Abstract
Streptococcus mutans is the primary etiological agent of human dental caries and, at times, of infective endocarditis. Within the oral cavity, the pathogen is subjected to conditions of stress. A well-conserved protein complex named ClpP (caseinolytic protease) plays a vital role in adaptation under stress conditions. To gain a better understanding of the global role of the ClpP protease in cellular homeostasis, a transcriptome analysis was performed using a ΔclpP mutant strain. The expression levels of more than 100 genes were up- or downregulated in the ΔclpP mutant compared to the wild type. Notably, the expression of genes in several genomic islands, such as TnSmu1 and TnSmu2, was differentially modulated in the ΔclpP mutant strain. ClpP deficiency also increased the expression of genes associated with a putative CRISPR locus. Furthermore, several stress-related genes and genes encoding bacteriocin-related peptides and many transcription factors were also found to be altered in the ΔclpP mutant strain. A comparative analysis of the two-dimensional protein profile of the wild type and the ΔclpP mutant strains showed altered protein profiles. Comparison of the transcriptome data with the proteomic data identified four common gene products, suggesting that the observed altered protein expression of these genes could be due to altered transcription. The results presented here indicate that ClpP-mediated proteolysis plays an important global role in the regulation of several important traits in this pathogen.
The low-GC-containing Gram-positive bacterium Streptococcus mutans, which is part of the normal flora of the human oral cavity, is considered to be the primary etiological agent in the formation of dental caries (31). Dental caries is a global health problem that affects 60 to 90% of school children, as well as many adults, in industrialized countries (23, 38, 39). While it is not fatal, dental caries is an infectious and costly disease, leading to annual expenditures of billions of dollars in the United States alone (44). S. mutans is also a leading causative agent of bacterial endocarditis, with greater than 14% of viridans streptococcus-induced endocarditis triggered by infections with S. mutans. S. mutans colonizes the oral cavity by metabolizing the dietary carbohydrates ingested by its host to produce glucan and to anchor itself to the tooth surface, forming densely populated microbial communities known as biofilms, commonly referred to as dental plaque, that include hundreds of other species of oral bacteria (44).
The environment in the oral cavity changes rapidly, and during oral colonization, S. mutans often encounters adverse environmental conditions, such as fluctuation in pH, variation in temperature, and alteration in oxygen tension or osmolarity (8). To cope with the environmental stresses, S. mutans has developed several strategies that allow it to grow under harsh conditions (for recent reviews, see references 27 and 28). For example, bacteria transiently induce the expression of a subset of proteins that are either molecular chaperones or proteases, which are collectively known as heat shock proteins (HSPs). Unfolded or misfolded proteins accumulate inside the cell due to stress, and molecular chaperones are required for proper folding and assembly of those aberrant proteins. Proteases are involved in the degradation of the proteins, not only under stress conditions, but also under normal growth conditions (13, 20).
In bacteria, there are several classes of cytoplasmic proteases that are responsible for the degradation of normal or abnormal proteins (19, 20, 25). For example, in the most studied Gram-negative bacterium, Escherichia coli, at least four classes of energy-dependent proteases, such as Lon, FtsH, HslUV, and Clp, are involved in protein turnover. While Clp and FtsH are widely present in bacteria, Lon and HslUV are absent in many Gram-positive bacteria, including all streptococcal species. In these bacteria, Clp-mediated proteolysis appears to be the most important for stress tolerance and global regulation (16).
The bacterial Clp proteolytic complex is structurally similar to the eukaryotic 26S proteasomal complex and is comprised of an AAA+ ATPase subunit and a proteolytic component known as ClpP (caseinolytic protease) (32, 49). The AAA+ ATPases typically recognize, denature, and translocate protein substrates into the proteolytic core of the ClpP peptidase for subsequent digestion (36). While all streptococci possess five Clp ATPases (ClpB, ClpC, ClpE, ClpL, and ClpX), they encode only one ClpP protease (3, 16). Interestingly, only three AAA+ ATPAses (ClpC, ClpE, and ClpX) have the distinct tripeptide sequence that is required for interaction with ClpP protease to form the Clp-proteolytic complex (16). Often adaptor proteins, which enhance and diversify the substrate spectra of their cognate AAA+ ATPases, modulate the function of the Clp-proteolytic complexes. The other two ATPases, ClpB and ClpL, do not interact with ClpP but are thought to cooperate with the HSP-70 system, which includes DnaJ, DnaK, and GrpE, to form a chaperone complex to disaggregate and refold denatured or misfolded proteins (18).
As in other bacteria, ClpP in S. mutans also plays an indispensable role in cellular protein quality control. Strains that are devoid of functional ClpP exhibit slow growth at elevated temperature, formation of long chains, increased clumping in liquid culture, reduced autolysis, and altered biofilm formation (11, 22, 29). We and others have previously demonstrated that clpP mutants also display reduced tolerance for several environmental stresses and that ClpP is required for adaptive response to toxic reagents, such as chlorhexadine and sodium fluoride (4, 11). Although ClpP-mediated proteolysis is undoubtedly very important for the proper display of virulence attributes by S. mutans, the molecular determinants responsible for the observed phenotypic defects are largely unknown.
To gain better insight into the “housekeeping” global role in protein degradation and how it relates to physiology and virulence, a previously constructed strain of S. mutans UA159 with clpP deleted was used (4), and global gene expression was studied by comparative DNA microarray analysis. Furthermore, to search for novel ClpP substrates, comparative proteomics analysis was also performed. We report here that deletion of clpP leads to alteration in the expression of genes belonging to bacteriocin production and the stress tolerance response. ClpP also differentially regulates the expression of several genes in genomic islands (GIs). Moreover, a mutant strain lacking clpP displayed a wide range of phenotypic defects, including reduced tolerance for DNA and cell wall-damaging agents and reduced mutacin production.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Streptococcus mutans UA159 and its derivatives were routinely grown in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY). E. coli strain Top10 was grown in Luria-Bertani (LB) medium. Antibiotics, such as ampicillin (100 μg/ml for E. coli), erythromycin (7.5 μg/ml for S. mutans and 400 μg/ml for E. coli), and kanamycin (50 μg/ml for E. coli), were used in growth media as required. IBS512, a ΔclpP strain derived from S. mutans UA159, was used for all our analyses (4). This ΔclpP strain was generated by a Cre-lox-mediated markerless gene deletion system (4).
For growth of S. mutans in the presence of DNA damage-inducing agents, mitomycin C (6 ng/ml) was added to THY agar medium before it was poured. Freshly grown overnight cultures were spun down, washed with 0.9% sterile saline solution, and finally resuspended in the same solution. The optical densities of the different S. mutans cultures were adjusted to the same level. Tenfold serial dilutions were prepared, and 7.5 μl was spread on THY agar plates supplemented or not with mitomycin C.
For growth of S. mutans under acidic and neutral pH conditions, the initial pHs of the THY broth media were adjusted to 5.5 and 7.0 with HCl prior to being autoclaved. Citrate-phosphate buffer (pH 5.5 or 7.0; 50 mM) was added to the sterilized media. Bacterial growth in these media was monitored using a Klett-Summerson colorimeter with a red filter (7).
Construction of complementing plasmid.
To express the clpP gene in trans, a fragment containing the clpP gene, with its ribosome binding sequence and upstream repeats, was amplified by PCR from S. mutans UA159 genomic DNA with the primers BamHI-clpP-F1 and HindIII-clpP-R2 (Table 1). The blunt-ended PCR product was cloned into the pCR4-Blunt-TOPO vector (Invitrogen) to generate pIBC3. A fragment containing clpP was isolated from pIBC3 by BamHI and EcoRI restriction and cloned into pIB184, an E. coli-S. mutans shuttle vector (6), using the same restriction sites, to create pIBC7.
TABLE 1.
