Abstract
Understanding the molecular basis of Clostridium difficile infection is a prerequisite to the development of effective countermeasures. Although there are methods for constructing gene-specific mutants of C. difficile, currently there is no effective method for generating libraries of random mutants. In this study, we developed a novel mariner-based transposon system for in vivo random mutagenesis of C. difficile R20291, the BI/NAP1/027 epidemic strain at the center of the C. difficile outbreaks in Stoke Mandeville, United Kingdom, in 2003 to 2004 and 2004 to 2005. Transposition occurred at a frequency of 4.5 (±0.4) × 10−4 per cell to give stable insertions at random genomic loci, which were defined only by the nucleotide sequence TA. Furthermore, mutants with just a single transposon insertion were generated in an overwhelming majority (98.3% in this study). Phenotypic screening of a C. difficile R20291 random mutant library yielded a sporulation/germination-defective clone with an insertion in the germination-specific protease gene cspBA and an auxotroph with an insertion in the pyrimidine biosynthesis gene pyrB. These results validate our mariner-based transposon system for use in forward genetic studies of C. difficile.
Clostridium difficile infection is widely recognized as the leading cause of health care-associated diarrhea in North America and Europe. Infection usually follows antibiotic treatment, which disrupts the native gastrointestinal microflora and thus allows C. difficile to proliferate. The emergence of so-called “epidemic” or “hypervirulent” strains of C. difficile over the last 5 to 10 years has compounded an already serious problem. Classed as BI/NAP1/027, these epidemic strains are believed to cause a more severe disease and lead to increased mortality and relapse rates (11, 20, 24).
Understanding the genetic and molecular basis of C. difficile infection will be a crucial step in the development of effective countermeasures. Methods for directed gene inactivation in C. difficile have recently been described (7, 21). This has opened the way for reverse genetic studies, in which the exact role of a specific gene, hypothesized to be important in a given phenotype, can be elucidated experimentally. By way of contrast, forward genetic studies aim to identify the genetic basis of a particular phenotype without making any assumptions about the genes involved. In forward genetic studies, transposons are often used to generate libraries of random insertion mutants. Libraries are then screened to identify mutants that are defective in a particular phenotype. Identification of the gene or genes which have been inactivated by transposon insertion then implicates them as having a role in that particular phenotype. Recently, just such an approach was used to identify a novel toxin-regulatory locus in Clostridium perfringens (29). This study elegantly demonstrated the power of forward genetic studies in bacterial pathogens.
A number of transposon mutagenesis systems have been described for Gram-positive bacteria (2, 3, 15, 16, 29, 32). Two different systems have recently been developed for use in C. perfringens (15, 29). Both are in vitro mutagenesis systems which rely on being able to transform the recipient organism. As such, they are not suitable for use in C. difficile because in the laboratory at present, recombinant DNA can be transferred into C. difficile only via conjugation. The conjugative transposons Tn916 and Tn5397 have been studied in C. difficile, but both have been found either to have a strong target site preference or to yield multiple insertions in individual clones (9, 30). Therefore, neither is well suited to generating libraries of random C. difficile mutants.
We reasoned that a mariner-based transposon mutagenesis system would be an effective tool for generating libraries of random C. difficile mutants. The mariner-transposable element Himar1 has been shown to insert randomly into the genomes of many bacterial species (3, 6, 16, 17, 32). The cognate Himar1 transposase is the only factor required for transposition, which occurs via a cut-and-paste mechanism (13, 14). The transposon itself is defined by inverted terminal repeats (ITRs) at either end and inserts into a TA target site. This is highly appropriate for an organism with a low-GC content such as C. difficile. In this study, we have developed a novel mariner-based transposon system for in vivo random mutagenesis of C. difficile. Moreover, we have demonstrated the system in C. difficile R20291, the BI/NAP1/027 epidemic strain at the center of the C. difficile outbreaks in Stoke Mandeville, United Kingdom, in 2003 to 2004 and 2004 to 2005. This new genetic tool opens the way for forward genetic studies of C. difficile.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmid transfer.
Escherichia coli TOP10 (Invitrogen) and E. coli CA434 (31) were cultured in Luria-Bertani (LB) medium, supplemented with erythromycin (50 μg/ml) or chloramphenicol (25 μg/ml), where appropriate. C. difficile R20291 was sourced from Jon Brazier (Anaerobe Reference Laboratory, Cardiff, United Kingdom). It is the BI/NAP1/027 epidemic strain isolated from the C. difficile outbreaks in Stoke Mandeville, United Kingdom, in 2003 to 2004 and 2004 to 2005. Routine culture of C. difficile R20291 was carried out in BHIS medium (brain heart infusion medium supplemented with 5 mg/ml yeast extract and 0.1% [wt/vol] l-cysteine) (26). Tryptose-yeast (TY) medium (3% [wt/vol] Bacto tryptose, 2% [wt/vol] yeast extract, and 0.1% [wt/vol] thioglycolate, adjusted to pH 7.4) was used to enhance expression from the tcdB promoter (PtcdB) of C. difficile (5). C. difficile medium was supplemented with d-cycloserine (250 μg/ml), cefoxitin (8 μg/ml), lincomycin (20 μg/ml), and/or thiamphenicol (15 μg/ml) where appropriate. For solid medium, agar was added to a final concentration of 1.0% (wt/vol). All C. difficile cultures were incubated in an anaerobic workstation at 37°C (Don Whitley, Yorkshire, United Kingdom).
