Abstract
Usher syndrome (USH) is a human hereditary disorder characterized by profound congenital deafness, retinitis pigmentosa and vestibular dysfunction. Myosin VIIa has been identified as the responsible gene for USH type 1B, and a number of missense mutations have been identified in the affected families. However, the molecular basis of the dysfunction of USH gene, myosin VIIa, in the affected families is unknown to date. Here we clarified the effects of USH1B mutations on human myosin VIIa motor function for the first time. The missense mutations of USH1B significantly inhibited the actin activation of ATPase activity of myosin VIIa. G25R, R212C, A397D and E450Q mutations abolished the actin-activated ATPase activity completely. P503L mutation increased the basal ATPase activity for 2-3 fold, but reduced the actin-activated ATPase activity to 50% of the wild type. While all the mutations examined, except for R302H, reduced the affinity for actin and the ATP hydrolysis cycling rate, they did not largely decrease the rate of ADP release from acto-myosin, suggesting that the mutations reduce the duty ratio of myosin VIIa. Taken together, the results suggest that the mutations responsible for USH1B cause the complete loss of the actin-activated ATPase activity or the reduction of duty ratio of myosin VIIa.
Usher syndrome (USH) is a clinically and genetically heterogeneous, autosomal recessive disorder characterized by sensorineural hearing loss and visual loss due to retinitis pigmentosa. Three clinical types of Usher syndrome have been described based on the severity of the hearing impairment, the age of onset of retinitis pigmentosa and the presence or absence of vestibular dysfunction. Usher syndrome type I (USH1), which accounts for 50 % of USH cases, is the most severe type characterized by profound congenital hearing loss, constant vestibular dysfunction and prepubertalal onset of retinitis pigmentosa. So far, at least six different loci have been mapped (USH1A-F) (1-6). USH1B is the most common subtype, accounting for 70-80 % of USH1. It was reported that the USH1B gene encodes myosin VIIa (7). In addition to human sensory disorder, myosin VIIa seems to be responsible for the sensory function in mice (8), zebrafish (9), and Drosophila (10).
Myosins are motor proteins that use energy from ATP hydrolysis to move along actin filaments. Based upon the structural analysis, several key structural elements that are critical for the mechanotransduction of myosin have been identified (11-13). The motor domain of myosin is composed of the three subdomains, i.e., N-terminal 25 kDa, central 50 kDa, and C-terminal 20 kDa. The central 50 kDa subdomain is separated by a deep cleft into the upper 50 kDa and the lower 50 kDa domains, which are involved in actin binding (11, 14). The ATP binding site consists of the three elements, i.e., “P-loop”, switch I and switch II helices. The actin binding sites are approximately 4 nm away from the nucleotide binding pocket, therefore, it is thought that the movement of switch-1/swtch-2 helices during ATP hydrolysis cycle, presumably Pi off from the active site, is critical for the communication between the catalytic pocket and the actin binding sites. The ATP binding induced conformational change in these switch I/switch II helices leads the closure of the cleft between the upper and lower 50 kDa domain and it is thought that this changes the myosin-actin binding. One of the elements connecting the upper and lower 50 kDa domain, termed “strut”, has been suggested to play an important role in controlling the cleft closure (13, 15).
Myosin VII is a member of the myosin superfamily that is classified based upon phylogenetic sequence comparisons into more than 20 classes (16-18). There are two subclasses of myosin VII identified in Drosophila and vertebrates, myosin VIIa and VIIb, which are encoded by independent genes. While mammalian myosin VIIa is found in a variety of tissues including lung, kidney, small intestine and testis (19, 20), its physiological function is best studied in the sensory hair cells of the inner ear and the retina. Myosin VIIa is present along the length of the stereocilia in the sensory hair cells, where it serves a role in regulating the tension of the stereocilia bundles (21, 22). Myosin VIIa is also found in the pericuticular necklace of the hair cells and might be involved in vesicle transport (21, 23). In the retina, myosin VIIa facilitates the transportation of opsin from the inner segment to outer segment of photoreceptor (24, 25). In the pigmented epithelial cells of the retina, myosin VIIa serves an important role of phagocytosis of shed outer segments (8, 19). These findings suggest that myosin VIIa plays a role in intracellular trafficking and production of force in localized cellular compartments.
