Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV)-infected cells express the latency-associated nuclear antigen (LANA) involved in the regulation of host and viral gene expression and maintenance of the KSHV latent episome. Performance of these diverse functions involves a 7-amino-acid chromatin-binding motif (CBM) situated at the amino terminus of LANA that is capable of binding directly to nucleosomes. LANA interacts with additional chromatin components, including methyl-CpG-binding protein 2 (MeCP2). Here, we show that the carboxy-terminal DNA-binding/dimerization domain of LANA provides the principal interaction with MeCP2 but that this association is modulated by the CBM. Both domains are required for LANA to colocalize with MeCP2 at chromocenters, regions of extensive pericentric heterochromatin that can be imaged by fluorescence microscopy. Within MeCP2, the methyl-CpG-binding domain (MBD) is the primary determinant for chromatin localization and acts together with the adjacent repression domains (the transcription repression domain [TRD] and the corepressor-interacting domain [CRID]) to redirect LANA to chromocenters. MeCP2 facilitates repression by LANA bound to the KSHV terminal repeats, a function that requires the MeCP2 C terminus in addition to the MBD and CRID/TRD. LANA and MeCP2 can also cooperate to stimulate transcription of the human E2F1 promoter, which lacks a LANA DNA-binding sequence, but this function requires both the N and C termini of LANA. The ability of LANA to establish multivalent interactions with histones and chromatin-binding proteins such as MeCP2 would enable LANA to direct regulatory complexes to specific chromosomal sites and thereby achieve stable reprogramming of cellular gene expression in latently infected cells.
Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8, is firmly linked to both the etiology and pathogenesis of Kaposi's sarcoma, primary effusion lymphoma, and variant multicentric Castleman's disease (reviewed in references 19 and 60). Most infection events lead to a semiquiescent state known as latency, in which the KSHV genome persists as a circularized episome that replicates in synchrony with the host DNA. A small number of viral proteins and microRNAs are expressed during latency and are used by the virus to immortalize the host cell, counter immune defenses, and extensively reprogram cellular gene expression.
The latency-associated nuclear antigen (LANA) is a >220-kDa viral protein encoded by open reading frame 73 (ORF73). LANA is expressed in all KSHV-infected cells and performs a variety of molecular functions essential for the establishment and continuous maintenance of the latent state (4, 18, 53, 65, 77). Initiation sites for episomal DNA replication lie within the KSHV terminal repeat (TR) sequences and consist of two LANA-binding sites and an adjacent replication element (6, 20, 29). The C terminus of LANA (LANAC) contains a sequence-specific DNA-binding and dimerization domain and is sufficient to initiate DNA replication in transient assays, albeit at reduced efficiency compared to the full-length protein (15). Recruitment of LANA to the TRs induces localized hyperacetylation of histones H3 and H4 and causes a strong bend in the DNA, characteristic of other initiator-origin interactions (63, 73). These changes may help to recruit the cellular origin recognition complex (ORC) to the origin, allowing the initiation of DNA synthesis during S phase (41, 63, 68). LANA also acts as a physical tether connecting viral episomes to the host chromosomes (5, 6, 14). This phenomenon is characteristic of other persistent viruses with episomal genomes, but the full functional significance of chromosomal tethering is not understood. One view is that physical linkage provides a partitioning function to ensure that the episomes are segregated equally into each daughter cell at cell division and are duly incorporated into the new nuclei. The situation may be more complex, because the expression of LANA alone is not sufficient to maintain TR-containing episomes for extended periods without drug selection (25, 66) and there is evidence from studies of papillomaviruses that tethering factors may also serve a gene-regulatory function (44).
Gene expression profiling of LANA-expressing cells has identified numerous cellular transcripts that are either activated or repressed by LANA (3, 54, 62, 69). There is evidence for several different mechanisms involving physical interactions with select components of the cellular transcriptional machinery. Interactions with glycogen synthase kinase 3β (GSK3β) and the retinoblastoma protein (pRB) promote activation of β-catenin- and E2F-regulated genes (3, 18, 53). Interactions with transcriptional activators, such as Brd2, Brd4, Sp1, AP-1, and CBP, and transcriptional inhibitors, such as heterochromatin protein 1 (HP1), DNA methyltransferase 3 (Dnmt3), and mSin3, have also been demonstrated previously (4, 33, 34, 39, 42, 52, 58, 62, 67, 78).
Chromosome tethering is important for LANA to stimulate transcription and support transient DNA replication (28, 38, 72). The principal tethering element has been mapped to a 7-amino-acid motif at the N terminus of LANA (8, 38, 72). Dubbed the chromatin-binding motif (CBM), this sequence is critical for association with host chromosomes throughout the cell cycle (72). CBM-containing fusion proteins uniformly coat metaphase chromosomes, implying an abundant target(s) (7, 8, 51, 72). This conjecture was substantiated by crystallographic and biochemical evidence that the CBM-containing peptide docks onto the surface of the nucleosome octamer, recognizing the interface between histone H2A and H2B subunits (8, 9). As indicated above, a number of additional chromatin-associated proteins interact with LANA, but their contribution to chromosome tethering is unknown (14, 33, 40, 52, 63, 78). One of these is methyl-CpG-binding protein 2 (MeCP2), a nuclear factor implicated in the transcriptional silencing of genes in CpG-methylated regions, activation of euchromatic genes, and mRNA splicing (13, 76, 79). A variety of mutations in the X-linked MeCP2 gene cause Rett syndrome, a progressive neurodevelopmental disorder (2). Early studies defined a discrete DNA-binding domain, the methyl-CpG-binding domain (MBD), capable of binding both methylated CpG dinucleotides and unmethylated sequences (22, 31). Transcriptional repression is attributed to separate domains known as the transcription repression domain (TRD) and the corepressor-interacting domain (CRID), which recruit additional corepressor proteins (27). MeCP2 may also alter higher-order chromatin architecture, independent of histone modifications or DNA methylation (22, 49).
Here, we show that MeCP2 interacts with LANAC but that this association is modulated by the CBM located at the N terminus. In agreement with the results of a prior study by Krithivas and colleagues (33), we find that coexpression of fluorescently tagged versions of LANA and MeCP2 in murine cells results in striking relocalization of LANA from diffuse nucleoplasmic distribution to concentration at discrete foci known as chromocenters, which correspond to major accumulations of pericentric heterochromatin. Optimal colocalization with MeCP2 requires the CBM, LANAC, and possibly some additional sequences within the N terminus. Although the MBD is sufficient to localize MeCP2 to chromocenters, relocalization of LANA requires both the MBD and TRD. Lastly, we show that MeCP2 enhances transactivation of the human E2F1 promoter by LANA and that this effect is dependent on the CBM and MBD. These findings indicate that multiple interactions are required for LANA to stably associate with chromatin and may occur as a two-step process in which nucleosome binding by the CBM facilitates the interaction of LANA with sequence- or context-specific cofactors such as MeCP2. These multivalent interactions may allow LANA to stabilize MeCP2 on low-affinity sites, enabling KSHV to reprogram select aspects of host cell gene expression.
MATERIALS AND METHODS
Expression plasmids.
Mammalian expression plasmids encoding full-length and truncated versions of LANA have been described previously (61, 72, 73). The open reading frame encoding full-length human MeCP2 (e2 isoform) was generated by PCR amplification with an Expand high-fidelity PCR system (Roche) using expressed sequence tag cDNA as the template and the following oligonucleotides: 5′-CGTCTAGAGTAGCTGGGATGTTAGGGCTC-3′ and 5′-GCGGATCCTAGCTAACTCTCTCGGTCACGG-3′ (added XbaI and BamHI sites are underlined). After sequencing, the fragment was subcloned into mammalian expression vectors adding a T7 epitope tag (pCGT) (71), a Flag tag (pCGFlag) (43), or red fluorescent protein (pCMV-DsRED; Clontech) to the N terminus of the product. Additional truncations and internal deletions were generated by PCR using custom primers and confirmed by DNA sequencing. For bacterial expression, sequences corresponding to LANA residues 2 to 31, 23 to 50, 2 to 50, and 936 to 1162 (LANAC) were amplified by PCR and subcloned into pET11c.ori+(−) (37).