Transposon mutants were screened for a sporulation/germination null (spo/ger−) phenotype following a 5-day incubation period in BHIS medium to allow sporulation to occur (27). Cultures were heat treated (at 60°C for 30 min) to kill vegetative cells and plated onto BHIS agar supplemented with 0.1% (wt/vol) sodium taurocholate (Sigma) to induce spore germination. Wild-type C. difficile R20291 was used as a sporulation/germination-positive (spo/ger+) control, and a spo0A knockout mutant was used as a spo/ger− control (7). Mutants with a spo/ger− phenotype were identified by failure to grow again after heat treatment.
Transposon mutants were screened for auxotrophy on C. difficile minimal medium. C. difficile minimal medium was made according to a recipe described by Karlsson et al. (10), with modifications made for ease of preparation. Briefly, separate stock solutions of amino acids (5×), salts (10×), glucose (20×), trace salts (50×), iron (100×), and vitamins (100×) were made by dissolving the appropriate components in distilled H2O (dH2O), as detailed in Table 1. Each stock solution was made fresh and filter sterilized (0.2-μm pore size) prior to use. C. difficile minimal medium was then made by mixing the appropriate volume of each stock solution together with sterile dH2O (Table 1). For solid C. difficile minimal medium, stock solutions were mixed together with molten agar in water (cooled to 50°C in a water bath following autoclaving), to give a final concentration of 1% (wt/vol) agar. Wild-type C. difficile R20291 was used as a nonauxotroph control, and a pyrF knockout mutant was used as an auxotroph control (7). Auxotrophic mutants were identified by failure to grow to grow on C. difficile minimal medium.
TABLE 1.
C. difficile minimal mediuma
| Stock solution component | Concn in stock solution (mg/ml) | Final concn in CDMM (mg/ml) |
|---|---|---|
| Amino acids (5×) | ||
| Casamino Acids | 50 | 10 |
| l-Tryptophan | 2.5 | 0.5 |
| l-Cysteine | 2.5 | 0.5 |
| Salts (10×) | ||
| Na2HPO4 | 50 | 5 |
| NaHCO3 | 50 | 5 |
| KH2PO4 | 9 | 0.9 |
| NaCl | 9 | 0.9 |
| Glucose (20×) | ||
| d-Glucose | 200 | 10 |
| Trace salts (50×) | ||
| (NH4)2SO4 | 2.0 | 0.04 |
| CaCl2·2H2O | 1.3 | 0.026 |
| MgCl2·6H2O | 1.0 | 0.02 |
| MnCl2·4H2O | 0.5 | 0.01 |
| CoCl2·6H2O | 0.05 | 0.001 |
| Iron (100×) | ||
| FeSO4·7H2O | 0.4 | 0.004 |
| Vitamins (100×) | ||
| d-Biotin | 0.1 | 0.001 |
| Calcium-d-pantothenate | 0.1 | 0.001 |
| Pyridoxine | 0.1 | 0.001 |
A 1-liter volume of C. difficile minimal medium (CDMM) was made by mixing 200 ml of 5× amino acids, 100 ml of 10× salts, 50 ml of 20× glucose, 20 ml of trace salts, 10 ml of 100× iron, and 10 ml of 100× vitamins with 610 ml sterile dH2O.
General molecular biology techniques.
Plasmids were isolated using a plasmid mini-prep kit (Qiagen). DNA was purified from agarose gels using a QIAquick gel extraction kit (Qiagen). Genomic DNA was isolated from C. difficile cultures using phenol-chloroform (25), following sequential treatment with lysozyme (10 mg/ml in phosphate-buffered saline [PBS] at 37°C for 30 min) and 10% (wt/vol) SDS (65°C for 30 min) to break down cell walls and lyse cells, respectively. Unless stated otherwise, enzymes were sourced from New England Biolabs (NEB), and PCRs were carried out using Phusion High-Fidelity DNA polymerase (NEB). All PCR primers used in this study are detailed in Table 2. E. coli strains were transformed by electroporation using a Gene-Pulser (Bio-Rad), as recommended by the manufacturer. Southern blot analysis was performed using a digoxigenin (DIG) High-Prime DNA labeling and detection kit (Roche) as instructed by the manufacturer. All DNA sequencing was carried out by Geneservice, United Kingdom.
TABLE 2.