Myosin VIIa consists of a conserved N-terminal motor domain, a neck domain with five IQ motifs that function as light chain binding sites, and a long tail domain (26). The tail domain consists of a putative coiled-coil domain, an SH3 (Src homology 3) domain, two MyTH4 (myosin tail homology 4) domains, and two FERM (band 4.1-ezrin-radixin-moesin) domains (Fig.1A). Although the function of the tail domain is obscure, it is thought that this domain serves as the binding site of the myosin VIIa partner molecules, thus important for cargo transportation. While the motor domain is conserved among the myosin superfamily members, each member has unique motor characteristics. One of the most important motor characteristics is “processivity” that describes the continuous movement of the motor molecules on actin filaments, thus, suitable as a cargo transporter (27). The motor activity of myosin VIIa was first determined in rat myosin VIIa (28). It was shown that the ATPase activity myosin VIIa is significantly activated by actin by more than 10 fold and myosin VIIa moves towards the plus-end of the actin filament, thus, a plus-end-directed motor. Recently, it was shown, with Drosophila myosin VIIa, that myosin VIIa is a high duty ratio motor in which the motor head strongly binds to actin for the majority of the cross-bridge cycling time (29) and consistently moves processively when myosin VIIa is tagged with a leucine zipper module to produce the forced two-headed molecules (30).
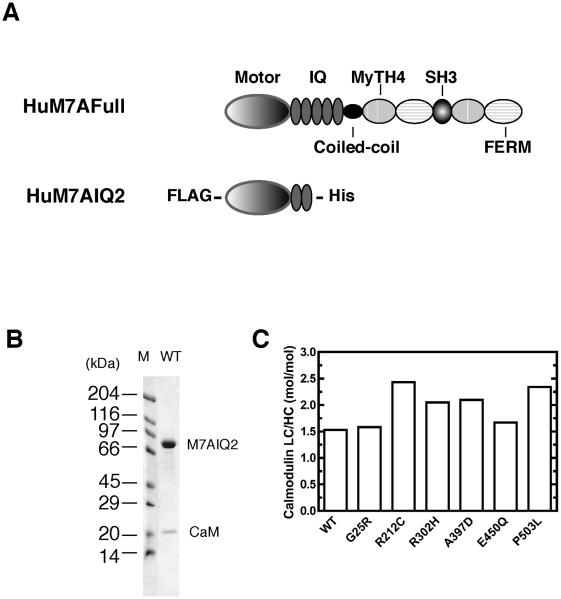
Figure 1. Human myosin VIIa construct.
A, Schematic diagram of the full-length human myosin VIIa (HuM7AFull) and the expressed truncated human myosin VIIa construct (HuM7AIQ2). B. SDS-PAGE of the purified wild-type HuM7AIQ2. The HuM7AIQ2 heavy chain was co-expressed with calmodulin (CaM). Lane M; molecular mass markers. C. The molar ratio of co-purified calmodulin light chain determined by densitometry analysis.