Isolation of HeLa nucleosomes.
Mononucleosomes were purified from HeLa cells by a small-scale procedure (72, 75). Briefly, 1.2 × 108 cells were collected by trypsinization, washed with phosphate-buffered saline (PBS), and resuspended in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol [DTT], and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). Triton X-100 (0.1% final concentration) was added, the cells were incubated on ice for 8 min, and nuclei were collected by centrifugation (5 min at 1,300 × g and 4°C). The nuclei were washed again in buffer A, collected by centrifugation (5 min at 1,700 × g and 4°C), and resuspended in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, and 0.5 mM PMSF). The insoluble chromatin fraction was separated from the soluble fraction by centrifugation (5 min at 1,700 × g and 4°C), washed once with buffer B, and resuspended in buffer C (10 mM Tris [pH 7.6], 10 mM KCl, 1 mM CaCl2). Micrococcal nuclease (Sigma) was dissolved in a mixture of 5 mM Tris (pH 7.5) and 0.01 mM CaCl2, and the solution was incubated with the chromatin fraction (2 U/ml) for 10 min at 37°C. Digestion was stopped by adding 1 mM EGTA, and the supernatant containing solubilized mononucleosomes was collected by centrifugation (5 min at 1,700 × g and 4°C). Nucleosome quality was assessed by agarose gel electrophoresis in the presence of ethidium bromide to detect DNA and by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Coomassie brilliant blue R-250 staining to detect core histones.
Nucleosome-binding assays.
The Rosetta(DE3) strain of Escherichia coli (Novagen) was transformed with plasmids encoding glutathione-S-transferase (GST) fusion proteins, and expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) overnight at room temperature. Cells were disrupted by a freeze-thaw cycle and sonication, and the clarified lysate was incubated with glutathione (GSH)-Sepharose beads for 2 h at 4°C with rotation. Beads were collected by centrifugation and washed three times in 0.1 M KCl-HEMG buffer (25 mM HEPES [pH 7.6], 0.1 M KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 20% glycerol, 0.1% NP-40, 5 mM DTT, 0.2 mM PMSF, and 0.2 mM sodium metabisulfite) before use. GSH-Sepharose beads were loaded with equal amounts of the fusion proteins, mixed with mononucleosomes in 0.15 M KCl-HEMG buffer, incubated overnight at 4°C, and then washed five times in 0.25 M KCl-HEMG buffer. Bound proteins were denatured by being boiled in SDS loading buffer, fractionated by SDS-PAGE, and stained with Coomassie blue.
In vitro protein-protein interaction assays.
Interactions between epitope-tagged polypeptides expressed in mammalian cells were detected by coimmunoprecipitation and immunoblotting. Transfected cells were incubated for 30 min on ice in either radioimmunoprecipitation assay (RIPA) 0.1% SDS buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, 1 mM NaF, 1 mM Na3VO4) or NP-40 buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 10% glycerol, 0.5 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 0.5 mM NaF, 1 mM Na3VO4, 1 mM PMSF). Cleared lysates were incubated with EZview red anti-Flag M2 affinity gel (Sigma) for 2 h at 4°C and washed five times in Tris-buffered saline (TBS). Beads were boiled in SDS loading buffer containing 10% β-mercaptoethanol, and recovered proteins were analyzed by SDS-PAGE followed by immunoblotting with anti-T7 (Novagen) or anti-Flag (Sigma) antibodies.
Additionally, full-length MeCP2 (MeCP2FL) or fragments spanning the MBD and TRD were expressed by in vitro transcription and translation in the presence of [35S]methionine by using a TNT T7 Quick system (Promega, Inc.). RNA encoding MeCP2FL was transcribed from a derivative of pCITE2a+ (Novagen). Templates encoding MeCP2 fragments were generated by PCR using an upstream primer that included a T7 RNA polymerase promoter and a rabbit β-globin translation initiation sequence (71). For GST pulldown assays, translation reaction mixtures (10 to 15 μl) were combined with GST-loaded beads in a total reaction volume of 50 μl, adjusted to contain 100 mM NaCl, and incubated at 4°C for 2 h. Beads were washed in the same buffer containing 250 mM NaCl, the bound proteins were denatured in SDS sample buffer and resolved by SDS-15% PAGE, and the labeled proteins were visualized by fluorography.
Immunofluorescence and green fluorescence microscopy and luciferase assays.
For subcellular localization studies, NIH 3T3 cells were seeded onto sterile coverslips in a 24-well plate and transfected with 100 ng of expression plasmids by using Lipofectamine 2000 (Invitrogen). Cells were fixed after 24 h and, if necessary, probed with antibodies as described previously (43). After drying, coverslips were applied to slides by using fluorescent mounting medium (Dako Corporation) and visualized by laser scanning confocal microscopy with a Zeiss LSM 510 META microscope. Images were captured using the Zeiss AIM software, and TIFF images were exported into Adobe Photoshop 7.0 for cropping and minor adjustments. Histones were detected using rabbit polyclonal anti-histone H2B (Upstate) diluted 1:2,000 and an Alexa Fluor 633-conjugated goat anti-rabbit antibody (heavy and light chains; Invitrogen) also diluted 1:2,000. For luciferase reporter assays, HeLa cells were transfected using Polyfect (Qiagen) and lysates were prepared and assayed after 24 h as described previously (61). Reporter plasmid pGL3p-7xTR (21) was a kind gift of Rolf Renne (University of Florida, Gainesville). pE2F1-luc has been described previously (72).
RESULTS
LANA is present in the nucleus of every KSHV-infected cell, where it plays critical roles in the modulation of gene expression and in the propagation of the viral episome, functions that are dependent on the physical association of LANA with host chromosomes. A steadily growing list of nuclear proteins have been shown to interact with LANA and, in general, have properties relevant to transcriptional regulation or chromatin binding. Of these cellular partners, MeCP2 is particularly interesting because of its ability to bind DNA, its expression in a wide variety of cell types (albeit at different levels), and its ability to modulate chromatin structure (33). How the MeCP2-LANA interaction contributes to KSHV latency is not known. To address this issue, we set out to characterize the regions of LANA and MeCP2 required for the interaction as a prelude to analyzing function.
LANAC interacts with MeCP2.
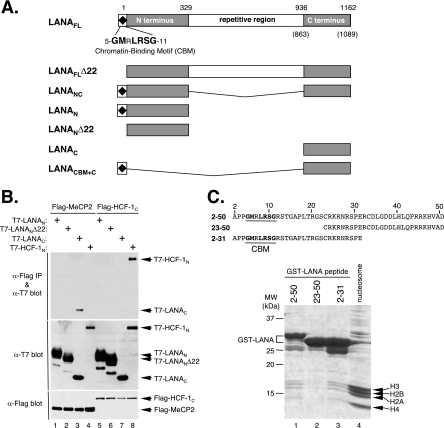
Based on the primary amino acid sequence, LANA can be divided into three discrete regions (Fig. 1A): a proline- and basic residue-rich N terminus (LANAN), a central region composed of a highly variable number of acidic repeats, and a C-terminal region (LANAC) that shows significant homology to orthologs from other gamma-2 herpesviruses (11, 61). The CBM lies at the extreme N terminus (7, 51, 72). To map the LANA-MeCP2 interaction, we subdivided LANA into fragments corresponding to N- and C-terminal regions and performed a coimmunoprecipitation assay using Flag epitope-tagged MeCP2 (Flag-MeCP2) and T7-tagged fragments corresponding to LANAN and LANAC (Fig. 1B, lanes 1 to 4). As reported previously, the central repetitive region is expressed very poorly and could not be analyzed on its own (36, 61; also data not shown). The tagged proteins were coexpressed in human 293T cells, immunoprecipitated with anti-Flag antibody-coupled beads, resolved by SDS-PAGE, and probed by immunoblotting with anti-T7 antibody. Under these conditions, T7-tagged LANAC (residues 936 to 1162) (Fig. 1B, lane 3) was readily coimmunoprecipitated by using Flag-MeCP2 but not by using a control protein, the Flag-tagged C terminus of HCF-1 (Flag-HCF-1C) (Fig. 1B, lane 7). T7-LANAN (residues 1 to 329) (Fig. 1B, lane 1) and a version lacking the first 22 residues, including the CBM (T7-LANANΔ22 [residues 23 to 329]) (Fig. 1B, lane 2), were not recovered, indicating that they do not stably associate with MeCP2 when expressed in isolation. As a control for the assay, we also included the T7-tagged HCF-1 N terminus (T7-HCF-1N) (Fig. 1B, lanes 4 and 8), which formed a robust and specific interaction with the control protein Flag-HCF-1C (Fig. 1B, lane 8) (70) but not with Flag-MeCP2 (Fig. 1B, lane 4). This result shows that MeCP2 interacts with LANAC and that this interaction does not require the N terminus or the central region.