Oligonucleotide primers
| Primer name | Sequence (5′-3′)a | Function [restriction site(s)] |
|---|---|---|
| PtcdB-F1 | tgcggccgcTTAATGAATTTAAAGAAATATTTACAATAGAAATCAAATTTTAGAATTAAC | Amplify tcdB promoter (NotI) |
| PtcdB-R1 | acatatgATTTTCTCCTTTACTATAATATTTTTATTGAATATTTTTACATCTAAATGC | Amplify tcdB promoter (NdeI) |
| HmrC9-F1 | tatgttcatatgGAAAAAAAGGAATTTCGTGTTTTGATAAAATACTGTTTTCTGAAGGG | Amplify Himar1 C9 transposase gene (NdeI)/PCR screening for Himar1 C9 transposase sequence |
| HmrC9-R1 | tatgttattaatTTATTCAACATAGTTCCCTTCAAGAGCGATACAACGATTATAACGACC | Amplify Himar1 C9 transposase gene (AseI)/PCR screening for Himar1 C9 transposase sequence |
| ITR1-F1 | tatgtttacgtataacaggttggctgataagtccccggtctgacacaattgTTTTTCGGCAAGTGTTCAAGAAGTTATTAAGTCGGGAGTGCAGTCGAAGTGGG | Amplify catP gene to yield transposon sequence (SnaBI/MfeI) |
| ITR-R1 | cagatgtttaaactaacaggttggctgataagtccccggtctaacaaaaaataagaagcctgcatttgcaggcttcttatttttaagcttAGACAAACCTGAAGTTAACTATTTATCAATTCCTGCAATTCGTTTACAAAACGG | Amplify catP gene to yield transposon sequence (PmeI/HindIII) |
| PtcdB-Fs1 | GATAGAGAAATATCAGTGAAATTAAAAATATCTAGACAAGCTGTTAATAAGG | PCR screening of chromosomal tcdB promoter |
| PtcdB-Rs1 | ATTGCTACATATTCATCTTCCTGAACACGAAATCTTACATTTGCC | PCR screening of chromosomal tcdB promoter |
| catP-F1 | GGCAAGTGTTCAAGAAGTTATTAAGTCGGGAGTGCAGTCGAAGTGG | PCR screening for transposon based catP gene and Southern probe synthesis |
| catP-R1 | TGAAGTTAACTATTTATCAATTCCTGCAATTCGTTTACAAAACGG | PCR screening for transposon based catP gene and Southern probe synthesis |
| catP-INV-F1 | TAAATCATTTTTAGCAGATTATGAAAGTGATACGCAACGGTATGG | Inverse PCR |
| catP-INV-R1 | TATTGTATAGCTTGGTATCATCTCATCATATATCCCCAATTCACC | Inverse PCR |
| catP-INV-R2 | TATTTGTGTGATATCCACTTTAACGGTCATGCTGTAGGTACAAGG | Sequencing of inverse PCR products |
Bases in capitals are complementary to the target sequence. Underlining indicates recognition sequences of the corresponding restriction endonucleases listed in the final column. Boldface indicates the mariner ITR sequences. Italics indicate the fdx terminator sequence of C. pasteurianum.
Conjugations.
Plasmids were transferred to C. difficile R20291 by conjugation as described previously (23), with minor modifications. Briefly, 1 ml of E. coli CA434 overnight culture harboring plasmid was washed in PBS and transferred to the anaerobic workstation. The E. coli pellet was resuspended in a 150-μl volume of overnight C. difficile culture and spotted onto BHIS agar. Following 24 h of incubation, the conjugation mating mixture was harvested into 500 μl of PBS and plated onto BHIS agar supplemented with antibiotics, which permitted only growth of C. difficile transconjugant clones. Transconjugant colonies were picked and restreaked following 48 to 72 h of incubation.
Plasmid stability assays.
Plasmid segregational stability was determined as described previously (8). Briefly, C. difficile transconjugants were cultured for 12 h in BHIS medium supplemented with antibiotic to select for the plasmid. The culture was washed twice in PBS to remove the antibiotic and then used to inoculate fresh, unsupplemented BHIS medium at 1% (vol/vol). This marked the start (i.e., 0 h) of the stability experiment. The unsupplemented culture was then diluted 1% (vol/vol) into fresh medium every 12 h. At 24, 48, and 72 h the culture was plated to enumerate total CFU and thiamphenicol-resistant (Tmr) CFU. Plasmid stability per generation was calculated as n√R, where R is the proportion of cells in the culture retaining the plasmid at the last time point it could be determined, and n is the number of C. difficile generations passed by this time in the absence of antibiotic selection. Assuming that cultures reached maximum cell density (i.e., 100%) in each 12-h period and given that an inoculum of 1% (vol/vol) was used for each subculture, we took the number of generations per 12 h period to be 6.64 (because 1 × 26.64 = 100).
Construction of transposon delivery vectors.