To date, a number of missense mutations of the myosin VIIa gene have been found in various USH1B families (7), which are predominantly located in the motor domain of myosin VIIa, suggesting that the motor function of myosin VIIa is critical for its roles in the inner ear and retina. However, it has never been demonstrated how these mutations affect the motor function of myosin VIIa, which is critical for understanding the correlation between the mutation and phenotype. Here we analyzed the effect of the myosin VIIa missense mutations (G25R, R212C, R302H, A397D, E450Q, P503L) found in USH1B patients, for the first time, on the mechanoenzymatic activities of human myosin VIIa. Gly25 is located in N-terminus region of the motor domain that is not conserved in myosin superfamily. Arg212 is in highly conserved switch I region in the ATP binding pocket. Arg302 is in the upper 50 kDa subdomian and not conserved among myosin family. Ala397 lies on the surface of the upper 50 kDa domain in the cleft of the motor domain. Glu450 is located in switch II region and highly conserved among all myosins. Pro503 is well conserved among the myosin superfamily and present on the outer surface of the motor domain that is thought to be one of the actin-binding sites in myosin II. All mutants examined, except for R302H, exhibited severe deficiency on the actin-activated ATPase activities. The results also indicated that the mutations significantly decrease the duty ratio of myosin VIIa.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes and modifying enzymes were purchased from New England Biolabs (Beverly, MA). Phosphoenolpyruvate and pyruvate kinase were obtained from Sigma (St. Louis, MO). Actin was prepared from rabbit skeletal muscle according to Spudich and Watt (31). Recombinant calmodulin was expressed in E. coli and purified as described previously (32).
Cloning, Expression and Purification of human myosin VIIA protein
Human myosin VIIA cDNA was obtained from Human kidney total RNA using RT-PCR. The nucleotide sequence was determined by direct DNA sequencing to confirm the authenticity of the DNA sequence of the clone. The cDNA fragment encoding Met1-Arg796 was subcloned into modified pFastBac1 baculovirus transfer vector containing a FLAG tag sequence and octa-histidine tag at 5′-end and 3′-end of the polylinker region, respectively. This construct (HuM7AIQ2) encodes the entire motor domain plus two IQ motifs of myosin VIIa. We used the Flag tag for purification, and the histidine tag for in vitro motility assays to attach the C-terminus of myosin to nitrocellulose surface coated with anti-hexa-histidine antibody (Rockland, Gilbertsville). Six point mutations (G25R, R212C, R302H, A397D, E450Q, P503L) were introduced into myosin VIIa cDNA individually using site-directed mutagenesis. To express the recombinant HuM7AIQ2 protein, Sf9 cells (about 1 × 109 cells) were co-infected with two viruses expressing the HuM7AIQ2 heavy chain and calmodulin. The infected cells were cultured for 3 days at 28 °C. Cells were harvested and washed with PBS and 5 mM EGTA. Cells were then lysed with sonication in 20 ml of lysis buffer (30 mM MOPS-KOH (pH 7.5), 150 mM KCl, 1 mM EGTA, 2 mM MgCl2, 1 mM MgATP, 0.5 mM dithiothreitol, 1 mM phenylmethanesulfonyl fluoride, 10 μg/ml leupeptin and 10 μg/ml trypsin inhibitor). After centrifugation at 100,000 × g for 30 min, the supernatant was incubated with 300 μl of anti-FLAG M2 affinity resin (Sigma) in a 50 ml conical tube on a rotating wheel for 1 hour at 4°C. The resin suspension was then loaded on a column (1× 10 cm) and washed with 30 ml buffer A (20 mM MOPS-KOH (pH 7.5), 100 mM KCl, 2 mM MgCl2, 1 mM EGTA, 5 μg/ml leupeptin and 0.5 mM dithiothreitol). HuM7AIQ2 was eluted with buffer A containing 0.1 mg/ml FLAG peptide. The HuM7AIQ2 was concentrated with a VIVASPIN concentrator (Vivascience) and dialyzed against buffer B (50 mM KCl, 20mM MOPS-KOH (pH 7.5), 2mM MgCl2, 1 mM EGTA and 1 mM dithiothreitol). The purified HuM7AIQ2 was stored on ice and used within 2 days. 50-100 μg of the purified protein was obtained from a standard infection. Protein concentration was determined by the densitometry of SDS-PAGE using smooth muscle myosin heavy chain as a standard, and analyzed with NIH ImageJ software. All experiments were done with at least 3 independent preparations.