FIG. 1.
MeCP2 interacts with the C-terminal domain of LANA. (A) Domain structure of KSHV LANA, illustrating the boundaries of engineered fragments used in this study. The amino acid numbering corresponds to the prototype sequence from the BC-1 cell line (57). Numbers in parentheses signify the actual coordinates of the LANA variant used here (GenBank accession no. AAB626557) (48). (B) Results from a coimmunoprecipitation assay using T7 epitope-tagged LANAN, LANANΔ22, and LANAC fragments and Flag-MeCP2FL. T7-HCF-1N serves as a positive control for association with Flag-HCF-1C (70). Human 293T cells were cotransfected with combinations of plasmids expressing the indicated constructs. After 24 h, protein extracts were prepared and immunoprecipitated (IP) using anti-Flag-coupled beads, resolved by SDS-6.5% PAGE, and immunoblotted with either anti-T7 antibody (α-T7) (top panels) or anti-Flag antibody (bottom panel). (C) The LANA CBM interacts with nucleosomes. Partially purified HeLa nucleosomes (lane 4) were incubated with GST fusion proteins corresponding to LANA residues 2 to 50 (lane 1), 23 to 50 (lane 2), and 2 to 31 (lane 3). The sequence of each LANA peptide is shown with the CBM highlighted. After extensive washing, bound complexes were heat denatured in SDS sample buffer, resolved by SDS-15% PAGE, and stained with Coomassie brilliant blue. Bands corresponding to each of the core histones are indicated.
CBM is required for nucleosome binding.
We also verified the interaction between the CBM and nucleosomes as reported previously by Barbera and colleagues (8, 9). For this purpose, solubilized HeLa mononucleosomes were incubated with beads carrying GST fused to peptides derived from the first 50 residues of LANA (Fig. 1C). After extensive washing, the bound protein complexes were denatured, fractionated by SDS-PAGE, and stained with Coomassie brilliant blue to detect bound proteins. The four core histones (H2A, H2B, H3, and H4) were readily retained by the two CBM-containing peptides (LANAN residues 2 to 50 fused to GST [GST-LANAN2-50] and GST-LANAN2-31) (Fig. 1C, lanes 1 and 3) but not by a truncated peptide that excludes the CBM (GST-LANAN23-50) (Fig. 1C, lane 2). This finding provides independent confirmation that the LANA CBM stably interacts with nucleosomes.
MeCP2 localizes LANA to pericentric heterochromatin.
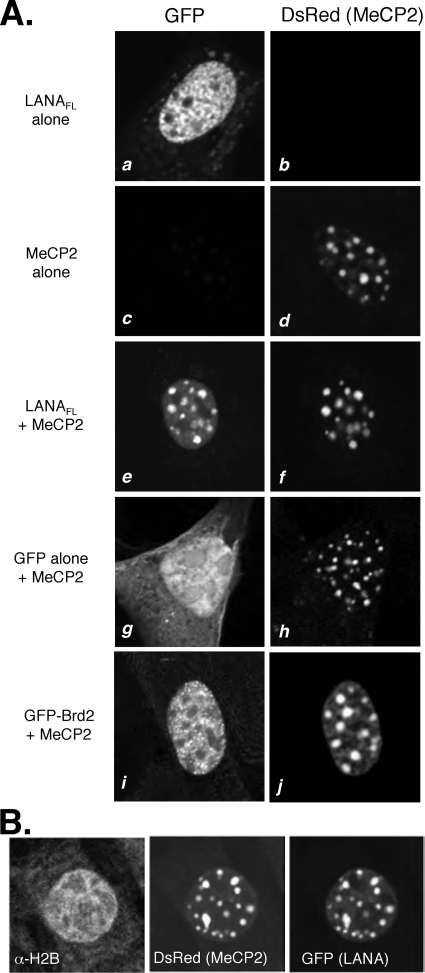
Although MeCP2 occupies sites throughout the euchromatic regions of the nucleus, the protein is most abundant in regions of pericentric heterochromatin (47). To characterize the LANA-MeCP2 association in vivo, we took advantage of the fact that, in murine NIH 3T3 cells, the large arrays of the major satellite repeats are organized into characteristic heterochromatic domains known as chromocenters and are readily visible by fluorescence microscopy (26, 55, 59). In human cells, the heterochromatic regions are more widely dispersed and MeCP2 shows more diffuse localization throughout the interphase nucleus. For ready detection, full-length LANA (LANAFL) was fused to green fluorescent protein (GFP) and MeCP2 was fused to Discosoma red fluorescent protein (DsRed). As illustrated by the representative cells shown in Fig. 2, when GFP-LANAFL was expressed alone, it was found in the nucleus (Fig. 2A, panel a), with a typical dispersed pattern and exclusion from the nucleoli. In the presence of DsRed-MeCP2, however, GFP-LANAFL was concentrated into well-defined foci that closely mirrored the accumulations of DsRed-MeCP2 in the chromocenters (Fig. 2A, compare panels e and f). Relocalization was specific because neither GFP alone nor GFP fused to bromodomain-containing protein 2 (GFP-Brd2) was found to colocalize with DsRed-MeCP2 (Fig. 2A, panels g to j). Thus, in murine cells, a GFP-LANA fusion protein can be relocalized to pericentric heterochromatin by coexpression with DsRed-MeCP2 fusion protein. We also probed transfected cells with a polyclonal antibody to histone H2B (Fig. 2B) and found no obvious correlation between histone density and the signal for DsRed-MeCP2 or GFP-LANAFL. From these data, we conclude that the relocalization of LANA is most likely a direct consequence of binding to MeCP2 rather than an indirect reflection of higher nucleosome densities due to MeCP2-induced chromatin compaction.
FIG. 2.
LANA colocalizes with MeCP2 in NIH 3T3 cells. (A) NIH 3T3 cells were transiently transfected with expression plasmids encoding GFP fused to full-length LANA (GFP-LANAFL) (61) (a, b, e, and f) and DsRed fused to full-length human MeCP2 (DsRed-MeCP2FL) (c to j). GFP alone (g and h) and GFP-Brd2 (i and j) serve as controls. Each image panel shows a single cell and is representative of the predominant pattern in that transfection experiment. (B) MeCP2-dependent relocalization of LANA is not mirrored by histones. NIH 3T3 cells were transfected to express cytomegalovirus lacZ, DsRed-MeCP2FL, and/or GFP-LANAFL. After 24 h, cells were fixed and probed with an antibody against total histone H2B (α-H2B).
Both the CBM of LANA and LANAC contribute to chromocenter localization.