Oligonucletides used in this study
| Name of primer | Primer sequence | Purpose |
|---|---|---|
| BamHI-clpP-F1 | 5′-GATCAGGATCCATCGCTTGTT-3′ | clpP cloning |
| HindIII-clpP-R2 | 5′-GCGCGAAGCTTTTATTTTAATTCATTATTTT-3′ | clpP cloning |
| smu137 F1 | 5′-ATGTATGCACATGACATTTT-3′ | RT-PCR |
| smu137 RTR | 5′-ACCTTCAATCGTATCTGCGATGGTT-3′ | RT-PCR |
| smu 140 F1 | 5′-GTGACTCTAGCCTGCAACTT-3′ | RT-PCR |
| smu140 RTR | 5′-TTCAAAGAAAGCATAGGCTTCATAA-3′ | RT-PCR |
| smu208c F1 | 5′-ATGGTGAGAAAGCTTTTTTA-3′ | RT-PCR |
| smu208c RTR | 5′-AGTAACCGTCCGTTTCTCTCTCTGT-3′ | RT-PCR |
| RNase HII RTF | 5′-ATGGCAACGATTAAGG-3′ | RT-PCR |
| RNase HII RTR | 5′-CCTGCCAACTTCATCAATAC-3′ | RT-PCR |
| clpE RTF | 5′-ATGCTCTGTCAAAATTGTAA-3′ | RT-PCR |
| clpE RTR | 5′-CCGGGACGGCGATTATTGCC-3′ | RT-PCR |
| smu947 RTF | 5′-ATGAACAATAAGCGAGAAAA-3′ | RT-PCR |
| smu947 RTR | 5′-CACCTTCTGCCTTAAACTGT-3′ | RT-PCR |
| smu1067c RTF | 5′-ATGACCATACTATATCATG-3′ | RT-PCR |
| smu1067c RTR | 5′-CCTTCTTCTTCCTTAGAAAG-3′ | RT-PCR |
| smu-bacA-F1 | 5′-GGGGCAAAGAATTATCAATTGAAGCTCCAACC-3′ | RT-PCR |
| smu-bacA-R1 | 5′-GCATTTGAAAATTGATCTACTTCAACTGGAGC-3′ | RT-PCR |
| smu bac C F | 5′-AACCTCAAAATAAAATCGAAGAAACGC-3′ | RT-PCR |
| smu bac C R | 5′-GCTGTGGAAGAAGGATTAAATAGCTGA-3′ | RT-PCR |
| psa RTF | 5′-CAGGAAATTGAAGCAGTAAAGCATG-3′ | RT-PCR |
| psa RTR | 5′-CCCTTTGTTGTTCACCTCCTGA-3′ | RT-PCR |
| smu1349 F1 | 5′-ATGACTAAGGGAACAAGGGAAAA-3′ | RT-PCR |
| smu1349 R1 | 5′-TTATTGGTTTCTCTTCTTTTTAT-3′ | RT-PCR |
| nlmA-F | 5′-ATGGATACACAGGCATTTGA-3′ | RT-PCR |
| nlmA-R | 5′-ATGGATCGAATGAGTCC-3′ | RT-PCR |
| nlmB-RT F | 5′-ATGGAATGGAGAATTAATACCATG-3′ | RT-PCR |
| nlmB-RT R | 5′-GTGTGGAAAAACTACAGATCCA-3′ | RT-PCR |
| nlmC-F | 5′-ATGAATACACAAGCATTTGA-3′ | RT-PCR |
| nlmC-R | 5′-CTAACCACAGGAATTAAGAG-3′ | RT-PCR |
| nlmT-F | 5′-TTAATCGTCATAGCCGTTAACATTCTCTTAGAGA-3′ | RT-PCR |
| nlmT-R | 5′-CCTTACTCATCCTAGTCACCTTAACTGAAGGAT-3′ | RT-PCR |
| comD-F | 5′-CGTAAATTTGTCGTTATTGTCTATC-3′ | RT-PCR |
| comD-R | 5′-GACAAGATACCCTTGACAGCATCG-3′ | RT-PCR |
| comE-F | 5′-TACAACAAGGACGTCTTGAAACCACCATTG-3′ | RT-PCR |
| comE-R | 5′-CAGAATCTCAGCAAAGGGACCTGAAACTG-3′ | RT-PCR |
| smu gyrA F1 | 5′-CTCTTCCAGATGTTCGCGATGG-3′ | RT PCR |
| smu gyrA Rev | 5′-GGCCATTCCAACAGCAATACCTGTCGCTCC-3′ | RT PCR |
Deferred antagonism assay.
Mutacin IV production by S. mutans was evaluated by a deferred antagonism assay (10). Briefly, S. mutans UA159 and its derivatives were stabbed into THY agar plates and allowed to grow under microaerophilic conditions at 37°C for 6 or 8 h. The plates were then overlaid with 10 ml of soft agar containing overnight-grown cultures (500 μl) of the indicator strains of choice (Streptococcus gordonii DL-1 and Streptococcus cristatus 5100). The plates were allowed to solidify at room temperature for 10 min. Then, they were incubated overnight at 37°C under microaerophilic conditions. The following day, the zones of inhibition of the indicator strains were evaluated for the different strains. Production of mutacin V (NlmC) was evaluated in a similar manner, except that Lactococcus lactis strain MG1363 was used as the indicator strain.
Antibiotic susceptibility stress.
Disc diffusion assays were performed to evaluate the antibiotic susceptibility of S. mutans UA159 and its derivatives as described previously (5). Briefly, antibiotic discs (6 mm in diameter; Becton Dickinson Laboratories) were placed on THY agar plates that were overlaid with 10 ml of soft THY agar containing 200 μl of the S. mutans strain of choice. The plates were incubated overnight at 37°C under microaerophilic conditions, and the zones of inhibition were measured. The antibiotics used for this study were bacitracin (10 U), cefaxolin (30 μg), cefixime (5 μg), cefoxitin (30 μg), imipenem (10 μg), levofloxacin (5 μg), polymixin B (300 U), trimethoprim (5 μg), and vancomycin (5 μg).
Isolation of RNA from bacterial cultures.
Total RNA was isolated from bacterial cultures according to the protocol described previously. Briefly, S. mutans UA159 and its derivatives were grown to mid-exponential phase (70 Klett units), and the cultures were harvested by centrifugation. The cell pellets were then suspended in 10 ml of RNAprotect bacterial reagent (Qiagen) and incubated at room temperature for 10 min. Total RNA was extracted using an RNeasy minikit (Qiagen) according to the manufacturer's instructions, with a modified bacterial-lysis step. The supernatants were loaded onto RNeasy minicolumns and purified using the manufacturer's protocol. The purified RNA samples were further treated with Turbo DNase (Ambion) according to the manufacturer's instructions to remove residual DNA contamination. The quality and quantity of the purified RNA samples were ascertained using a spectrophotometer and agarose gel electrophoresis.
Semiquantitative RT-PCR.
Total RNA (DNA free) was used to prepare cDNA, using Superscript II reverse transcriptase (Invitrogen). Briefly, RNA samples (500 ng) were mixed with random primer (100 ng) and deoxynucleotide triphosphates (dNTP) (0.5 mM), and the cocktail was heated at 65°C for 5 min, followed by quick chilling on ice. First-strand buffer (SuperScript II reverse transcriptase [Invitrogen]), 10 mM dithiothreitol (DTT), and RNase inhibitor (40 U; Roche) were added to the cocktail and incubated at room temperature for 2 min, and reverse transcriptase was added to the reaction mixture. The reaction mixture was further incubated at room temperature for 10 min, followed by incubation at 42°C for 50 min to synthesize cDNA. The reaction mixture was heat inactivated at 70°C for 15 min. To degrade the DNA-RNA hybrid, RNase H (2 U; Invitrogen) was added to the reaction mixture and incubated at 37°C for 45 min. Finally, the cDNA was purified using a PCR purification kit (Qiagen), and the cDNA concentration was determined using a UV spectrophotometer (Shimadzu). Five to 20 ng of cDNA was used to carry out the second-step PCR, using ReadyMix Taq PCR mix with MgCl2 (Sigma). Twenty to 24 PCR cycles were carried out to amplify the cDNA products of interest. The amplified PCR products were then electrophoresed on a 2% agarose gel. As an internal control, the gyrA gene was used to ensure that equal amounts of cDNA were used in each reverse transcription (RT)-PCR.
Whole-genome microarray.
A high-density microarray from NimbleGen Systems, Inc. (Madison, WI), was used for transcriptome analysis. The array contained 19 replicates of each 60-mer oligonucleotide probe designed for the 1,960 open reading frames in the genome of S. mutans UA159 (3). Further, the whole genome was represented five times on each chip (i.e., five technical replicates per chip), so that there was a total of 95 probes per gene per chip. Total RNA samples from bacterial cultures (harvested at the mid-log phase of growth; 70 Klett units) were isolated for labeling and hybridization. For each strain, two independently isolated RNA samples were analyzed using the array chips, generating two sets of data for each strain. Gene expression values were calculated with ArrayStar 3.0 software (DNAStar) and normalized with the algorithm available with the software. The moderated t test was applied to the data set, and the P value (from the t test), assigned as ≤0.05, was used to show the significance of the difference between the experimental pairs. The genes with a cutoff of 1.5 or greater were selected for further analysis.
Two-dimensional gel electrophoresis and spot identification.
S. mutans strains UA159 and IBS512 were grown to mid-exponential growth phase, and the cells were harvested. Total cellular lysates were prepared in a buffer containing 10 mM Tris-Cl, pH 7.4, and 0.3% sodium dodecyl sulfate (SDS), and the protein concentrations in the samples were determined using a bicinchoninic acid (BCA) assay. Protein samples were sent to Kendrick Laboratories, Inc. (Madison, WI), for two-dimensional gel electrophoresis, which was performed according to the carrier ampholine method of isoelectric focusing (IEF) (35). IEF was carried out in glass tubes with 12.5 μg of each sample, which was spiked with 50 ng of tropomyosin (molecular weight [MW], 33,000; pI, 5.2), as an internal standard. Electrophoresis with a 10% SDS slab gel was carried out as the second dimension. The gels were stained with a special silver stain that omitted the glutaraldehyde step and that was compatible with mass spectroscopy. Two independent samples were run for each strain. The differences between two similar spots were calculated from spot percentages (the individual spot density divided by the total density of all measured spots). Spots that showed at least 1.7-fold difference between IBS512 and UA159 with a value of ≤0.05 (t test) were extracted, and their identities were determined by ESI (electrospray ionization) mass spectrometry.
RESULTS
Characterization of a ΔclpP mutant strain.
In S. mutans, ClpP, the major intracellular protease, is responsible for mounting an adaptive stress response under thermal and acid stresses by degrading the misfolded proteins that accumulate under stress conditions (15, 16, 27, 29). However, the role of ClpP in intracellular protein quality control under normal growth conditions or in other stress responses is not well known. To understand the function of ClpP in the regulation of protein turnover, we used a strain with clpP deleted, IBS512, which was previously constructed in our laboratory (4). In this strain, the clpP gene was deleted by a Cre-loxP system without introducing any antibiotic resistance gene, and this deletion construction did not generate any polar effect, as judged by RT-PCR analysis of the downstream genes (reference 4 and data not shown). In order to complement the clpP deficiency, the entire clpP open reading frame, along with the promoter region, was cloned into pIB184 (6), an E. coli-streptococcal shuttle plasmid, to generate pIBC7. IBS512 was transformed with pIBC7 to generate a complementing strain, while both IBS512 and UA159 were transformed with the vector plasmid pIB184, so that all three strains could be grown under similar growth conditions. We first verified the stress-sensitive phenotypes of the three strains. As expected, the growth of the strain with clpP deleted was severely affected at high temperature compared to the growth of the wild-type strain (data not shown). Since clpP mutants in other bacteria are also sensitive to acid stress, to elucidate the effect of a clpP null mutation on the acid stress response, a growth kinetics study was carried out for all three strains in THY broth (pH 5.5 or 7.0) supplemented with erythromycin, as described in Materials and Methods. As expected, the growth of IBS512(pIB184) at pH 5.5 was also severely affected relative to that of UA159(pIB184) (data not shown). This growth defect was partially complemented in IBS512(pIBC7). Thus, it appears that ClpP is also involved in the acid stress tolerance response in S. mutans.