The mariner transposon, consisting of the catP gene and the transcriptional terminator sequence from the ferrodoxin (fdx) gene of Clostridium pasteurianum, flanked by inverted terminal repeats, was constructed by PCR using primers ITR-F1 and ITR-R1 with plasmid pMTL5402F (7) as a template. The resulting product was cloned as an SnaBI/PmeI fragment into EcoICRI (Promega)-digested pMTL80241 (8) and sequenced in situ, using the M13 universal sequencing primers, to give plasmid pMTL80241::miniTn(catP). The hyperactive Himar1 C9 transposase gene (12) was PCR amplified without a promoter using primers HmrC9-F1 and HmrC9-R1 with plasmid pMarA (16) as a template. The resulting product was cloned as an NdeI/AseI fragment into NdeI-digested pMTL80241::miniTn(catP) and sequenced in situ using primers M13R, HmrC9-F1, and HmrC9-R1, to give pMTL80241::Himar1 C9-miniTn(catP). The transposase gene and catP transposon sequences were then excised together on a single PstI restriction fragment and cloned into SbfI-digested pMTL82250 (8) to give pMTL-SC0. Finally, pMTL-SC1 was generated by cloning the tcdB promoter of C. difficile R20291 into pMTL-SC0 in order to drive expression of Himar1 C9. To do this, a 326-bp fragment comprising the intergenic sequence between tcdD and tcdB was amplified using primers PtcdB-F1 and PtcdB-R1 with R20291 genomic DNA as a template. The resulting PCR product was cloned as an NotI/NdeI fragment into similarly digested pMTL-SC0 and sequenced using the M13R primer, thus giving rise to pMTL-SC1 (Fig. 1).
FIG. 1.
Vector map of plasmid pMTL-SC1. Expression of the hyperactive mariner transposase gene Himar1 C9 was driven by the C. difficile toxin B promoter, PtcdB. The control plasmid pMTL-SC0 was identical, except that there was no promoter driving expression of the transposase gene. The plasmid backbone consisted of the pBP1 replicon of C. botulinum (repA and orf2), the macrolide-lincosamide-streptogramin B antibiotic resistance gene ermB, the Gram-negative replicon ColE1, and the conjugal transfer function traJ. The whole mariner element (i.e., transposase gene and catP mini-transposon) can be excised as an SbfI fragment. The transcriptional terminators (Ω) are identical in sequence to those found immediately downstream of the fdx gene of Clostridium pasteurianum and the CD0164 open reading frame of C. difficile 630. This vector conforms to the pMTL80000 modular system for Clostridium shuttle plasmids (8).
Isolation of transposon mutants.
The mariner plasmids were transferred into C. difficile R20291 by conjugation. Transconjugants were initially selected on BHIS medium supplemented with cycloserine, cefoxitin, and lincomycin and then picked and restreaked onto TY medium supplemented with the same antibiotics in order to enhance expression from the tcdB promoter, which was driving expression of the Himar1 C9 transposase in pMTL-SC1. After 72 h, all growth was harvested into PBS, and serial dilutions were made and plated onto BHIS medium supplemented with cycloserine, cefoxitin, and thiamphenicol to select for the transposon-based catP marker. Individual colonies, visible after 12 to 16 h, were picked and restreaked onto the same medium twice for further analysis and/or phenotypic screening.
Inverse PCR and DNA sequence analysis.
Genomic DNA was isolated from individual transposon mutants and digested overnight with HindIII at a concentration of 200 ng/μl. The HindIII restriction endonuclease was heat inactivated (65°C for 30 min), and DNA was diluted to a concentration of 5 ng/μl in a reaction with T4 DNA ligase to favor self-ligation (and thus circularization) of restriction fragments. Ligation reaction mixtures were incubated at ambient temperature for 1 h, and then the T4 ligase was heat inactivated (65°C for 30 min). Inverse PCRs were carried out in 50-μl volumes using the KOD Hot Start DNA polymerase Master Mix kit (Novagen), with 100 ng of ligated DNA and primers catP-INV-F1 and catP-INV-R1, which face out from the transposon-based catP sequence. Inverse PCR products were run out on a 0.8% (wt/vol) agarose gel, purified with the QIAquick gel purification kit (Qiagen), and sequenced using primer catP-INV-R2 (Table 2). To identify the genomic location of transposon insertions, sequence data were analyzed using GENtle (http://gentle.magnusmanske.de/) and compared to the genome sequence of C. difficile R20291 (Refseq number NC_013316; GenBank accession number FN545816) (28) using Artemis (http://www.sanger.ac.uk/Software/Artemis/).
RESULTS AND DISCUSSION
Construction of a mariner-based transposon system for C. difficile.
As a first step toward constructing a transposon mutagenesis system, it was necessary to identify a suitable vehicle for delivering a transposon into the chromosome of C. difficile. In other bacteria, both suicide (33) and conditional (3, 16, 17, 32) plasmid vectors have been used for this purpose. However, the low frequency of DNA transfer achieved by conjugation from E. coli to C. difficile means that use of a suicide vector would be unfeasible for constructing mutant libraries, and no conditional vectors have been described for C. difficile to date.