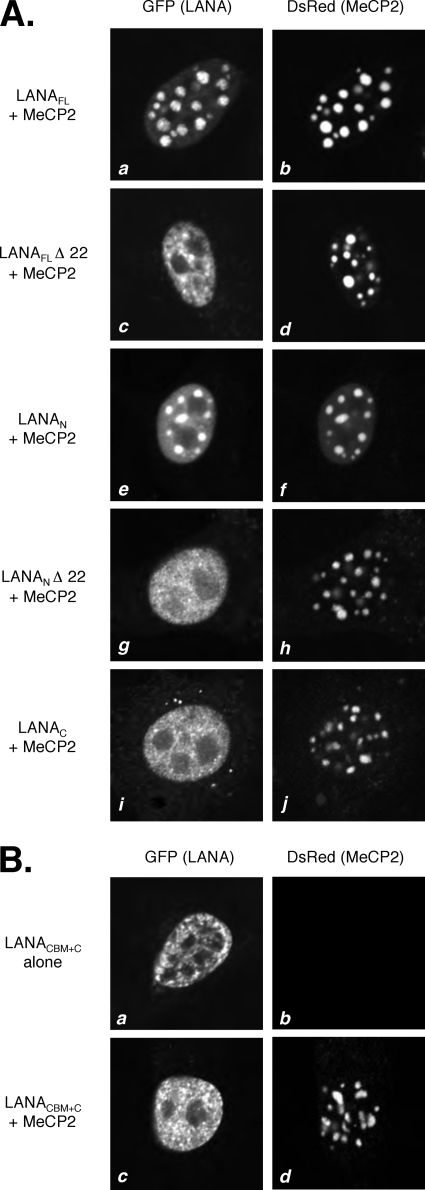
Using this NIH 3T3 cell relocalization assay, we next tested a series of LANA truncations to identify the functional domains required for colocalization with DsRed-MeCP2 (Fig. 3A). As before, GFP-LANAFL (Fig. 3A, panel a) was localized predominantly to the chromocenters, coincident with DsRed-MeCP2 (Fig. 3A, panel b). Deletion of the CBM from GFP-LANAFL, yielding GFP-LANAFLΔ22 (Fig. 3A, panel c), resulted in much more diffuse distribution throughout the nucleoplasm, with a significant reduction in the extent of colocalization with DsRed-MeCP2. The distribution of DsRed-MeCP2 did not appear to change. Interestingly, the isolated LANA N terminus (GFP-LANAN) (Fig. 3A, panel e) showed an intermediate pattern of distribution, with obvious chromocenter foci superimposed upon a stronger background signal. Removal of the CBM from this fragment (yielding GFP-LANANΔ22) resulted in a complete loss of chromocenter localization (Fig. 3A, panel g). Lastly, the isolated C terminus (GFP-LANAC) (Fig. 3A, panel g) showed a diffuse nuclear distribution pattern similar to that of GFP-LANANΔ22 (Fig. 3A, compare panels g and i). It should be noted that although the principal nuclear localization signal (NLS) for LANA is in the N terminus, there is a secondary NLS in the C terminus and this is sufficient for nuclear accumulation of LANAC (51, 61). These results demonstrate that multiple regions of LANA are required for optimal relocalization of GFP-LANA (61) to pericentric heterochromatin in the presence of DsRed-MeCP2. Even though LANAC is sufficient for coimmunoprecipitation of MeCP2, N-terminal functions, including that of the CBM, are required for effective colocalization with DsRed-MeCP2.
FIG. 3.
Colocalization with MeCP2 requires multiple regions of LANA. (A) NIH 3T3 cells were transfected with plasmids encoding DsRed-MeCP2FL and the following GFP-LANA derivatives: GFP-LANAFL (a and b), GFP-LANAFLΔ22 (c and d), GFP-LANAN (e and f), GFP-LANANΔ22 (g and h), and GFP-LANAC (i and j). (B) Fusion of the CBM to LANAC is not sufficient for colocalization with MeCP2. Cells were transfected to express GFP-LANACBM+C on its own (a and b) or with DsRed-MeCP2FL (c and d).
To explore these requirements further, we fused the first 22 residues of LANA, including the CBM, directly onto GFP-LANAC. As expected, the miniature LANA protein (GFP-LANACBM+C) was nuclear (Fig. 3B, panel a) but was not relocalized to the chromocenters in the presence of DsRed-MeCP2 (Fig. 3B, panel c). It is possible that the close juxtapositioning of the CBM and C-terminal domain interferes with binding to MeCP2 or to nucleosomes. Alternatively, the fusion may have removed sequences within the N terminus that are required to stabilize one or both interactions. Further studies are needed to distinguish these and other possibilities.
The MBD is required for localization of MeCP2 to chromocenters.
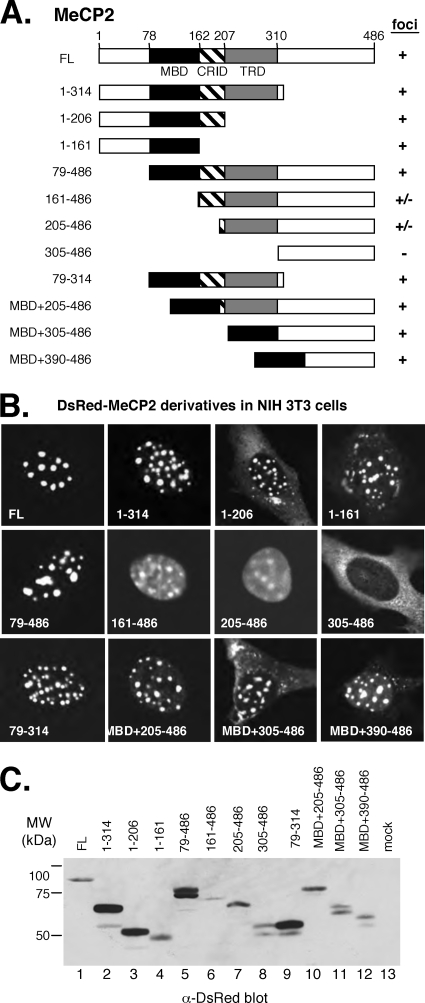
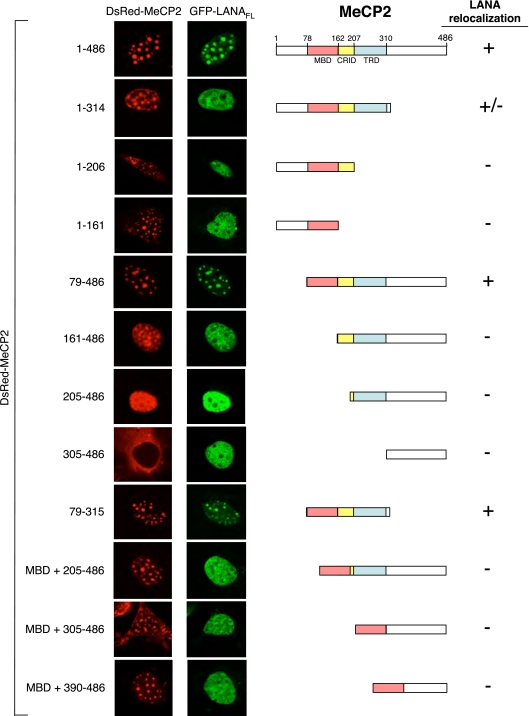
To map the regions of MeCP2 necessary for localization to chromocenters, we prepared a series of MeCP2 truncations in the context of the DsRed-MeCP2 fusion (Fig. 4A) and expressed the constructs individually in NIH 3T3 cells (Fig. 4B). Two major functional domains in MeCP2 have been identified: the MBD (residues 78 to 162), which specifically targets MeCP2 to methylated DNA sequences (45), and the TRD (residues 207 to 310) (46). A less clearly defined CRID (residues 162 to 206) lies between the MBD and TRD (30). Our analysis revealed that all the truncation proteins that retained the MBD were localized predominantly to chromocenters. Two deletion constructs (DsRed-MeCP2161-486 and DsRed-MeCP2205-486) that lacked the MBD but retained the TRD showed more diffuse distribution throughout the nucleoplasm but with some concentration in the chromocenters. A deletion that removed the MBD and TRD (yielding DsRed-MeCP2305-486) resulted in complete loss of nuclear localization, consistent with findings of previous mapping studies (47). Immunoblotting using an anti-DsRed antibody confirmed that all of the fusion proteins were expressed, albeit with some variation in abundance (Fig. 4C).
FIG. 4.
Localization of MeCP2 to chromocenters is specified by multiple domains. (A) Domain structures of human MeCP2 and the truncation proteins used in this study. (B) The MBD is required for localization of MeCP2 to chromocenters. Representative NIH 3T3 cells expressing each of the DsRed-MeCP2 derivatives are shown. The abilities of MeCP2 derivatives to localize (+) the DsRed signal to chromocenters (foci) are summarized in panel A. (C) Immunoblot analyzed using anti-DsRed antibody to detect expression of DsRed-MeCP2 fusion proteins in the cell extracts.