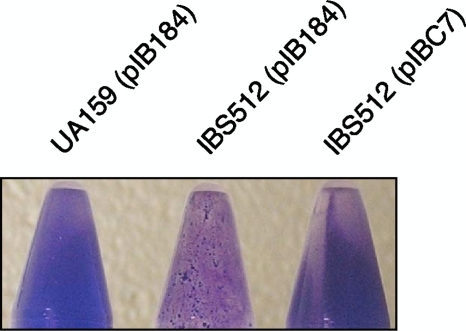
The abilities of the UA159(pIB184), IBS512(pIB184), and IBS512(pIBC7) strains to form biofilm were tested on a polypropylene surface in CDM medium (complete defined medium) supplemented with 0.5% sucrose. In contrast to the study reported by Deng et al. (11), where increased biofilm formation by a clpP-deficient S. mutans strain in the presence of sucrose was observed, we found that biofilm formation by IBS512 was drastically reduced when the polypropylene surface was used as a solid support (Fig. 1); however, under identical growth conditions, the planktonic growth of IBS512 was similar to that of the wild-type strain (data not shown). The biofilm formed by IBS512 was patchy in nature, with large aggregates, while the biofilms appeared to be uniform in both the wild-type [UA159(pIB184)] and the complementing [IBS512(pIBC7)] strains. Thus, our results suggest that ClpP is required for optimum biofilm formation by S. mutans.
FIG. 1.
Inactivation of clpP causes reduction of biofilm formation by S. mutans. Biofilms were grown in CDM medium containing sucrose on a polypropylene surface. After 72 h of growth, biofilms of UA159 (wild type), IBS512 (ΔclpP mutant), and IBS512(pIBC7) (ΔclpP or clpP+) were stained with crystal violet. Experiments were repeated at least two times, and representative biofilms are shown.
Transcriptome analysis of a ΔclpP mutant strain reveals an altered global transcriptional profile.
The phenotypic and morphological changes in the ΔclpP mutant suggested that ClpP plays a fundamental regulatory role in the expression of various virulence properties and cellular physiology. To explore the global regulatory role of ClpP in S. mutans, we performed a whole-genome transcriptome analysis by comparing the gene expression pattern of the exponentially grown (70-Klett unit) cultures of IBS512 and UA159 using a high-density NimbleGen microarray chip. Exponentially grown cultures were chosen because clpP transcription appears to be optimum at this growth phase (52). A change in the relative transcript amount of ≥1.5-fold was selected for differential gene expression. Transcriptome analysis revealed that the transcription of 105 genes was downregulated while the transcription of 124 genes was upregulated in the ΔclpP strain. Several differentially expressed genes could be clustered into many groups (Tables 2 and 3), such as genes associated with bacteriocin production and genes associated with a stress response pathway.
TABLE 2.
Important genes of S. mutans differentially expressed in the ΔclpP strain (IBS512)
| Gene name | Description and/or putative function | Fold change relative to WTa |
|---|---|---|
| Stress related | ||
| SMU.22 | Putative peptidoglycan hydrolase; GbpB | 1.5 |
| SMU.81 | Heat shock protein HSP-70 cofactor; GrpE | 2.1 |
| SMU.82 | Molecular chaperone; DnaK | 1.9 |
| SMU.83 | Heat shock protein HSP-40; DnaJ | 2.1 |
| SMU.140 | Glutathione reductase | −3.2 |
| SMU.561 | Nudix hydrolase (DNA repair) | 1.6 |
| SMU.562 | ATP-dependent protease; ClpE | 1.9 |
| SMU.949 | ATP-dependent protease; ClpX | 1.5 |
| SMU.991 | Putative ribonucleotide reductase | 1.5 |
| SMU.1954 | Chaperonin GroEL | 1.7 |
| SMU.1955 | Cochaperonin GroES | 1.6 |
| SMU.2084 | Transcriptional regulator SpxA | 1.8 |
| SMU.2164 | Serine protease HtrA | 2.0 |
| GIs | ||
| GI-4 | ||
| SMU.100 | Putative sorbose PTS system, IIB | −2.7 |
| SMU.101 | Putative sorbose PTS system, IIC | −1.6 |
| SMU.107 | Hypothetical protein | 1.5 |
| SMU.109 | Putative permease | 1.5 |
| GI-6 (TnSmu1) | ||
| SMU.191 | Putative integrase | 1.9 |
| SMU.193 | Hypothetical protein | 1.8 |
| SMU.194 | Bacteriophage P2 associated | 2.0 |
| SMU.195 | Bacteriophage SPP1 ORF5 related | 1.7 |
| SMU.196 | Putative transfer protein with CHAP | 1.8 |
| domain | ||
| SMU.197 | Hypothetical protein | 2.1 |
| SMU.198 | Putative conjugative transposon protein | 2.6 |
| (helicase) | ||
| SMU.199 | Putative permease/transporter | 3.0 |
| SMU.200 | Hypothetical protein | 3.4 |
| SMU.201 | Putative transposon-related protein | 3.6 |
| SMU.202 | Hypothetical protein | 3.6 |
| SMU204 | Hypothetical protein | 3.2 |
| SMU.205 | Hypothetical protein | 3.0 |
| SMU.206 | Hypothetical protein | 3.6 |
| SMU.207 | XRE family-like protein | 2.9 |
| SMU.208 | ABC transporter | 3.1 |
| SMU.209 | Hypothetical protein | 2.7 |
| SMU.210 | Hypothetical protein | 2.6 |
| SMU.211 | Hypothetical protein | 2.7 |
| SMU.212 | Hypothetical protein | 3.2 |
| SMU.213 | Hypothetical protein | 2.4 |
| SMU.214 | Hypothetical protein | 3.8 |
| SMU.215 | Hypothetical protein | 2.6 |
| SMU.216 | Hypothetical protein | 2.8 |
| SMU.217 | Hypothetical protein | 2.3 |
| SMU.218 | Putative transcriptional regulator | 1.4 |
| SMU.222 | Putative integrase | 1.5 |
| SMU.223 | Hypothetical protein | 1.6 |
| GI-12 (TnSmu2) | ||
| SMU.1339 | Smt operon: BacD; bacitracin synthase | −5.3 |
| SMU.1340 | Smt operon: BacA2; surfactin synthase | −5.8 |
| SMU.1341 | Smt operon: Grs; gramicidin S synthase | −6.7 |
| SMU.1342 | Smt operon: BacA; bacitracin synthase 1 | −6.2 |
| SMU.1343 | Smt operon: PksC; polyketide synthase | −6.5 |
| SMU.1344 | Smt operon: FabD; acyl carrier protein | −0.4 |
| transacylase | ||
| SMU.1345 | Smt operon: ItuA; peptide synthase | −6.9 |
| similar to MycA | ||
| SMU.1346 | Smt operon: BacT; putative thioesterase | −6.0 |
| SMU.1347 | Smt operon: YlbB; ABC transporter | −4.3 |
| permease | ||
| SMU.1348 | Smt operon: PsaA; ABC transporter | −4.4 |
| SMU.1349 | Transcriptional regulator; TetR/AcrR | 1.9 |
| family | ||
| SMU.1365 | ABC transporter permease | −4.1 |
| (= SMU.11347) | ||
| SMU.1366 | ABC transporter (= SMU.1348) | −4.5 |
| CRISPR associated | ||
| SMU.1752 | Hypothetical protein | 1.6 |
| SMU.1753 | CRISPR-associated Cas2 family protein | 1.6 |
| SMU.1754 | CRISPR-associated DNA polymerase | 1.6 |
| SMU.1755 | CRISPR-associated Cas1 family protein | 1.6 |
| SMU.1757 | CRISPR-associated Cas1 family protein | 1.5 |
| SMU.1758 | CRISPR-associated Cas4 family protein | 1.7 |
| SMU.1760 | CRISPR-associated Csd2 family protein | 1.7 |
| SMU.1761 | CRISPR-associated RAMP Csd1 family | 2.1 |
| protein | ||
| SMU.1762 | CRISPR-associated RAMP Csd1 family | 1.8 |
| protein | ||
| SMU.1763 | CRISPR-associated protein Cas5 | 2.0 |
| SMU.1764 | CRISPR-associated helicase Cas3 | 2.7 |
| Bacteriocin related | ||
| SMU.150 | Nonlantibiotic mutacin IV: peptide | −2.3 |
| NlmB | ||
| SMU.151 | Nonlantibiotic mutacin IV: peptide | −2.0 |
| NlmB | ||
| SMU.152 | Putative bacteriocin immunity protein | −2.0 |
| SMU.423 | Putative bacteriocin peptide; BsmC | −1.9 |
| SMU.1897 | ABC transport system CslAB | −1.5 |
| SMU.1898 | −2.0 | |
| SMU1899 | −2.0 | |
| SMU.1900 | −2.2 | |
| SMU.1902 | Putative bacteriocin peptide; BsmK | −2.9 |
| SMU.1905 | Putative bacteriocin peptide; BsmL | −2.1 |
| SMU.1906 | Putative bacteriocin peptide; BsmB | −1.8 |
| SMU.1909 | Putative bacteriocin immunity protein | −1.7 |
| SMU.1910 | Putative bacteriocin immunity protein | −1.5 |
| SMU.1913 | BlpL-like immunity protein | −1.7 |
| SMU.1914 | Mutacin V (NlmC or BsmA) | −1.7 |
WT, wild type. P values are given in Table S1 in the supplemental material.