Autonomously replicating, but segregationally unstable plasmids have been used as “pseudo-suicide vectors” in gene-directed inactivation methods for C. difficile (7, 21). Therefore, we proposed to use a similar approach. To identify a suitable pseudo-suicide vector to deliver our mariner transposon system, we assessed the conjugation frequency and segregational stability of four Gram-positive plasmid replicons in C. difficile R20291, all of which are readily available and have been reported previously (8). Each replicon was tested in an identical shuttle vector context, consisting of the chloramphenicol/thiamphenicol resistance gene catP, the Gram-negative replicon ColE1, the conjugal transfer function traJ, and a lacZα gene harboring a multiple cloning site. The plasmid based on the C. difficile replicon pCD6 transferred with the highest frequency and displayed the greatest stability in R20291, followed by the plasmids based on the Clostridium botulinum replicon pBP1 and the Clostridium butyricum replicon pCB102, respectively (Table 3). We were unable to transfer the plasmid based on the Bacillus subtilis pIM13 replicon. It was notable that cells of C. difficile R20291 harboring either the pBP1- or the pCB102-based plasmid took 48 to 72 h to form colonies on thiamphenicol plates, whereas those with the pCD6-based plasmid formed visible colonies after 24 h. Although pCB102 was the most unstable replicon of those we were able to transfer, we selected the pBP1 replicon for transposon delivery because, even though it was slightly more stable than the pCB102 replicon (1.3% per generation), it could be conjugated into R20291 at a frequency almost 8-fold greater than that of pCB102.
TABLE 3.
Plasmid replicon performance in C. difficile R20291
| Gram-positive replicon | Source of replicon |
Replicon contexta | Conjugation frequency into R20291b | Segregational stability in R20291c | |
|---|---|---|---|---|---|
| Organism | Reference(s) | ||||
| pBP1 | C. botulinum | 8 | pMTL82151 | (2.61 ± 0.04) × 10−7 | 57.2 (± 0.7) |
| pCB102 | C. butyricum | 4, 18 | pMTL83151 | (3.40 ± 1.90) × 10−8 | 55.9 (± 1.1) |
| pCD6 | C. difficile | 23 | pMTL84151 | (4.48 ± 0.47) × 10−7 | 76.0 (± 0.7) |
| pIM13 | B. subtilis | 1, 19 | pMTL85151 | —d | —d |
Each Gram-positive replicon was tested in an identical shuttle vector context consisting of the chloramphenicol/thiamphenicol resistance gene catP, the Gram-negative replicon ColE1, the conjugal transfer function traJ, and a lacZα multiple cloning site (8).
Conjugation frequencies were calculated as the number of transconjugant colonies per CFU of E. coli donor.
Segregational stabilities were calculated as percent stability per generation as described in Materials and Methods.
The pIM13-based plasmid pMTL85151 could not be transferred into C. difficile R20291.
Having selected the pBP1 replicon as our transposon delivery vehicle, we constructed the mariner plasmids pMTL-SC0 and pMTL-SC1 as described in Materials and Methods. These plasmids are identical except that expression of the Himar1 C9 transposase gene is driven by the C. difficile toxin B promoter in pMTL-SC1, whereas there is no promoter driving its expression in pMTL-SC0 (Fig. 1). As such, pMTL-SC0 served as the no-transposase control plasmid. In addition to the pBP1 pseudo-suicide replicon (repA and orf2), the mariner plasmids pMTL-SC0 and pMTL-SC1 each harbor the antibiotic resistance gene ermB, the Gram-negative replicon ColE1, and the conjugal transfer function traJ in their backbones. The transposon itself consists of the antibiotic resistance gene catP and a transcriptional terminator (Ω), flanked by inverted terminal repeats (ITR1 and ITR2). The whole mariner element (i.e., the Himar1 C9 transposase and the catP transposon) can be excised as an SbfI fragment, so it is easily transferred to alternative vector contexts.
Isolation and analysis of transposon mutants.
The mariner plasmids pMTL-SC0 and pMTL-SC1 were transferred separately into C. difficile R20291 by conjugation. Transconjugants were selected on BHIS medium supplemented with cycloserine, cefoxitin, and lincomycin and then subcultured on TY medium as described in Materials and Methods. This was done in an attempt to enhance expression from the tcdB promoter, which was driving expression of the Himar1 C9 transposase in pMTL-SC1 (5). Transconjugant clones were finally subcultured onto BHIS medium under selection for the transposon-based catP marker. After 12 to 16 h of incubation thiamphenicol resistant (Tmr) colonies were visible at a frequency of 4.5 (±0.4) × 10−4 (calculated as the ratio of Tmr CFU to total CFU) for the pMTL-SC1 transconjugant cultures. In contrast, no Tmr colonies were visible for the pMTL-SC0 (no-transposase control) transconjugant cultures. We postulated that the pMTL-SC1-derived Tmr colonies were the result of one or more independent transposition event(s). To test this hypothesis 17 randomly selected Tmr colonies, all derived from the same conjugation, were isolated for further analysis.