The CRID/TRD region of MeCP2 is required for association with LANA.
To identify domains within MeCP2 responsible for relocalization of LANA to chromocenters, we cotransfected cells with GFP-LANAFL and each of the DsRed-MeCP2 truncation constructs (Fig. 5). Consistent with the behavior of MeCP2FL, the subcellular localization of each truncation protein was essentially unaltered by the presence of GFP-LANAFL (compare Fig. 4 and 5). Only three MeCP2 derivatives (DsRed-MeCP21-314, DsRed-MeCP279-486, and DsRed-MeCP279-315) retained the ability to effectively recruit GFP-LANAFL to the chromocenters. In addition to the MBD, all three of these fragments retained the CRID and TRD. Proteins with truncations that individually removed the TRD (DsRed-MeCP21-206), the MBD (DsRed-MeCP2161-486), or the CRID (DsRed-MeCP2MBD + 205-486) were unable to relocalize GFP-LANAFL, indicating that all three domains are required.
FIG. 5.
The CRID/TRD region of MeCP2 is required to relocalize LANA to chromocenters. Cells were cotransfected with plasmids expressing GFP-LANAFL and full-length or truncated versions of DsRed-MeCP2. Single cells displaying the predominant pattern for each transfection experiment are shown.
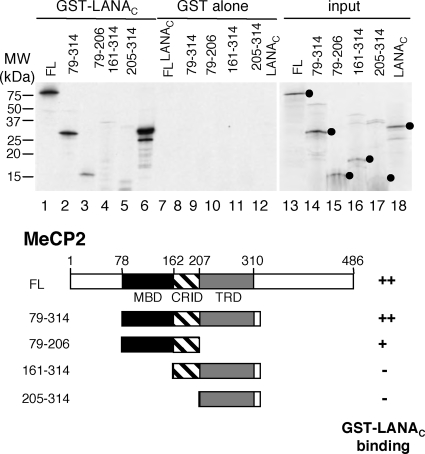
This analysis does not tell us whether the MBD is needed simply to localize the MeCP2-LANA complex to chromocenters or whether it may actually contribute to the stability of the interaction. To address this issue, we used a GST pulldown assay to map the domains required for binding rather than localization. In vitro-translated MeCP2 was incubated with agarose beads loaded with GST-LANAC or GST alone (Fig. 6). As expected, MeCP2FL was recovered with the GST-LANAC beads only (Fig. 6, compare lanes 1 and 7). A fragment spanning the MBD, CRID, and TRD (MeCP279-314) was recovered with similar efficiency (Fig. 6, lane 2), and a further truncation protein that lacked the TRD (MeCP279-206) retained some binding activity (Fig. 6, lane 3). Fragments that lacked the MBD (MeCP2161-314 and MeCP2205-314) showed no specific interaction (Fig. 6, lanes 4 and 5). In vitro-translated LANAC (Fig. 6, lanes 6 and 12) served as a positive control for binding, reflecting the self-oligomerization properties of LANAC (61). Combining the results of these in vivo and in vitro experiments, we conclude that LANAC interacts with the central MBD/CRID/TRD region of MeCP2 and that this interaction is sufficient to relocalize LANA to the chromocenters.
FIG. 6.
LANA binds to the multiple domains within MeCP2. In vitro binding analysis using recombinant GST-LANAC purified from bacteria and 35S-labeled full-length or truncated MeCP2 (shown schematically below) synthesized by in vitro translation. Binding reactions were performed with 100 mM NaCl, and mixtures were washed in 250 mM NaCl. 35S-labeled LANAC (lanes 6, 12, and 18) served as a positive control for binding.
The MeCP2 C terminus contributes to LANA-mediated repression.
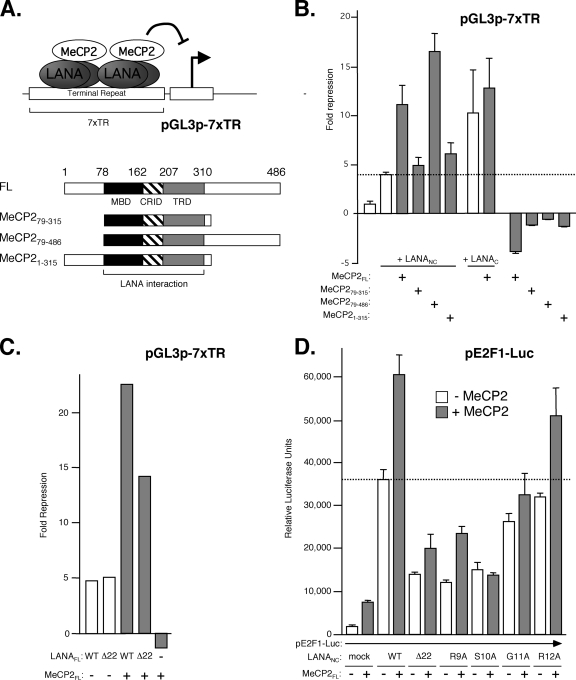
LANA binds to DNA via its C terminus, recognizing an imperfect 20-bp palindrome found in the TRs, and in this context it represses transcription, especially when there are multiple TRs as found in the KSHV genome (21). We asked if MeCP2 can contribute to the transcriptional properties of LANA by measuring the activity of a reporter construct (pGL3p-7xTR) consisting of the constitutively active simian virus 40 early promoter, a luciferase reporter gene, and seven copies of the 300-bp TR inserted downstream (Fig. 7A). In HeLa cells, the reporter was active but was repressed fourfold by the addition of a fragment consisting of the N and C termini of LANA without the central repetitive region (LANANC) (Fig. 7B). The level of repression was increased to 12-fold by coexpression of LANANC and MeCP2FL. In the absence of LANA, MeCP2 increased promoter activity slightly, suggesting that repression is dependent strictly on LANA. Expression of the MeCP2 central MBD/CRID/TRD region alone (MeCP279-315) (Fig. 7A), which is sufficient for recruitment of LANA to chromocenters, failed to enhance TR-mediated repression, but repression (16-fold) was restored by the inclusion of the MeCP2 C terminus (MeCP279-486). Synergy was not observed with the addition of the N terminus (MeCP21-315).
FIG. 7.
LANA and MeCP2 cooperate to regulate transcription. (A) LANA bound to reiterated copies of the KSHV TR sequence can repress a constitutively active promoter located on the same plasmid. The schematic shows MeCP2 truncations, indicating the three domains (MBD, CRID, and TRD) required for association with LANA. (B) HeLa cells were cotransfected with the pGL3p-7xTR reporter and expression plasmids encoding derivatives of LANA and/or MeCP2. Luciferase activity was measured after 24 h, and the degree of repression (n-fold) was calculated relative to expression in cells with the reporter alone. Values represent the means and standard deviations of results from three independent assays. The dotted line indicates the level of repression by LANANC alone. Note that expression of MeCP2 in the absence of LANA results in modest stimulation. (C) Results of a repression assay comparing the LANAFL wild type (WT) and the CBM deletion mutant (Δ22) in the presence or absence of MeCP2. (D) Stimulation of the human E2F1 promoter by LANA and MeCP2 is sensitive to mutations in the LANA CBM. Cells were cotransfected with the pE2F1-luc reporter and plasmids expressing WT and mutant versions of LANANC in the presence or absence of MeCP2FL. The dotted line corresponds to reporter activity in the presence of LANANC alone.
Consistent with the findings described above, isolated LANAC was capable of repressing transcription from the pGL3p-7xTR reporter (21) but showed no significant synergy with MeCP2 (Fig. 7B), in keeping with the lack of colocalization to chromocenters (Fig. 3A). Likewise, deletion of the CBM from full-length LANA (yielding LANAFLΔ22) had no effect on repression by LANA alone (Fig. 7C) but reduced the extent of synergy with MeCP2, again consistent with the poor colocalization to the chromocenters.