TABLE 3.
Transcriptional regulators differentially expressed in the ΔclpP strain (IBS512)
| Gene name | Description and/or putative function | COGa | Fold change relative to WTb |
|---|---|---|---|
| SMU.61 | Putative transcriptional regulator PlcR | COG1396 | 1.5 |
| SMU.80 | Heat-inducible transcriptional regulator HcrA | COG1420 | 1.8 |
| SMU.135 | Putative transcriptional regulator; MleR related | COG0583 | 1.5 |
| SMU.136 | Putative transcriptional regulator | COG1476 | −1.7 |
| SMU.218 | Putative transcriptional regulator | COG1396 | 1.5 |
| SMU.424 | Transcriptional repressor CopY | COG3682 | −1.5 |
| SMU.1349 | Transcriptional regulator; TetR/AcrR family | COG1309 | 1.9 |
| SMU.1398 | Putative transcriptional regulator | COG2932 | −1.7 |
| SMU.1409 | Putative transcriptional regulator | COG2207 | −1.6 |
| SMU.1728 | Transcription elongation factor GreA | COG0782 | 1.5 |
| SMU.1924 | Response regulator CovR | COG0745 | 1.7 |
| SMU.1969 | Putative transcriptional regulator | COG1846 | −1.6 |
| SMU.2084 | Transcriptional regulator SpxA | COG1393 | 1.8 |
COG, clusters of orthologous groups.
P values are given in Table S1 in the supplemental material.
The expression of 15 genes related to bacteriocin production was downregulated in the ΔclpP mutant strain. S. mutans UA159 predominantly produces two types of bacteriocins, mutacin IV, which is encoded by the nlmA and nlmB genes, and mutacin V, which is encoded by nlmC. The expression of the nlm genes was decreased between 1.7- and 2.3-fold in IBS512. However, the expression of an ABC transport system, encoded by nlmT (SMU.286) and nlmE (SMU.287), which is required for the biogenesis and secretion of mutacin IV in UA159, was not altered in the IBS512 strain (data not shown). On the other hand, the expression of another ABC transport system, encoded by the cslAB locus, was downregulated about 2.0-fold in the strain with clpP deleted (Table 2).
Several genes that are associated with general stress responses were found to be upregulated in IBS512. Genes such as dnaK, dnaJ, groEL, and groES were previously shown to be regulated by ClpP in S. mutans and other bacteria (29, 34, 40). Here, it was found that grpE, which encodes a cofactor of heat shock protein 70, was upregulated in the strain with clpP deleted. Two Clp ATPase-encoding genes, clpE and clpX, were also upregulated in the mutant. A previous microarray study of Streptococcus pneumoniae suggested that ClpP upregulates htrA, which encodes an extracellular serine protease (40). This microarray analysis also found that htrA gene expression was increased about 2.0-fold in the strain with clpP deleted. GbpB, a glucan binding protein, is known to be induced during various stress responses (9). We found that gbpB gene expression was increased in the mutant strain (Table 2). The only stress-related gene that was found to be downregulated in the mutant was SMU.140, which encodes a putative glutathione reductase (Table 2).
The genome of S. mutans contains at least 11 genomic islands, which are thought to have been acquired by the organism by horizontal gene transfer. We found that genes within three different genomic islands were differentially regulated in the clpP mutant (Table 1). Among them were two genomic islands, TnSmu1 and TnSmu2, whose distribution among the different isolates had been previously studied (50, 51). TnSmu1 corresponds to a large region of 23 kb spanning from SMU.191 to SMU.226 (50, 51). TnSmu1, which lies beside a cluster of tRNA genes, encodes many predicted transposases, integrases, transporter proteins, and hypothetical proteins. Most of the genes within TnSmu1 were upregulated in the strain with clpP deleted (Table 2). At least seven genes were induced 3.0-fold or more in the strain with clpP deleted compared to the wild-type strain, UA159.
In contrast to TnSmu1 genes, several genes within TnSmu2 were downregulated in the clpP mutant strain. TnSmu2 is the largest genomic island (57 kb) found in the S. mutans genome, and it contains about 47 genes organized into several operons (50). The largest operon within this genomic island is the smt operon (I. Biswas, unpublished data), which is approximately 32 kb in length. The smt operon includes 10 genes, which show high homology to several secondary-metabolite biosynthesis genes. Expression of all 10 of the genes in the smt operon (SMU.1339 to SMU.1348) was drastically reduced in the clpP mutant strain (Table 2). In fact, the fold difference in the expression of these genes was the highest of all the genes that were differentially regulated in the mutant. TnSmu2 also contains two genes, SMU.1365 and SMU.1366, that are generated due to gene duplication of SMU.1347 and SMU.1348, respectively. Expression of SMU.1365 and SMU.1366 was also drastically decreased (more than 4.0-fold) in the mutant strain. A putative transcriptional regulator of the TetR/AcrR family, SMU.1349, is present just upstream of the smt operon and is transcribed divergently. In contrast to the smt operon genes, expression of SMU.1349 was increased about 2-fold in the clpP mutant strain (Table 2).
The expression of four genes in another genomic island, GI-4, was also differentially regulated in the clpP mutant strain. GI-4 is a small genomic island less than 10 kb in length and includes about 12 genes, two of which encode a sorbose phosphotransferase (PTS) gene cluster (SMU.100 and SMU.101). The expression of these two genes was downregulated in the mutant. The expression of two other genes, SMU.107 and SMU.109, in the same genomic island was upregulated (Table 2). Thus, taken together, it appears that the expression levels of several horizontally transferred genes were differentially affected by the clpP deletion.
Arrays of clustered, regularly spaced short palindromic repeats (CRISPR) are found in many bacteria, and they generally confer immunity against invasion by foreign DNA elements, such as phages (45). The genome of S. mutans includes two CRISPR loci designated CRISPR1 and CRISPR2 (46). CRISPR1 is located between SMU.1400 and SMU.1398 and is preceded by five CRISPR-associated genes (cas) (SMU.1402 to SMU.1405) that are needed for CRISPR function. CRISPR2 is located between SMU.1752 and SMU.1753, and the cas genes are SMU.1754 to SMU.1764, located just upstream of CRISPR2. We found that the cas genes associated with the CRISPR2 locus were all upregulated in the clpP mutant strain (Table 2); however, the expression levels of the CRISPR1 cas genes were unaltered in the clpP mutant.
We also observed that the expression levels of several transcription factors were differentially regulated in the clpP mutant (Table 3). However, the fold differences in the expression levels between the mutant and the wild-type strains were not very great; the greatest difference was 1.9-fold for the SMU.1349 gene. None of the transcriptional regulators in S. mutans are characterized, except HrcA, which acts as a transcriptional repressor of heat shock response genes (21). The expression of hrcA was upregulated about 1.8-fold in the clpP mutant. The expression of another transcriptional regulator, spxA, was also upregulated in the clpP mutant, while the expression of five transcription factors was decreased in the clpP mutant strain. Therefore, the results suggest that ClpP plays an important role in overall gene expression in S. mutans by modulating the expression of several transcriptional regulators.
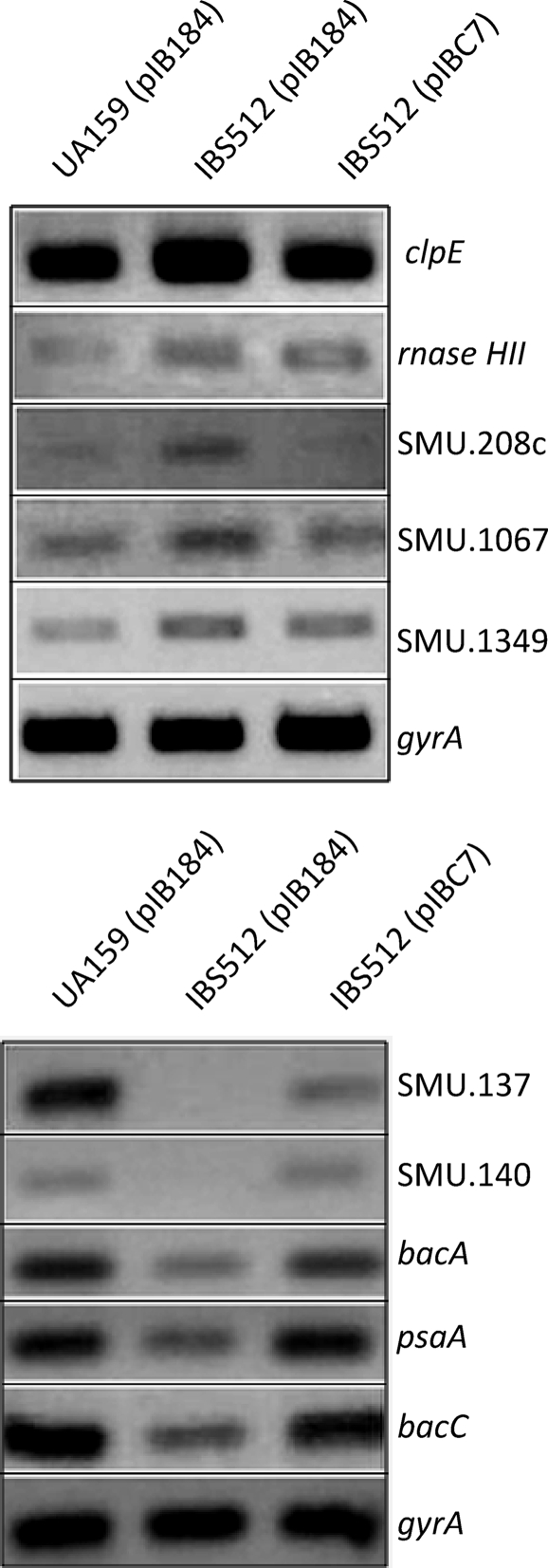
In an attempt to validate the microarray results, semiquantitative RT-PCR (sqRT-PCR) analyses were carried out. Toward this end, RNA isolated from exponentially grown cultures of UA159, IBS512, and IBS512(pIBC7) was used for sqRT-PCRs with primers listed in Table 1. Ten genes that were differentially regulated according to the microarray data (Table 2; see Table S1 in the supplemental material) were selected, and their expression was analyzed. The genes chosen were clpE, rnaseHII, SMU.137, SMU.140, SMU.208, SMU.1067, SMU.1339 (bacC), SMU.1342 (bacA), SMU.1348 (psaA), and SMU.1349. The results are shown in Fig. 2 and are consistent with the observed microarray results. For example, the expression of clpE, rnaseHII, SMU.208c, SMU.1067, and SMU.1349 was increased in the ΔclpP strain (IBS512), while the expression of bacA, bacC, psaA, SMU.137, and SMU.140 was decreased in IBS512. The complementing strain, IBS512(pIBC7), which was included in the sqRT-PCR analyses, showed the expected complementation. Thus, ClpP indeed regulates the expression of these genes at the transcriptional level.