PCR analysis with primers catP-F1 and catP-R1 (Table 2) revealed that the transposon-based catP sequence was still present in the genomic DNA of all 17 clones isolated for further analysis (Fig. 2A). In contrast, PCR analysis with primers HmrC9-F1 and HmrC9-R1 (Table 2) revealed that the plasmid-based Himar1 C9 transposase gene was no longer present in any of the 17 clones (Fig. 2B). This indicated that the transposon had mobilized from the plasmid and that pMTL-SC1 (harboring the Himar1 C9 transposase) had subsequently been lost from the cells, thus immobilizing the transposon in situ. To ensure that pMTL-SC1 had not integrated into the C. difficile R20291 chromosome via homologous recombination at the PtcdB locus, a third PCR was carried out on the same genomic DNA templates, using primers PtcdB-Fs1 and PtcdB-Rs1. These primers flank the chromosome-based PtcdB sequence which is common with pMTL-SC1. The results revealed that no such integration event had occurred (Fig. 2C), providing further evidence that one or more independent transposition events had occurred.
FIG. 2.
PCR screens of 17 randomly selected pMTL-SC1-derived Tmr clones. Genomic DNA prepared from each clone was screened for the transposon-based catP gene (A), the plasmid-based Himar1 C9 transposase gene (B), and an uninterrupted chromosomal tcdB promoter sequence (C). Lane M, 1-kb ladder (Promega); lane P, pMTL-SC1; lane wt, wild-type C. difficile R20291; lanes 1 to 17, pMTL-SC1-derived Tmr clones 1 to 17.
To establish whether each of the 17 pMTL-SC1-derived Tmr clones was the result of an independent transposition event, Southern blot analysis was carried out. Genomic DNA was isolated from wild-type C. difficile R20291 and each of the Tmr clones, digested with HindIII, resolved on a 0.8% (wt/vol) agarose gel, and transferred to a Hybond H+ nitrocellulose membrane. Probing the membrane for the transposon-based catP sequence revealed that the transposon was present on a different size restriction fragment in each of the 17 Tmr clones and confirmed its absence in wild-type R20291 genomic DNA (Fig. 3). Furthermore, 16 of the 17 clones analyzed had a single transposon insertion while only 1 clone (clone 12) appeared to have a double insertion. These results indicated that each of the 17 Tmr clones did, indeed, arise from independent transposition events and suggested that the mariner-based catP transposon inserted randomly into the genome of C. difficile R20291.
FIG. 3.
Southern hybridization analysis of pMTL-SC1-derived Tmr clones. Genomic DNA samples were digested with HindIII. The membrane was probed for the transposon-based catP sequence. Lane wt, wild-type C. difficile R20291; lanes 1 to 17, pMTL-SC1-derived Tmr clones 1 to 17.
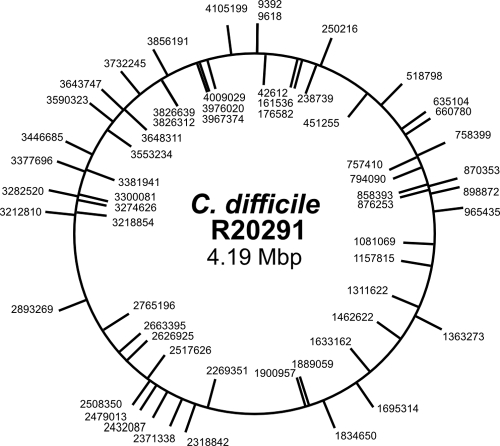
To further test the randomness of our mariner-based transposon, we successfully sequenced 60 independent transposon insertions, including those of the 17 clones which had been analyzed by Southern blotting. All of the Tmr clones sequenced had a single transposon insertion, with the exception of one (clone 12), which had already been found to have a double insertion by Southern blotting. Transposon insertions were distributed throughout the genome of C. difficile R20291, with no evidence for a preferred target site (Fig. 4). Furthermore, insertions were found to be stable through at least 10 serial subcultures in the absence of selection (data not shown). Characteristic of Himar1-based transposons, all insertions occurred at a TA dinucleotide target site, which was duplicated at the point of insertion. Overall, there were 28 insertions in the plus strand and 32 in the minus strand. Moreover, 45 of the 60 insertions sequenced (75%) were located within protein coding sequences. This is within the range that would be expected for a random mutagen, considering that 81% of the C. difficile R20291 genome is protein coding. Collectively, these data provide good evidence that our mariner-based transposon system is an effective tool for generating libraries of random C. difficile mutants.
FIG. 4.