As we have shown previously (72), a reporter construct consisting of the human E2F1 promoter fused to a luciferase gene can be stimulated severalfold by the expression of LANANC, which lacks the central repetitive region (Fig. 7D). Similar results were obtained with LANAFL but were reduced in magnitude because of inherently lower expression levels of the full-length protein (data not shown). Transactivation was enhanced by the addition of MeCP2FL. In the absence of LANA, MeCP2FL had only a small effect on the reporter activity. As in the repression assay, the stimulatory effect of MeCP2FL was sensitive to mutations in the LANA CBM. Deletion of the CBM (Δ22) significantly reduced the LANA- and MeCP2-dependent increases in promoter activity, although it did not abolish them entirely. Likewise, single-residue substitutions within the CBM (R9A and S10A) that have been shown previously to severely compromise chromatin binding (72) produced results similar to those of the deletion of the entire CBM, whereas mutations of less critical flanking residues (G11A and R12A) had more moderate effects. Thus, MeCP2 can enhance the ability of LANA to stimulate E2F1 promoter activity and requires a functional CBM in LANA.
DISCUSSION
Studies of LANA often divide the protein into three discrete regions: the N terminus, a central repetitive region, and the C terminus (51, 61). This surgical approach has been quite successful, leading to the identification of the autonomous DNA-binding and dimerization domain in LANAC and to the N-terminal CBM, which serves as the major determinant of chromosome binding (15, 61). Deletion of the central repetitive region is advantageous because this removes sequences that suppress LANA mRNA translation, thereby increasing the expression of the other functional domains (36). For several LANA functions, however, it appears that multiple regions of the protein are required, and together with the results of the present study, this observation argues that N- and C-terminal sequences may be brought into proximity to each other in the native LANA protein (12, 17). The studies reported here show that optimal colocalization of LANA and MeCP2 at the major sites of pericentric heterochromatin (chromocenters) involves sequences from both the N- and C-terminal regions of LANA, and yet the results from coimmunoprecipitation and in vitro binding studies point to LANAC as the principal region of interaction with MeCP2 (Fig. 1B and 6A). We readily acknowledge that this conclusion runs contrary to the earlier reports from Hayward and colleagues, implicating the first 15 residues of LANA (i.e., the CBM) as the major determinant for binding to MeCP2 (33, 62). The reason for this discrepancy is not clear, but it is worth noting that a portion of the C terminus of herpesvirus saimiri (HVS) LANA (analogous to residues to 1063 to 1134 of KSHV LANA) was also found previously to be sufficient for interaction with MeCP2 (24). One possibility is that there are indeed multiple points of contact between LANA and MeCP2 and that subtle differences in the experimental conditions used in each study emphasize one interaction more strongly than the other. Support for this idea comes from the facts that LANAN showed partial colocalization with MeCP2 at the chromocenters (Fig. 3A, panel e) and that LANAC alone was not sufficient for relocalization to chromocenters (Fig. 3A, panel i). One caveat is that it is difficult to exclude the contributions of other cellular proteins such as HP1 or SUV39H1, which have been found to interact with both MeCP2 and LANA and may therefore confound the analysis (58). This concern is tempered by the fact that we are overexpressing LANA and MeCP2. Interestingly, we found that directly fusing the CBM to LANAC did not restore localization of LANA to the chromocenters in the presence of MeCP2, suggesting that additional N-terminal sequences might also be important or that the artificial fusion lacks sufficient flexibility to make the necessary interactions. Analyses of deletions within LANAN may speak to these alternative possibilities.
Accumulation of MeCP2 at the chromocenters of murine cells is well documented (1, 10, 32, 35). Equivalent structures are hard to discern in human cells due to the more dispersed nature of the satellite repeats. Recruitment of LANA to murine chromocenters in the presence of ectopic MeCP2 was first reported by Krithivas and colleagues, who also found that deletion of the CBM restored the diffuse pattern seen in the absence of MeCP2 (33). Our analysis showed that the MBD was the principal determinant for targeting MeCP2 to the chromocenters and that fragments lacking the MBD, but retaining the TRD, were localized throughout the nucleoplasm with only weak potential for focus formation (Fig. 4 and 5). In the Krithivas study, the morphology of the chromocenters seems to have been disturbed in cells expressing a version of LANA lacking the CBM, whereas we found the localization of MeCP2 to be essentially unaffected by the presence or absence of LANA, implying that the MBD is a dominant determinant for chromocenter localization. Whether the same is true for more scattered heterochromatic regions or sites within the euchromatic portions of the nucleus cannot be determined from static imaging studies and will require other methods. Photobleaching experiments utilizing similar fusion proteins have revealed a surprisingly dynamic association between MeCP2 and chromatin, with individual molecules rapidly exchanging between the dense heterochromatin of the chromocenter regions and more open and less heavily CpG-methylated sites elsewhere in the nucleus (35). This pattern suggests that the association with LANA may significantly alter the dynamics of MeCP2 movement rather than localization per se. By providing additional nucleosomal contact through the CBM, LANA has the potential to stabilize MeCP2 at its preferred binding sites or, alternatively, enable MeCP2 to occupy low-affinity sites that would not otherwise retain the MBD. Indeed, it has been reported previously that introduction of LANA into murine L cells results in a striking redistribution of endogenous MeCP2, away from the chromocenters into smaller foci of condensed chromatin (64).
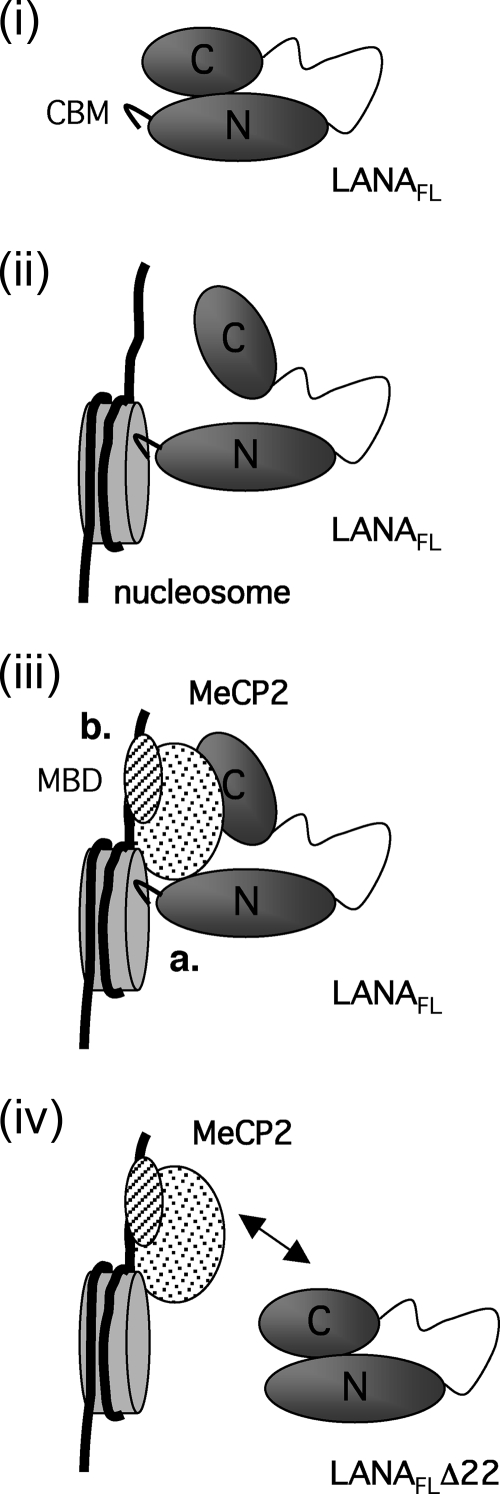
The interplay between the N- and C-terminal domains of LANA suggests that in the absence of chromatin binding, LANA adopts a conformation in which the C-terminal MeCP2-interacting surface is relatively inaccessible to other proteins (illustrated in Fig. 8, panel i). Formation of LANA dimers may also contribute to this masking effect. It is conceivable that docking of the CBM onto a nucleosome renders the C-terminal domain more accessible (Fig. 8, panel ii) and thereby facilitates complex formation with sequence- or context-specific accessory factors such as MeCP2 (Fig. 8, panel iii). Such complexes would be stabilized through the combined protein-protein and protein-DNA contacts. Deletion of the CBM not only would reduce contact with nucleosomes but also may interfere with the interaction between LANAC and MeCP2 (Fig. 8, panel iv), consistent with the more widespread distribution of LANAFLΔ22 (Fig. 3A, panel c). It will be interesting to determine if other chromatin-associated proteins, such as Brd2 and Brd4, that also interact primarily with LANAC are similarly influenced by the engagement (or removal) of the N-terminal CBM. In this regard, docking of the CBM onto nucleosomes may serve as a signal indicating that LANA is functioning in a chromatin context, thereby limiting unproductive interactions with nonchromatin partner proteins.