FIG. 2.
Semiquantitative RT-PCR. Total RNA was harvested from UA159(pIB184), IBS512(pIB184), and IBS512(pIBC7) and subjected to cDNA synthesis. Five nanograms of cDNA from each strain was used for semiquantitative RT-PCR. Ten genes (clpE, rnaseHII, SMU.208c, SMU.1067, SMU.1349, SMU.137, SMU.140, bacA, psaA, and bacC) were subjected to semiquantitative RT PCR. The gyrA gene was included to ensure that equal amounts of RNA were used for all reactions. (Top) Upregulated genes. (Bottom) Downregulated genes.
ClpP is involved in protein turnover under nonstress conditions.
To understand the role of ClpP in protein turnover in the cell under nonstress conditions, a two-dimensional gel electrophoresis approach was used to compare the proteomes of UA159 and IBS512. Crude cellular lysates from UA159 and IBS512 were prepared from mid-exponential-phase cultures and subjected to proteomic analysis. A total of 339 well-resolved protein spots could be detected in the pI range of 5.0 to 8.2 by silver staining (data not shown). Comparison of the proteomes from UA159 and IBS512 revealed that at least 53 proteins had levels of expression altered by a change of 1.7-fold or greater, with a P value of ≤0.05. Among the differentially expressed proteins, 33 were downregulated while 21 were upregulated in strain IBS512 (ΔclpP) relative to those in the wild-type strain grown under the same conditions. This result is fairly consistent with an earlier report, which suggested that about 27 protein spots were altered (12 upregulated and 15 downregulated) in a clpP mutant strain (29). However, in that study, except for two spots, the identities of the spots were not confirmed (29). In an effort to identify the proteins, the spots were excised from the gels and their identities were determined by mass fingerprinting. The identities of only 30 spots could be determined, and the results are shown in Table 4 with the putative functional roles of the proteins. The upregulated proteins include several transcriptional regulators (SMU.38, SMU.491, and SMU.640), a cell division protein (FtsZ), a bacteriocin operon component protein (SMU.1810), an ABC transporter protein (SMU.923), and proteins with unassigned functions (SMU.475, SMU.605, and SMU.2075). Among the upregulated proteins, three independent spots corresponded to SMU.640. The relative abundances of two of the three spots were the highest (9.5- and 16.5-fold) among all the spots. Three other spots were upregulated more than 5-fold; however, the identity of only one spot could be determined (SMU.605). Proteins that were downregulated in strain IBS512 include ABC transporters (SMU.878), transcriptional regulators (SMU.1509), a stress response protein (SMU.2067), competence-specific proteins (CoiA amd ComX), a transposon protein (SMU.1032), and a putative bacitracin synthetase (BacA). Two spots that were identified as SMU.650 (alanyl-tRNA synthetase) and SMU.1657 were the most downregulated proteins (12- and 15-fold, respectively). The alternate sigma factor ComX was also highly downregulated (about 8-fold) in IBS512 compared to UA159.
TABLE 4.
Protein spots identified by two-dimensional gel analysis
| Gene name | Protein | No. of peptide matches | Fold change (IBS512 vs. UA159) | P (t test) (IBS512 vs. UA159) | Alteration in IBS512 vs. UA159a |
|---|---|---|---|---|---|
| SMU.548 | UDP-N-acetylmuramoyl-l-alanyl-d-glutamate synthetase | 4/2 | −2.0/−1.7 | 0.043/0.032 | Downregulated |
| SMU.644 | Putative competence protein CoiA | 3 | −6.4 | 0.001 | Downregulated |
| SMU.650 | Alanyl-tRNA synthetase | 10 | −11.9 | 0.008 | Downregulated |
| SMU.858 | Aspartate carbamoyltransferase subunit | 5 | −11.9 | 0.008 | Downregulated |
| SMU.878 | Multiple sugar binding ABC transporter | 7 | −2.3 | 0.045 | Downregulated |
| SMU.910 | Glucosyl-S-transferase | 8/11/15 | −1.8/−2.1/−4.3 | 0.031/0.042 | Downregulated |
| SMU.962 | Putative dehydrogenase | 2 | −2.1 | 0.042 | Downregulated |
| SMU.1032 | Putative transposon integrase | 9 | −2.4 | 0.021 | Downregulated |
| SMU.1293 | Hypothetical protein | 3 | −1.9 | 0.017 | Downregulated |
| SMU.1334 | Putative phosphopantetheinyl transferase | 7 | −6.9 | 0.019 | Downregulated |
| SMU.1342 | Putative bacitracin synthetase 1; BacA | 10 | −4.6 | 0.031 | Downregulated |
| SMU.1462 | Putative oxidoreductase | 3 | −1.9 | 0.045 | Downregulated |
| SMU.1500 | Putative exonuclease RexB | 4 | −4.6 | 0.031 | Downregulated |
| SMU.1509 | Putative transcriptional regulator | 3 | −1.8 | 0.030 | Downregulated |
| SMU.1657 | Putative nitrogen regulatory protein PII | 2 | −14.8 | 0.001 | Downregulated |
| SMU.1949 | Putative membrane carboxypeptidase, penicillin-binding protein 2A | 6 | −1.9 | 0.017 | Downregulated |
| SMU.1997 | Putative ComX1; transcriptional regulator | 3 | −7.7 | 0.002 | Downregulated |
| SMU.2067 | Putative stress response protein; possible cell wall biogenesis protein | 2/4 | −2.5/−6.4 | 0.034/0.001 | Downregulated |
| SMU.38 | Putative transcriptional regulator | 1 | 6.0 | 0.179 | Upregulated |
| SMU.82 | Molecular chaperone DnaK | 15 | 2.5 | 0.011 | Upregulated |
| SMU.374 | Putative oxidoreductase | 3 | 2.2 | 0.007 | Upregulated |
| SMU.475 | Hypothetical protein | 3 | 3.8 | 0.029 | Upregulated |
| SMU.491 | Putative DeoR-type transcriptional regulator | 1 | 3.4 | 0.062 | Upregulated |
| SMU.552 | Cell division protein FtsZ | 3 | 16.5 | 0.001 | Upregulated |
| SMU.605 | Hypothetical protein | 3 | 6.0 | 0.179 | Upregulated |
| SMU.640 | Putative transcriptional regulator (GntR family) | 3/2/3 | 1.7/9.5/16.5 | 0.029 | Upregulated |
| SMU.923 | Putative ABC transporter; ATP-binding protein | 3 | 2.0 | 0.031 | Upregulated |
| SMU.1241 | Exonuclease ABC subunit C (UvrC; nucleotide excision repair) | 6 | 3.1 | 0.058 | Upregulated |
| SMU.1810 | Putative bacteriocin operon component | 1 | 1.7 | 0.004 | Upregulated |
| SMU.2075 | Hypothetical protein | 5 | 3.1 | 0.034 | Upregulated |
The up- and downregulated proteins were identified in S. mutans IBS512 and UA159, respectively.
Comparison of the transcriptome results with the proteome results identified only four common gene products: DnaK (SMU.82), SMU.1293, SMU.1342, and SMU.1949. Interestingly, although several transcriptional regulators were identified by proteome analysis, these regulators were different from those that were identified by the transcriptome study. This suggests that the transcriptional regulators identified by the proteome study do not autoregulate their own expression; however, they may be involved in the regulation of some of the genes that were identified in the microarray study.
clpP deficiency leads to increased sensitivity to trimethoprim.