Genetic map of mariner transposon insertions. Sixty independent transposon insertions were sequenced. Insertions in the plus orientation are marked on the circle exterior. Insertions in the minus orientation are marked on the circle interior. Numbers indicate the precise point of insertion according to genome sequence data for C. difficile R20291 (Refseq number NC_013316; GenBank accession number FN545816) (28).
Phenotypic screens and identification of transposon insertions.
Finally, to demonstrate the use of our mariner-based transposon system for forward genetic studies, we generated and screened a C. difficile R20291 mutant library for sporulation/germination mutants (spo/ger−) and auxotrophic mutants. We identified one spo/ger− mutant that failed to grow on BHIS medium supplemented with 0.1% (wt/vol) taurocholate following heat treatment (60°C for 30 min) and one auxotroph that failed to grow on C. difficile minimal medium. Inverse PCR was carried out to identify the genes which had been interrupted in these mutants (Table 4). The spo/ger− mutant was found to have an insertion in a germination-specific protease gene (cspBA) which has been shown to be essential for spore germination in C. perfringens (22). The auxotroph mutant was found to have an insertion in the gene encoding the aspartate carbomoyltransferase catalytic chain (pyrB). A search of the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/kegg2.html) revealed that this mutant is auxotrophic for uracil. These results validate the use of our mariner-based transposon system for forward genetic studies of C. difficile.
TABLE 4.
Transposon insertion sites in C. difficile R20291 mutants
| Phenotype | Transposon insertion sitea | Location in genome (nt)b | ORF interruptedc | Description |
|---|---|---|---|---|
| spo/ger− | TGGGAACACGTATGCAACTA— Tn—TATAACTGGTACAGCAGCAG | 2517626 | cspBA (CDR20291_2147) | Putative germination specific protease |
| Auxotroph | TCCCTTTATAGCTTCTTTTA— Tn—TATATAATCTGGCATTTTTA | 238739 | pyrB (CDR20291_0185) | Aspartate carbomoyltransferase catalytic chain |
The Tn insertion is indicated by dashes on either side, and the target site duplication is shown in boldface.
nt, nucleotide.
ORF, open reading frame.
In summary, we have successfully developed a novel mariner-based transposon system for in vivo random mutagenesis of C. difficile and demonstrated its use in the epidemic BI/NAP1/027 strain R20291. The transposon inserted into the genome in a random fashion, generating mutants with just a single insertion in an overwhelming majority (98.3% in this study). This is superior to the conjugative transposons Tn916 and Tn5397, both of which either display a strong target site preference or yield multiple insertions with a high frequency in C. difficile (9, 30). This new genetic tool opens the way for forward genetic studies of C. difficile.
Acknowledgments
We thank John Heap for useful discussions throughout this work and Laura Whitehorn for technical assistance.
We acknowledge the financial support of the MRC (G0601176) and the European Union (HEALTH-F3-2008-223585).
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Azeddoug, H., J. Hubert, and G. Reysset. 1992. Stable inheritance of shuttle vectors based on plasmid pIM13 in a mutant strain of Clostridium acetobutylicum. J. Gen. Microbiol. 138:1371-1378. [DOI] [PubMed] [Google Scholar]
- 2.Bae, T., A. K. Banger, A. Wallace, E. M. Glass, F. Aslund, O. Schneewind, and D. M. Missiakas. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. U. S. A. 101:12312-12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao, M., A. P. Bitar, and H. Marquis. 2007. A mariner-based transposition system for Listeria monocytogenes. Appl. Environ. Microbiol. 73:2758-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, M. E., J. D. Oultram, and M. Young. 1985. Identification of restriction fragments from two cryptic Clostridium butyricum plasmids that promote the establishment of a replication-defective plasmid in Bacillus subtilis. J. Gen. Microbiol. 131:2097-2105. [DOI] [PubMed] [Google Scholar]
- 5.Dupuy, B., and A. L. Sonenshein. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 27:107-120. [DOI] [PubMed] [Google Scholar]
- 6.Gao, L. Y., R. Groger, J. S. Cox, S. M. Beverley, E. H. Lawson, and E. J. Brown. 2003. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect. Immun. 71:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heap, J. T., O. J. Pennington, S. T. Cartman, G. P. Carter, and N. P. Minton. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452-464. [DOI] [PubMed] [Google Scholar]
- 8.Heap, J. T., O. J. Pennington, S. T. Cartman, and N. P. Minton. 2009. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78:79-85. [DOI] [PubMed] [Google Scholar]
- 9.Hussain, H. A., A. P. Roberts, and P. Mullany. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J. Med. Microbiol. 54:137-141. [DOI] [PubMed] [Google Scholar]
- 10.Karlsson, S., L. G. Burman, and T. Akerlund. 1999. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology 145:1683-1693. [DOI] [PubMed] [Google Scholar]
