonnefoy N., Neupert W., Herrmann J. M. (2003) EMBO J. 22, 6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herrmann J. M., Funes S. (2005) Gene 354, 43–52 [DOI] [PubMed] [Google Scholar]
- 45.Dong Y., Palmer S. R., Hasona A., Nagamori S., Kaback H. R., Dalbey R. E., Brady L. J. (2008) J. Bacteriol. 190, 2458–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiefer D., Kuhn A. (2007) Int. Rev. Cytol. 259, 113–138 [DOI] [PubMed] [Google Scholar]
- 47.Kohler R., Boehringer D., Greber B., Bingel-Erlenmeyer R., Collinson I., Schaffitzel C., Ban N. (2009) Mol. Cell 34, 344–353 [DOI] [PubMed] [Google Scholar]
- 48.Funes S., Nargang F. E., Neupert W., Herrmann J. M. (2004) Mol. Biol. Cell 15, 1853–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore M., Goforth R. L., Mori H., Henry R. (2003) J. Cell Biol. 162, 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klostermann E., Droste Gen Helling I., Carde J. P., Schünemann D. (2002) Biochem. J. 368, 777–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasch J. C., Nickelsen J., Schünemann D. (2005) Appl. Microbiol. Biotechnol. 69, 440–447 [DOI] [PubMed] [Google Scholar]
- 52.Eissenberg J. C. (2001) Gene 275, 19–29 [DOI] [PubMed] [Google Scholar]
- 53.Fischle W., Wang Y., Jacobs S. A., Kim Y., Allis C. D., Khorasanizadeh S. (2003) Genes Dev. 17, 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandrasekar S., Chartron J., Jaru-Ampornpan P., Shan S. O. (2008) J. Mol. Biol. 375, 425–436 [DOI] [PubMed] [Google Scholar]
- 55.Stengel K. F., Holdermann I., Wild K., Sinning I. (2007) FEBS Lett. 581, 5671–5676 [DOI] [PubMed] [Google Scholar]
- 56.Marty N. J., Rajalingam D., Kight A. D., Lewis N. E., Fologea D., Kumar T. K., Henry R. L., Goforth R. L. (2009) J. Biol. Chem. 284, 14891–14903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Claros M. G., von Heijne G. (1994) Comput. Appl. Biosci. 10, 685–686 [DOI] [PubMed] [Google Scholar]
- 58.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 59.Siegel L. M., Monty K. J. (1966) Biochim. Biophys. Acta 112, 346–362 [DOI] [PubMed] [Google Scholar]