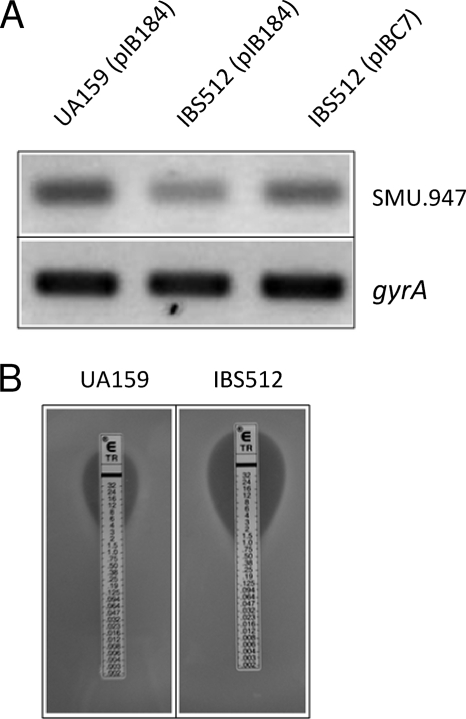
The expression of SMU.947 was decreased about 1.6-fold in IBS512 compared to the wild-type strain, UA159 (see Table S1 in the supplemental material). SMU.947 encodes the dihydrofolate reductase enzyme, which converts dihydrofolate into tetrahydrofolate, a methyl group shuttle enzyme required for de novo biosynthesis of purines, thymidylic acid, and certain amino acids. To verify whether ClpP modulates the expression of SMU.947, sqRT-PCR analysis was performed to directly measure the transcript amount. As shown in Fig. 3, the amount of SMU.947 transcript was decreased about 2-fold in IBS512 compared to the wild-type strain, UA159. As expected, the transcript amount was increased in the complementing strain [IBS512(pIBC7)]; however, the level of complementation was only partial (increased ∼1.4-fold compared to IBS512). A dihydrofolate reductase inhibitor, trimethoprim, blocks the conversion of dihydrofolate to tetrahydrofolate and acts as a bacteriostatic reagent. To verify whether the clpP mutant strain IBS512 is more susceptible to trimethoprim, we performed an E-strip test to determine the MIC of trimethoprim. As shown in Fig. 3, the MIC was 4-fold lower for IBS512 than for UA159 (1 μg/ml versus 4 μg/ml, respectively). Thus, ClpP is involved in regulating the purine biosynthesis pathway by modulating dihydrofolate reductase activity.
FIG. 3.
clpP deficiency leads to increased sensitivity to trimethoprim. (A) Expression of dihydrofolate reductase transcript in UA159(pIB184), IBS512(pIB184), and IBS512(pIBC7). RNA was extracted from all three cultures and subjected to cDNA synthesis. Semiquantitative RT-PCR was then used to measure the levels of dihydrofolate reductase transcript in all three cultures. The expression of gyrA was included as an internal control to ensure that equal amounts of RNA were analyzed for each culture. (B) Trimethoprim-containing E strips were aseptically placed on UA159 and IBS512 lawns. The plates were incubated under microaerophilic conditions at 37°C overnight. The MICs for the two cultures were noted.
Mutacin production is affected in the clpP mutant strain.
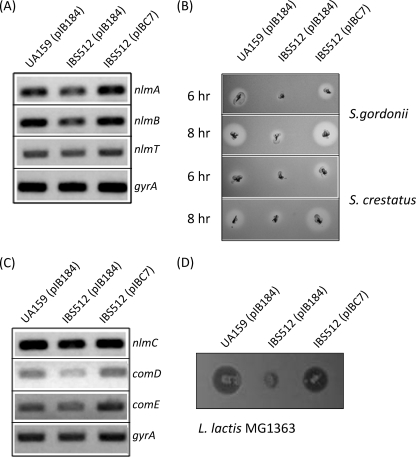
The microarray data revealed that the expression of nlmA, nlmB, and nlmC was reduced in the strain with clpP deleted (Table 2). To confirm the microarray results, transcriptions were measured directly by sqRT-PCR analysis. As expected, the nlmA, nlmB, and nlmC transcript levels in the mid-exponential phase were more than 2.0-fold lower in strain IBS512, while the complemented strain, IBS512(pIBC7), produced amounts of transcript equal to those in the wild-type strain, UA159 (Fig. 4).
FIG. 4.
Mutacin production is modulated by ClpP. (A) Semiquantitative RT-PCR was carried out for mutacin IV-encoding (nlmA and nlmB) and transporter (nlmT) genes. To ensure that equal quantities of RNA were used for all three cultures, gyrA was used as an internal control. (B) A deferred antagonism assay for mutacin IV production was carried out by pouring lawns of the indicator strains of S. gordonii and S. crestatus on plates stabbed with the three experimental strains at two different times, 6 and 8 h after patching. (C) Semiquantitative RT-PCR analysis was employed for the mutacin V-encoding gene (nlmC) and the two-component signal transduction system gene (comDE). The transcript of gyrA was used as an internal control. (D) A deferred antagonism assay for mutacin V was performed as described for panel B, except that the indicator strain employed was L. lactis MG1363.
To measure the production of mutacins directly, IBS512 and the isogenic parental strain, UA159, were subjected to deferred antagonism assays using S. cristatus (5100), S. gordonii (DL-1), and L. lactis (MG1363) as indicator strains. As shown in Fig. 4, the production levels of both mutacin IV (assayed by growth inhibition of S. cristatus and S. gordonii) and mutacin V (assayed by growth inhibition of L. lactis) were drastically reduced in IBS512, but not in the other strains. To verify that this effect was due to the inactivation of clpP, the mutacin production levels in IBS512(pIBC7) were also measured. In the complemented strain, the level of mutacin production was fully restored to the wild-type level. Thus, ClpP appears to regulate mutacin production in S. mutans by modulating the expression of nlm genes.
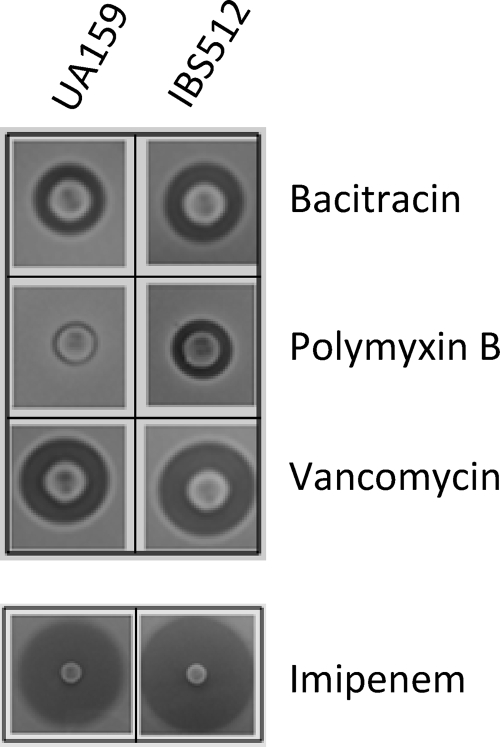
ClpP protects the cell from cell wall-damaging agents.
Our microarray results showed that several cell division-related genes, such as mreD and mreC, were upregulated in the ΔclpP mutant strain (see Table S1 in the supplemental material). Furthermore, our proteomics study indicated that FtsZ, the major bacterial cytoskeletal protein, was also increased in the ΔclpP mutant strain (Table 4). This observation led us to believe that ClpP probably plays a role in the proper maintenance of cell wall structure; therefore, the ΔclpP strain should be more susceptible to cell wall-damaging antibiotics. To verify if deletion of clpP has any effect on protecting the cells from cell wall-damaging antibiotics, a total of eight antibiotics with various cell wall biosynthesis targets were used in a disc diffusion assay, as described in Materials and Methods. The ΔclpP S. mutans strain, IBS512, showed increased sensitivity to treatment with all the cell wall-targeting antibiotics compared to the wild-type strain, UA159 (Fig. 5). However, the degree of increase in sensitivity varied depending on the nature of the antibiotics used. On the other hand, this increased sensitivity of IBS512 to antibiotics, such as rifampin, ciprofloxacin, and clindamycin, that do not target cell wall development was not observed (data not shown). Thus, these results suggest that ClpP is required for proper maintenance of cell wall structure in streptococci.
FIG. 5.
Antibiotic sensitivity assay. Bacitracin-, polymyxin B-, vancomycin-, and imipenem-containing discs were placed on UA159 and IBS512 lawns. The plates were then incubated under microaerophilic conditions at 37°C for 16 h. The inhibitory-zone diameters for both cultures were measured and compared.
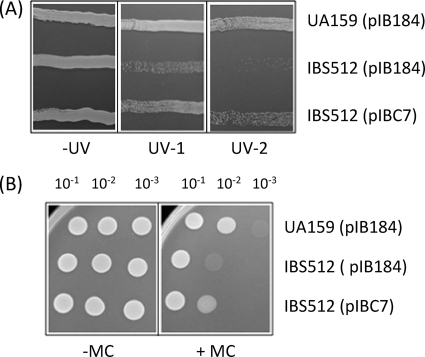
ClpP is involved in the DNA damage response.
The proteomics study suggested that RexB protein expression was downregulated in the ClpP mutant (Table 4). RexB is a component of the RexAB complex, which is a paralog of E. coli RecBCD that is involved in double-strand DNA break (DSB) repair and homologous recombination (12). Although the role of ClpP in various stress responses is well understood, its role in the DNA damage response is not well studied. In some Gram-positive organisms, ClpP appears to play a role in tolerance of exposure to DNA-damaging agents (33, 41). Therefore, to verify the role of ClpP in double-strand DNA break repair, the S. mutans strain with clpP deleted, IBS512, was evaluated for its ability to withstand exposure to UV irradiation and mitomycin C. The latter agent alkylates double-stranded DNA and blocks DNA replication to cause double-stranded DNA breaks. In order to minimize the variation, both the wild-type (UA159) and the clpP-deficient (IBS512) strains were transformed with the vector plasmid pIB184, and the IBS512 strain was also transformed separately with the complementing plasmid pIBC7. As shown in Fig. 6, both the UV irradiation and mitomycin C treatments adversely affected the growth of IBS512(pIB184), but not the growth of the UA159(pIB184) and IBS512(pIBC7) strains. These results clearly show that in S. mutans, ClpP is involved in the repair of damaged DNA; however, the activity of ClpP in S. mutans appears to be the opposite of what is seen in Staphylococcus aureus (33).
FIG. 6.
ClpP is essential for tolerance of DNA damage-induced stress. (A) Effects of UV exposure. UA159(pIB184), IBS512(pIB184), and IBS512(pIBC7) were streaked onto THY plates supplemented with erythromycin. Two such plates, each containing all three cultures, were transiently exposed to UV irradiation at two different doses (0.25 × 102 μJ/cm2 [UV-1] and 0.5 × 102 μJ/cm2 [UV-2]). A nonirradiated plate (−UV) containing the cultures was used as a control. (B) Exposure to mitomycin C (6 ng/ml) impairs the growth of the ΔclpP mutant. UA159(pIB184), IBS512(pIB184), and IBS512(pIBC7) were serially diluted and spotted on THY-erythromycin plates in the presence (+MC) and absence (−MC) of mitomycin C. After incubation under microaerophilic conditions at 37°C, the changes in the growth pattern were noted and photographed.