FIG. 8.
Schematics of possible multivalent interactions among LANA, core histones, and MeCP2. (i) Evidence suggests that LANA may be folded such that LANAN, which includes the CBM, is brought into proximity to LANAC. For simplicity, only one molecule of LANA is shown; however, LANA most likely exists as a stable dimer. (ii) Engagement of the CBM with a nucleosome may alter the conformation, making the C-terminal domain more accessible to secondary interactions. (iii) Interaction of LANAC with MeCP2 forms a relatively stable complex mediated by contacts between the CBM and the nucleosome core (a) and DNA and the MeCP2 MBD (b). As a consequence of these multivalent interactions, LANA accumulates with MeCP2 at the chromocenters. (iv) In the absence of the CBM, the LANA protein (LANAFLΔ22) shows reduced retention at the chromocenters. This may occur because the deletion alters the accessibility of the LANAC domain or increases the rate at which LANA exchanges with other interaction partners located in more euchromatic regions of the genome.
The interplay between the N- and C-terminal regions of LANA was also evident in the analysis of MeCP2-mediated repression (Fig. 7B and C). In line with findings from earlier studies by Garber and colleagues, LANAC was sufficient to bind to reiterated TRs and repress the constitutively active promoter driving luciferase (21). Interestingly, the N terminus and more specifically the CBM were necessary for further repression in the presence of MeCP2. This finding mirrors the ability of LANA and MeCP2 to colocalize to chromocenters and suggests that the interaction of MeCP2 with LANAC observed in vitro is insufficient in vivo. The still poorly characterized C terminus of MeCP2 is critical for the amplification of LANA-mediated repression. This region contains a highly conserved polyhistidine stretch and two WW motifs embedded within a proline-rich region shown to bind the sequence-independent DNA-binding factor HMGB1 (16, 27). Furthermore, it is known that the C terminus is required for the chromatin compaction function associated with MeCP2, and this feature may be directly relevant to repression of the reporter (50).
Although we have used the localization of LANA to pericentric heterochromatin as a tool to explore the structural requirements for the LANA-MeCP2 interaction, the LANA-MeCP2 complexes are most likely also recruited to dispersed euchromatic sites. In neuroblastoma cells, MeCP2 can be detected at a large number of locations within euchromatic regions of the genome, including numerous active promoters, intergenic regions, and introns of active genes, by chromatin immunoprecipitation-on-chip (ChIP-chip) (76). Strikingly, the majority of the MeCP2-occupied promoters are not heavily CpG methyated, and levels of associated mRNA transcripts were typically increased rather than decreased by MeCP2 overexpression. The most parsimonious explanation for these provocative findings is that MeCP2 functions as both a negative and a positive regulator of gene expression rather than as a global repressor as first believed. Similar conclusions have been drawn from careful comparisons of transcript levels in the hypothalamuses of wild-type and MeCP2 knockout mice (13). How MeCP2 stimulates transcription is currently unknown but may involve the recruitment of positive coactivators in place of the well-characterized corepressors. This type of exchange would not be without ample precedent; a variety of regulatory factors, including members of the nuclear hormone receptor superfamily, are known to switch their transcriptional properties in this manner (23, 56, 74). Indeed, MeCP2 has been shown previously to associate with the transcriptional activator CREB1 at the activated somatostatin promoter in neural tissues, but not with a repressed target gene (13). We have shown that activation of the E2F1 promoter by LANA is further enhanced by coexpression with MeCP2. Although MeCP2 is itself a bona fide chromatin-binding protein, it could not compensate for deleterious mutations in the LANA CBM, underscoring the importance of histone association for LANA functions. In broader terms, it is likely that the association of LANA with MeCP2 provides latent KSHV with a means to alter the expression of select host cell genes and possibly those of the viral genome itself. Identifying LANA-MeCP2-regulated genes will shed new light on the maintenance and establishment of viral latency and may also offer fresh insight into the function of MeCP2 in nonneuronal cell types.
Acknowledgments
Naoko Tanese provided reagents and valuable comments on the manuscript; this work could not have been completed without her considerable help. We also thank Gregory David, Nicole Sunseri, Jennie Chung, Søren Ottosen, and Rolf Renne for insightful comments, plasmids, or assistance with the microscopy.
S.M. is the recipient of a postdoctoral fellowship from the Rett Syndrome Research Foundation, and the project was supported by grants (no. GM 61139-04 and S10 RR017970-01) from the NIH.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.Agarwal, N., T. Hardt, A. Brero, D. Nowak, U. Rothbauer, A. Becker, H. Leonhardt, and M. C. Cardoso. 2007. MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 35:5402-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amir, R. E., I. B. Van den Veyver, M. Wan, C. Q. Tran, U. Francke, and H. Y. Zoghbi. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23:185-188. [DOI] [PubMed] [Google Scholar]
- 3.An, F. Q., N. Compitello, E. Horwitz, M. Sramkoski, E. S. Knudsen, and R. Renne. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-asociated herpesvirus modulates cellular gene expression and protects lymphoid cells from P16INK4A-induced cell cycle arrest. J. Biol. Chem. 280:3862-3874. [DOI] [PubMed] [Google Scholar]
- 4.An, J., Y. Sun, and M. B. Rettig. 2004. Transcriptional coactivation of c-Jun by the KSHV-encoded LANA. Blood 103:222-228. [DOI] [PubMed] [Google Scholar]
- 5.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 6.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbera, A. J., M. E. Ballestas, and K. M. Kaye. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856-861. [DOI] [PubMed] [Google Scholar]
- 9.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, K. Luger, and K. M. Kaye. 2006. Kaposi's sarcoma-associated herpesvirus LANA hitches a ride on the chromosome. Cell Cycle 5:1048-1052. [DOI] [PubMed] [Google Scholar]
- 10.Brero, A., H. P. Easwaran, D. Nowak, I. Grunewald, T. Cremer, H. Leonhardt, and M. C. Cardoso. 2005. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 169:733-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnside, K. L., J. T. Ryan, H. Bielefeldt-Ohmann, A. Gregory Bruce, M. E. Thouless, C. C. Tsai, and T. M. Rose. 2006. RFHVMn ORF73 is structurally related to the KSHV ORF73 latency-associated nuclear antigen (LANA) and is expressed in retroperitoneal fibromatosis (RF) tumor cells. Virology 354:103-115. [DOI] [PubMed] [Google Scholar]
- 12.Cai, Q. L., J. S. Knight, S. C. Verma, P. Zald, and E. S. Robertson. 2006. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chahrour, M., S. Y. Jung, C. Shaw, X. Zhou, S. T. Wong, J. Qin, and H. Y. Zoghbi. 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320:1224-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 15.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 16.Dintilhac, A., and J. Bernues. 2002. HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. J. Biol. Chem. 277:7021-7028. [DOI] [PubMed] [Google Scholar]
- 17.Fujimuro, M., J. Liu, J. Zhu, H. Yokosawa, and S. D. Hayward. 2005. Regulation of the interaction between glycogen synthase kinase 3 and the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen. J. Virol. 79:10429-10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 19.Ganem, D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1:273-296. [DOI] [PubMed] [Google Scholar]
- 20.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 21.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgel, P. T., R. A. Horowitz-Scherer, N. Adkins, C. L. Woodcock, P. A. Wade, and J. C. Hansen. 2003. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 278:32181-32188. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, S., G. Akopyan, H. Garban, and B. Bonavida. 2006. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25:1125-1142. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths, R., and A. Whitehouse. 2007. Herpesvirus saimiri episomal persistence is maintained via interaction between open reading frame 73 and the cellular chromosome-associated protein MeCP2. J. Virol. 81:4021-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grundhoff, A., and D. Ganem. 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Invest. 113:124-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guenatri, M., D. Bailly, C. Maison, and G. Almouzni. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166:493-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hite, K. C., V. H. Adams, and J. C. Hansen. 2009. Recent advances in MeCP2 structure and function. Biochem. Cell Biol. 87:219-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu, J., and R. Renne. 2005. Characterization of the minimal replicator of Kaposi's sarcoma-associated herpesvirus latent origin. J. Virol. 79:2637-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura, H., and K. Shiota. 2003. Methyl-CpG binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 278:4806-4812. [DOI] [PubMed] [Google Scholar]