- 11.Kuijper, E. J., F. Barbut, J. S. Brazier, N. Kleinkauf, T. Eckmanns, M. L. Lambert, D. Drudy, F. Fitzpatrick, C. Wiuff, D. J. Brown, J. E. Coia, H. Pituch, P. Reichert, J. Even, J. Mossong, A. F. Widmer, K. E. Olsen, F. Allerberger, D. W. Notermans, M. Delmee, B. Coignard, M. Wilcox, B. Patel, R. Frei, E. Nagy, E. Bouza, M. Marin, T. Akerlund, A. Virolainen-Julkunen, O. Lyytikainen, S. Kotila, A. Ingebretsen, B. Smyth, P. Rooney, I. R. Poxton, and D. L. Monnet. 2008. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill. 13:pii=18942. [PubMed] [Google Scholar]
- 12.Lampe, D. J., B. J. Akerley, E. J. Rubin, J. J. Mekalanos, and H. M. Robertson. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. U. S. A. 96:11428-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lampe, D. J., M. E. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 14.Lampe, D. J., T. E. Grant, and H. M. Robertson. 1998. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149:179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanckriet, A., L. Timbermont, L. J. Happonen, M. I. Pajunen, F. Pasmans, F. Haesebrouck, R. Ducatelle, H. Savilahti, and F. Van Immerseel. 2009. Generation of single-copy transposon insertions in Clostridium perfringens by electroporation of phage mu DNA transposition complexes. Appl. Environ. Microbiol. 75:2638-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Breton, Y., N. P. Mohapatra, and W. G. Haldenwang. 2006. In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl. Environ. Microbiol. 72:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maier, T. M., R. Pechous, M. Casey, T. C. Zahrt, and D. W. Frank. 2006. In vivo Himar1-based transposon mutagenesis of Francisella tularensis. Appl. Environ. Microbiol. 72:1878-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minton, N. P., and J. Morris. 1981. Isolations and partial characterization of three cryptic plasmids from strains of Clostridium butyricum. J. Gen. Microbiol. 127:325-331. [Google Scholar]
- 19.Monod, M., C. Denoya, and D. Dubnau. 1986. Sequence and properties of pIM13, a macrolide-lincosamide-streptogramin B resistance plasmid from Bacillus subtilis. J. Bacteriol. 167:138-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Connor, J. R., S. Johnson, and D. N. Gerding. 2009. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology 136:1913-1924. [DOI] [PubMed] [Google Scholar]
- 21.O'Connor, J. R., D. Lyras, K. A. Farrow, V. Adams, D. R. Powell, J. Hinds, J. K. Cheung, and J. I. Rood. 2006. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 61:1335- 1351. [DOI] [PubMed] [Google Scholar]
- 22.Paredes-Sabja, D., P. Setlow, and M. R. Sarker. 2009. The protease CspB is essential for initiation of cortex hydrolysis and dipicolinic acid (DPA) release during germination of spores of Clostridium perfringens type A food poisoning isolates. Microbiology 155:3464-3472. [DOI] [PubMed] [Google Scholar]
- 23.Purdy, D., T. A. O'Keeffe, M. Elmore, M. Herbert, A. McLeod, M. Bokori-Brown, A. Ostrowski, and N. P. Minton. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46:439-452. [DOI] [PubMed] [Google Scholar]
- 24.Redelings, M. D., F. Sorvillo, and L. Mascola. 2007. Increase in Clostridium difficile-related mortality rates, United States, 1999-2004. Emerg. Infect. Dis. 13:1417-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., and W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Smith, C. J., S. M. Markowitz, and F. L. Macrina. 1981. Transferable tetracycline resistance in Clostridium difficile. Antimicrob. Agents Chemother. 19:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorg, J. A., and A. L. Sonenshein. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stabler, R. A., M. He, L. Dawson, M. Martin, E. Valiente, C. Corton, T. D. Lawley, M. Sebaihia, M. A. Quail, G. Rose, D. N. Gerding, M. Gibert, M. R. Popoff, J. Parkhill, G. Dougan, and B. W. Wren. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidal, J. E., J. Chen, J. Li, and B. A. McClane. 2009. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13. PLoS One 4:e6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, H., M. C. Smith, and P. Mullany. 2006. The conjugative transposon Tn5397 has a strong preference for integration into its Clostridium difficile target site. J. Bacteriol. 188:4871-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams, D. R., D. I. Young, and M. Young. 1990. Conjugative plasmid transfer from Escherichia coli to Clostridium acetobutylicum. J. Gen. Microbiol. 136:819-826. [DOI] [PubMed] [Google Scholar]
- 32.Wilson, A. C., M. Perego, and J. A. Hoch. 2007. New transposon delivery plasmids for insertional mutagenesis in Bacillus anthracis. J. Microbiol. Methods 71:332-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, J. K., M. A. Pritchett, D. J. Lampe, H. M. Robertson, and W. W. Metcalf. 2000. In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivorans C2A using a modified version of the insect mariner-family transposable element Himar1. Proc. Natl. Acad. Sci. U. S. A. 97:9665-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]