DISCUSSION
ClpP-mediated proteolysis play a significant role in cellular protein homeostasis, protein quality control, and certain cellular regulatory and differentiation processes. ClpP-mediated proteolysis is essential to maintain cellular integrity under three different scenarios (26). The most important and the best studied is the ClpP-mediated proteolysis that is induced by severe thermal and other stresses that lead to concomitant protein aggregation. On the other hand, bacterial entry into stationary phase, which does not produce significant amounts of protein aggregates, also triggers ClpP-mediated proteolysis (26). ClpP is also essential for intracellular protein quality control; if ClpP is absent, newly synthesized proteins are highly prone to aggregation, even at permissive temperature (26). Most of the previous studies were focused on the role of ClpP in stress-related protein degradation, while very little is known about the function of ClpP in protein quality control under nonstress conditions (14-16, 29). We report here that under nonstress conditions, ClpP plays pleiotropic and complex roles in S. mutans global gene regulation, protein turnover, and expression of important virulence traits. Our genomic and proteomics studies have also revealed several new findings that are discussed below.
Among the new findings that emerged from this study is the involvement of ClpP in the regulation of genes within the genomic islands. At least 11 GIs are included in the UA159 genome (3), and we found that genes in three GIs were differentially expressed in the ΔclpP strain (Table 1). GI-6, which is also known as TnSmu1 (51), includes about 34 genes, and we found that 28 of these genes were upregulated in the ΔclpP strain. On the other hand, at least 13 genes in GI-12, also known as TnSmu2 (50), were downregulated in the ΔclpP strain. Interestingly, the difference in the expression of these genes between the wild type and the mutant was among the highest. TnSmu2 includes an operon, smt, which is thought to be involved in the biosynthesis of secondary metabolites. There are 10 genes (SMU.1339 to SMU.1348) within the smt operon, and all 10 genes were downregulated in the clpP deletion strain. Furthermore, one gene product of the smt operon, BacA, was also identified by our proteomics analysis. However, the expression of SMU.1349, a TetR/AcrR family transcription factor, was upregulated in the clpP deletion strain. At present, we do not know the exact mechanisms by which ClpP modulates the expression of these GI genes. We recently studied the expression of the smt operon genes and found that the genes are regulated by CovR, an orphan response regulator, and probably by a histone-like protein (HLP) (Biswas, unpublished). It appears that this operon is repressed by HLP, which binds to the promoter region of the operon. On the other hand, CovR, which has much higher binding affinity than HLP for the smt promoter, displaces HLP from the promoter and allows transcription to occur. Therefore, CovR acts as an antisilencer of the smt operon. To verify whether ClpP is involved in the degradation of CovR or stabilization of HLP, we performed Western blot analysis on the crude cell lysates isolated from exponentially grown wild-type or ΔclpP strains using CovR- or HLP-specific antibodies (kindly provided by V. Pancholi). We found that the amounts of both the CovR and HLP proteins were not significantly different in the ΔclpP strain than in the wild-type strain (data not shown). Therefore, ClpP probably employs other mechanisms to regulate the expression of genes in the smt operon. For example, the putative transcription factor SMU.1349 might be involved in the repression of smt operon genes, and the role of ClpP is to maintain a certain level of SMU.1349 protein in the cell. In the absence of ClpP, the SMU.1349 protein level increases, and it further represses the smt operon. Similarly, for the TnSmu1 genes, SMU.218, another putative transcriptional regulator, might be involved in gene expression. Experiments are under way to evaluate the roles of SMU.218 and SMU.1349 in ClpP-mediated gene regulation.
The expression of only one of the two CRISPR loci that are present in S. mutans, the CRISPR2 locus, was altered in the ΔclpP strain. Although CRISPR2 is present in about 30% of S. mutans isolates (46), its exact function is not known. Furthermore, it has been proposed that the CRISPR2 locus might be inactive in S. mutans strain UA159 due to premature truncation of two cas genes (46). Thus, it is surprising to see that the CRISPR2 locus was upregulated in the ΔclpP mutant. It is currently unknown how the expression levels of the CRISPR locus genes are regulated in bacteria. Our results simply suggest that ClpP plays a role in the transcription of some of the CRISPR-associated genes; however, the exact mechanism by which ClpP regulates CRISPR expression is still not understood.
In oral streptococci, the major carbohydrate transport system is the PTS system. PTS system components are not only involved in sugar transport, but also influence biofilm development, gene regulation, and acid stress tolerance (1, 28). The microarray analysis reported here showed that at least three PTS system components were downregulated in the ΔclpP strain. They are SMU.100 (a putative sorbose PTS system IIB component) (Table 2), SMU.1185 (a mannose-specific PTS system component) (see Table S1 in the supplemental material), and SMU.1600 (a cellobiose-specific PTS IIB component) (see Table S1 in the supplemental material). Interestingly, UA159 can neither produce acid from sorbose nor grow on semidefined media containing sorbose as the sole carbon source, indicating that the genes are probably nonfunctional in S. mutans (51). At present, it is not clear why ClpP appears to regulate these PTS systems, and this warrants further analysis.
In accordance with the earlier studies, the expression levels of several stress-related genes and proteins were found to be upregulated in the clpP mutant strain (29, 33, 40). For example, transcription of genes encoding HSPs, such as DnaJ, DnaK, and the HSP-70 cofactor GrpE, were induced in the clpP mutant strain. Furthermore, the transcription of genes encoding chaperone proteins, such as GroEL and GroES, were also upregulated in the clpP mutant. We also found that the amount of DnaK protein was increased in the clpP mutant strain. Both GroELS and DnaK are very important chaperones, and mutations inactivating the genes encoding these chaperones display pleiotropic phenotypes, including defects in nucleic acid biosynthesis, proteolysis, and cell division (17). In group B streptococci (GBS), the absence of ClpP leads to the accumulation of oxidized DnaK, indicating that oxidized DnaK is a preferred substrate for ClpP-mediated degradation (34). Oxidized proteins lose their conformation, which causes enzyme inactivation, and the amounts of oxidized proteins increase proportionally with time (43). In accordance with this, Kock et al. (26) have shown that in Bacillus subtilis, ClpP is indeed involved in the degradation of the GroESL and DnaK chaperones in stationary-phase cells. We believe that these chaperones and other proteins are continuously oxidized during bacterial growth and that one of the functions of ClpP is to remove the unwanted inactivated proteins from the cell.
Another important finding is the role of ClpP in the protection of S. mutans cells against DNA-damaging agents. In E. coli, there are several proteins that are involved in the DSB repair pathway (12); among them, RecA and RecBCD, which is composed of three subunits, are the principal repair proteins. Gram-positive bacteria do not contain RecBCD; instead, they encode the RexAB system. Since the amount of RexB was reduced in the strain with clp deleted, it is possible that the observed sensitivity to DSB was due to the reduction of RexB protein in the cell. Since the transcription of rexB was unaffected in the strain with clp deleted, ClpP probably regulates RexB expression at the posttranslational step. There is another possibility for the observed defect in DSB repair. In E. coli, a transcriptional repressor, LexA, regulates genes involved in the repair of DNA damage by a process known as the SOS response (24). Gram-positive bacteria do not contain LexA; instead, the HdiR protein serves as a functional homolog of LexA (41). In L. lactis, HdiR is a bona fide substrate for ClpP-mediated proteolysis, and mutant strains lacking functional ClpP show increased sensitivity to DNA-damaging agents (41). Thus, it is possible that in S. mutans, HdiR is also a substrate for ClpP-mediated degradation. However, in L. lactis and Streptococcus uberis, HdiR-mediated DNA damage repair is primarily mediated by the Y-family polymerase UmuC (48). While many streptococci contain genes encoding UmuC homologs with identities over 50%, several streptococci, including S. mutans and S. pneumoniae, do not contain any gene encoding an UmuC homolog (48). Therefore, the mechanisms by which HdiR is involved in DNA damage repair in S. mutans and other streptococci that do not contain UmuC might be different from that in the streptococci that contain UmuC. Furthermore, a recent transcriptome analysis has shown that in S. aureus several putative HdiR-regulated genes are upregulated in a clpP-deficient strain; however, sensitivity to DNA-damaging agents was not evaluated in that study (33).
Both our microarray and proteomics analyses identified several cell division-related genes or proteins whose expression levels were altered in the clpP mutant strain. Consistent with the findings, we also observed that during growth at high temperature, the clpP mutant strain displayed aberrant cell morphology due to the presence of large cells, indicating a defect in proper cell division (data not shown). A recent study of GBS also demonstrated a similar finding, where the authors observed large multiseptate cells containing nonparallel septa in a clpP mutant strain grown at elevated temperature. We also found that the ΔclpP mutant strain showed increased sensitivity to several cell wall-targeting antibiotics, indicating that ClpP deficiency also interferes with cell wall biosynthesis. Although how ClpP interferes with cell wall biosynthesis remains to be deciphered, we speculate that several cell wall biosynthesis components could be subject to ClpP proteolysis. For example, bacitracin is known to block dephosphorylation of undecaprenyl pyrophosphate (which acts as a lipid carrier during peptidoglycan biosynthesis [42]), causing depletion of the amount of lipid carrier that envelopes the nascent polymers of peptidoglycan during its biogenesis. The S. mutans dgk gene encodes the undecaprenol kinase activity (30), and it is possible that the gene product is a substrate of ClpP-mediated degradation.