- 31.Klose, R. J., S. A. Sarraf, L. Schmiedeberg, S. M. McDermott, I. Stancheva, and A. P. Bird. 2005. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell 19:667-678. [DOI] [PubMed] [Google Scholar]
- 32.Koch, C., and W. H. Stratling. 2004. DNA binding of methyl-CpG-binding protein MeCP2 in human MCF7 cells. Biochemistry 43:5011-5021. [DOI] [PubMed] [Google Scholar]
- 33.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar, A., S. Kamboj, B. M. Malone, S. Kudo, J. L. Twiss, K. J. Czymmek, J. M. LaSalle, and N. C. Schanen. 2008. Analysis of protein domains and Rett syndrome mutations indicate that multiple regions influence chromatin-binding dynamics of the chromatin-associated protein MECP2 in vivo. J. Cell Sci. 121:1128-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwun, H. J., S. R. da Silva, I. Shah, N. Blake, P. S. Moore, and Y. Chang. 2007. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics Epstein-Barr virus EBNA1 immune evasion through central repeat domain effects on protein processing. J. Virol. 81:8225-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai, J. S., and W. Herr. 1997. Interdigitated residues within a small region of VP16 interact with Oct-1, HCF, and DNA. Mol. Cell. Biol. 17:3937-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim, C., C. Choi, and J. Choe. 2004. Mitotic chromosome-binding activity of latency-associated nuclear antigen 1 is required for DNA replication from terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus. J. Virol. 78:7248-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim, C., Y. Gwack, S. Hwang, S. Kim, and J. Choe. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016-31022. [DOI] [PubMed] [Google Scholar]
- 40.Lim, C., D. Lee, T. Seo, C. Choi, and J. Choe. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J. Biol. Chem. 278:7397-7405. [DOI] [PubMed] [Google Scholar]
- 41.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, F., L. Day, S. J. Gao, and P. M. Lieberman. 2006. Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi's sarcoma-associated herpesvirus lytic transcription. J. Virol. 80:5273-5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahajan, S. S., M. M. Little, R. Vazquez, and A. C. Wilson. 2002. Interaction of HCF-1 with a cellular nuclear export factor. J. Biol. Chem. 277:44292-44299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McPhillips, M. G., J. G. Oliveira, J. E. Spindler, R. Mitra, and A. A. McBride. 2006. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 80:9530-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nan, X., R. R. Meehan, and A. Bird. 1993. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 21:4886-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 47.Nan, X., P. Tate, E. Li, and A. Bird. 1996. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 16:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1998. Human herpesvirus 8—the first human Rhadinovirus. J. Natl. Cancer Inst. Monogr. 23:73-77. [DOI] [PubMed] [Google Scholar]
- 49.Nikitina, T., R. P. Ghosh, R. A. Horowitz-Scherer, J. C. Hansen, S. A. Grigoryev, and C. L. Woodcock. 2007. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J. Biol. Chem. 282:28237-28245. [DOI] [PubMed] [Google Scholar]
- 50.Nikitina, T., X. Shi, R. P. Ghosh, R. A. Horowitz-Scherer, J. C. Hansen, and C. L. Woodcock. 2007. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol. Cell. Biol. 27:864-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 54.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rice, J. C., S. D. Briggs, B. Ueberheide, C. M. Barber, J. Shabanowitz, D. F. Hunt, Y. Shinkai, and C. D. Allis. 2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12:1591-1598. [DOI] [PubMed] [Google Scholar]
- 56.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20:1405-1428. [DOI] [PubMed] [Google Scholar]
- 57.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakakibara, S., K. Ueda, K. Nishimura, E. Do, E. Ohsaki, T. Okuno, and K. Yamanishi. 2004. Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen. J. Virol. 78:7299-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulz, T. F. 2006. The pleiotropic effects of Kaposi's sarcoma herpesvirus. J. Pathol. 208:187-198. [DOI] [PubMed] [Google Scholar]
- 61.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shamay, M., A. Krithivas, J. Zhang, and S. D. Hayward. 2006. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. U. S. A. 103:14554-14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stedman, W., Z. Deng, F. Lu, and P. M. Lieberman. 2004. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J. Virol. 78:12566-12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stuber, G., K. Mattsson, E. Flaberg, E. Kati, L. Markasz, J. A. Sheldon, G. Klein, T. F. Schulz, and L. Szekely. 2007. HHV-8 encoded LANA-1 alters the higher organization of the cell nucleus. Mol. Cancer 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang, J., G. M. Gordon, M. G. Muller, M. Dahiya, and K. E. Foreman. 2003. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen induces expression of the helix-loop-helix protein Id-1 in human endothelial cells. J. Virol. 77:5975-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ueda, K., S. Sakakibara, E. Ohsaki, and K. Yada. 2006. Lack of a mechanism for faithful partition and maintenance of the KSHV genome. Virus Res. 122:85-94. [DOI] [PubMed] [Google Scholar]
- 67.Verma, S. C., S. Borah, and E. S. Robertson. 2004. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 78:10348-10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verma, S. C., T. Choudhuri, R. Kaul, and E. S. Robertson. 2006. Latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats. J. Virol. 80:2243-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watanabe, T., M. Sugaya, A. M. Atkins, E. A. Aquilino, A. Yang, D. L. Borris, J. Brady, and A. Blauvelt. 2003. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen prolongs the life span of primary human umbilical vein endothelial cells. J. Virol. 77:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson, A. C., M. J. Boutros, K. M. Johnson, and W. Herr. 2000. HCF amino- and carboxy-terminal subunit association through two separate sets of interaction modules: involvement of fibronectin type 3 repeats. Mol. Cell. Biol. 20:6721-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson, A. C., R. N. Freiman, H. Goto, T. Nishimoto, and W. Herr. 1997. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol. Cell. Biol. 17:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong, L. Y., G. A. Matchett, and A. C. Wilson. 2004. Transcriptional activation by the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen is facilitated by an N-terminal chromatin-binding motif. J. Virol. 78:10074-10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wong, L. Y., and A. C. Wilson. 2005. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen induces a strong bend on binding to terminal repeat DNA. J. Virol. 79:13829-13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wysocka, J., P. T. Reilly, and W. Herr. 2001. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 21:3820-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yasui, D. H., S. Peddada, M. C. Bieda, R. O. Vallero, A. Hogart, R. P. Nagarajan, K. N. Thatcher, P. J. Farnham, and J. M. Lasalle. 2007. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc. Natl. Acad. Sci. U. S. A. 104:19416-19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye, F. C., F. C. Zhou, S. M. Yoo, J. P. Xie, P. J. Browning, and S. J. Gao. 2004. Disruption of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol. 78:11121-11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.You, J., V. Srinivasan, G. V. Denis, W. J. Harrington, Jr., M. E. Ballestas, K. M. Kaye, and P. M. Howley. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 80:8909-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Young, J. I., E. P. Hong, J. C. Castle, J. Crespo-Barreto, A. B. Bowman, M. F. Rose, D. Kang, R. Richman, J. M. Johnson, S. Berget, and H. Y. Zoghbi. 2005. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl. Acad. Sci. U. S. A. 102:17551-17558. [DOI] [PMC free article] [PubMed] [Google Scholar]