Perhaps the most significant finding was the role of ClpP in the expression of nlm genes and other bacteriocin-related genes whose exact functions are currently unknown (47). The expression of nlmAB is regulated by at least three different two-component systems (LiaRS, CiaRH, and ComDE) (5). Furthermore, ComDE also regulates nlmC transcription (47). Our microarray and sqRT-PCR analyses suggested that transcription of all nlm genes was downregulated in the strain with clpP deleted. Moreover, we found that the expression of both comE and comD was significantly reduced in the clpP mutant. In S. pneumoniae, the ComDE system positively autoregulates its own transcription (37); however this positive autoregulation has not been demonstrated in S. mutans. We speculate that comDE expression is also autoregulated and that ComE, the response regulator component, is a substrate for ClpP degradation. We are currently evaluating whether ClpP can degrade the ComE protein in vivo.
In conclusion, our global analysis revealed a broad impact of the S. mutans ClpP protease on several virulence-related pathways, such as biofilm formation, stress response, DNA repair, antibiotic defense, and mutacin production. In streptococci, ClpXP, ClpCP, or ClpEP complexes generally mediate regulatory proteolysis that requires certain amino acid sequences, which serve as degradation signals. The only degradation signal that has been identified in streptococci is the SsrA tag; substrates containing this tag are degraded by the ClpXP complex (2). Although the exact targets of ClpP protease were not identified in this study, our proteomics study has identified several candidates. Future analysis of these candidate substrates may identify degradation signals that are recognized by various Clp complexes.
Supplementary Material
Acknowledgments
This work was supported by an NIDCR grant (DE016686) and an NCRR COBRE project grant (P20RR01644) to I.B.
Footnotes
Published ahead of print on 28 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abranches, J., M. M. Candella, Z. T. Wen, H. V. Baker, and R. A. Burne. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 188:3748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahlawat, S., and D. A. Morrison. 2009. ClpXP degrades SsrA-tagged proteins in Streptococcus pneumoniae. J. Bacteriol. 191:2894-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee, A., and I. Biswas. 2008. Markerless multiple-gene-deletion system for Streptococcus mutans. Appl. Environ. Microbiol. 74:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas, I., L. Drake, D. Erkina, and S. Biswas. 2008. Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. J. Bacteriol. 190:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas, I., J. K. Jha, and N. Fromm. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas, S., and I. Biswas. 2006. Regulation of the glucosyltransferase (gtfBC) operon by CovR in Streptococcus mutans. J. Bacteriol. 188:988-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlsson, J. 1997. Bacterial metabolism in dental biofilms. Adv. Dent. Res. 11:75-80. [DOI] [PubMed] [Google Scholar]
- 9.Chia, J. S., Y. Y. Lee, P. T. Huang, and J. Y. Chen. 2001. Identification of stress-responsive genes in Streptococcus mutans by differential display reverse transcription-PCR. Infect. Immun. 69:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong, P., L. Drake, and I. Biswas. 2008. LiaS regulates virulence factor expression in Streptococcus mutans. Infect. Immun. 76:3093-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, D. M., J. M. ten Cate, and W. Crielaard. 2007. The adaptive response of Streptococcus mutans towards oral care products: involvement of the ClpP serine protease. Eur. J. Oral Sci. 115:363-370. [DOI] [PubMed] [Google Scholar]
- 12.Dillingham, M. S., and S. C. Kowalczykowski. 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72:642-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrmann, M., and T. Clausen. 2004. Proteolysis as a regulatory mechanism. Annu. Rev. Genet. 38:709-724. [DOI] [PubMed] [Google Scholar]
- 14.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445-1462. [DOI] [PubMed] [Google Scholar]
- 15.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol. Microbiol. 31:79-87. [DOI] [PubMed] [Google Scholar]
- 16.Frees, D., K. Savijoki, P. Varmanen, and H. Ingmer. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63:1285-1295. [DOI] [PubMed] [Google Scholar]
- 17.Georgopoulos, C., K. Liberek, M. Zylicz, and D. Ang. 1994. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response, p. 209-249. In A. T. R. I. Morimoto, and C. Georgopoulos (ed.), The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Glover, J. R., and S. Lindquist. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94:73-82. [DOI] [PubMed] [Google Scholar]
- 19.Gottesman, S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19:565-587. [DOI] [PubMed] [Google Scholar]
- 20.Hengge, R., and B. Bukau. 2003. Proteolysis in prokaryotes: protein quality control and regulatory principles. Mol. Microbiol. 49:1451-1462. [DOI] [PubMed] [Google Scholar]
- 21.Jayaraman, G. C., J. E. Penders, and R. A. Burne. 1997. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25:329-341. [DOI] [PubMed] [Google Scholar]
- 22.Kajfasz, J. K., A. R. Martinez, I. Rivera-Ramos, J. Abranches, H. Koo, R. G. Quivey, Jr., and J. A. Lemos. 2009. Role of Clp proteins in expression of virulence properties of Streptococcus mutans. J. Bacteriol. 191:2060-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaste, L. M., R. H. Selwitz, R. J. Oldakowski, J. A. Brunelle, D. M. Winn, and L. J. Brown. 1996. Coronal caries in the primary and permanent dentition of children and adolescents 1-17 years of age: United States, 1988-1991. J. Dent. Res. 75:631-641. [DOI] [PubMed] [Google Scholar]
- 24.Kelley, W. L. 2006. Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol. Microbiol. 62:1228-1238. [DOI] [PubMed] [Google Scholar]
- 25.Kirstein, J., N. Moliere, D. A. Dougan, and K. Turgay. 2009. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 7:589-599. [DOI] [PubMed] [Google Scholar]
- 26.Kock, H., U. Gerth, and M. Hecker. 2004. The ClpP peptidase is the major determinant of bulk protein turnover in Bacillus subtilis. J. Bacteriol. 186:5856-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemos, J. A., J. Abranches, and R. A. Burne. 2005. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 7:95-107. [PubMed] [Google Scholar]
- 28.Lemos, J. A., and R. A. Burne. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemos, J. A., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lis, M., and H. K. Kuramitsu. 2003. The stress-responsive dgk gene from Streptococcus mutans encodes a putative undecaprenol kinase activity. Infect. Immun. 71:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurizi, M. R., W. P. Clark, Y. Katayama, S. Rudikoff, J. Pumphrey, B. Bowers, and S. Gottesman. 1990. Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 265:12536-12545. [PubMed] [Google Scholar]
- 33.Michel, A., F. Agerer, C. R. Hauck, M. Herrmann, J. Ullrich, J. Hacker, and K. Ohlsen. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188:5783-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair, S., C. Poyart, J. L. Beretti, H. Veiga-Fernandes, P. Berche, and P. Trieu-Cuot. 2003. Role of the Streptococcus agalactiae ClpP serine protease in heat-induced stress defense and growth arrest. Microbiology 149:407-417. [DOI] [PubMed] [Google Scholar]
- 35.O'Farrell, P. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 36.Ogura, T., and A. J. Wilkinson. 2001. AAA+ superfamily ATPases: common structure—diverse function. Genes Cells 6:575-597. [DOI] [PubMed] [Google Scholar]
- 37.Pestova, E. V., L. S. Havarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 38.Peterson, P. 2003. The world oral health report 2003. WHO, Geneva, Switzerland.
- 39.Pitts, N. B., I. G. Chestnutt, D. Evans, D. White, B. Chadwick, and J. G. Steele. 2006. The dentinal caries experience of children in the United Kingdom, 2003. Br. Dent. J. 200:313-320. [DOI] [PubMed] [Google Scholar]
- 40.Robertson, G. T., W. L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savijoki, K., H. Ingmer, D. Frees, F. K. Vogensen, A. Palva, and P. Varmanen. 2003. Heat and DNA damage induction of the LexA-like regulator HdiR from Lactococcus lactis is mediated by RecA and ClpP. Mol. Microbiol. 50:609-621. [DOI] [PubMed] [Google Scholar]
- 42.Siewert, G., and J. L. Strominger. 1967. Bacitracin: an inhibitor of the dephosphorylation of lipid pyrophosphate, an intermediate in the biosynthesis of the peptidoglycan of bacterial cell walls. Proc. Natl. Acad. Sci. U. S. A. 57:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stadtman, E. R. 1992. Protein oxidation and aging. Science 257:1220-1224. [DOI] [PubMed] [Google Scholar]
- 44.Tanzer, J. M., J. Livingston, and A. M. Thompson. 2001. The microbiology of primary dental caries in humans. J. Dent. Educ. 65:1028-1037. [PubMed] [Google Scholar]
- 45.van der Oost, J., M. M. Jore, E. R. Westra, M. Lundgren, and S. J. Brouns. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34:401-407. [DOI] [PubMed] [Google Scholar]
- 46.van der Ploeg, J. R. 2009. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155:1966-1976. [DOI] [PubMed] [Google Scholar]
- 47.van der Ploeg, J. R. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187:3980-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varhimo, E., K. Savijoki, J. Jalava, O. P. Kuipers, and P. Varmanen. 2007. Identification of a novel streptococcal gene cassette mediating SOS mutagenesis in Streptococcus uberis. J. Bacteriol. 189:5210-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, J., J. A. Hartling, and J. M. Flanagan. 1997. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91:447-456. [DOI] [PubMed] [Google Scholar]
- 50.Waterhouse, J. C., and R. R. Russell. 2006. Dispensable genes and foreign DNA in Streptococcus mutans. Microbiology 152:1777-1788. [DOI] [PubMed] [Google Scholar]
- 51.Waterhouse, J. C., D. C. Swan, and R. R. Russell. 2007. Comparative genome hybridization of Streptococcus mutans strains. Oral Microbiol. Immunol. 22:103-110. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, J., A. Banerjee, and I. Biswas. 2008. Transcription of clpP is enhanced by a unique tandem repeat sequence in Streptococcus mutans. J. Bacteriol. 191:1056-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.