Steady-state ATPase Assay
The steady-state ATPase activity was measured in the presence of ATP regeneration system (20 units/ml pyruvate kinase and 2 mM phosphoenolpyruvate) at 25 °C. The actin-activated ATPase assay was carried out in buffer B with 0.2 mg/ml calmodulin. Ca2+ATPase activity was measured in the buffer containing 600 mM KCl, 30 mM Tris-HCl (pH 8.5), 10 mM MgCl2, 0.2 mg/ml calmodulin and 2 mM ATP. The liberated pyruvate was determined as described (33).
Stopped-flow Measurements
Kinetic measurements were performed in buffer B with 0.2 mg/ml calmodulin at 25 °C using a KinTek SF-2001 apparatus (KinTek Co., Clarence, PA) as described previously (29). The rate of ADP release from acto-myosin was measured by monitoring the light-scattering change at 420 nm after mixing 0.5 μM acto-HuM7AIQ2 and 0.1 mM MgADP with 2 mM MgATP.
Actin cosedimentation assay
Prior to the assay, HuM7AIQ2 was centrifuged at 300,000 X g for 10 min to remove any potential aggregates, and the supernatant was used in the actin cosedimentation assay. One mg/ml actin was mixed with 0.5 μM HuM7AIQ2 in buffer B in the presence or absence of ATP regeneration system and 2 mM MgATP, and incubated for 10 min at room temperature. Then, the reaction mixtures were centrifuged at 300,000 X g for 10 min. The supernatants and dissolved pellets were subjected to the densitometry analysis of SDS-PAGE.
In Vitro Motility Assay
The actin-translocating velocity was measured by in vitro actin-translocating assay as described (34). A glass surface was coated with nitrocellulose. Anti-hexa-histidine monoclonal antibody was applied to the flow cell, followed by BSA blocking. Then, HuM7AIQ2 was applied to the flow cells and incubated for 30 min at room temperature. The unbound HuM7AIQ2 was washed out with buffer B. The movement of the rhodamin-labeled actin filaments was observed in buffer C (25 mM KCl, 25 mM MOPS (pH 7.5), 2 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 36 μg/ml catalase, 4.5 mg/ml glucose, 216 μg/ml glucose oxidase, 0.1 mg/ml calmodulin and 4 mM MgATP) at 25 °C. Actin filament velocity was calculated from the movement distance and the elapsed time in successive snapshots. Student's t test was used for statistical comparison of mean values. A value of p < 0.01 was considered to be significant.
RESULTS
Expression and purification of human myosin VIIa
In order to study the effect of missense mutations found in Usher Syndrome Type IB on the motor activity of myosin VIIa, it is critical to use human myosin VIIa. We succeeded in isolating functionally active human myosin VIIa for the first time. To avoid the complexity as a result from the potential interaction between the two head domains, or a tail domain and a head domain like myosin V and VI (35-38), we decided to use the single-headed construct (HuM7AIQ2) for the functional analysis of myosin VIIa mutants (Fig. 1A). The wild-type and all the USB1B mutants were co-expressed with calmodulin as a light chain. The purified proteins were co-purified with calmodulin, indicating that calmodulin serves as subunits as previously reported (28, 39, 40) (Fig.1B). The low molecular mass calmodulin band showed its characteristic Ca2+ dependent shift in mobility on an SDS-PAGE (date not shown). The number of calmodulin associated with the wild-type heavy chain was estimated to be 1.53 ± 0.06 by densitometry of an SDS-PAGE. The number of co-purified calmodulin light chain for each mutant myosin VIIa heavy chain is shown in Fig. 1C. The result was consistent with the expected number of calmodulin light chain of the construct having two IQ motifs. All experiments were performed in the presence of exogenous calmodulin (0.2 mg/ml) to ensure that the light chain binding IQ motifs were occupied with calmodulin.
Steady-state ATPase activity
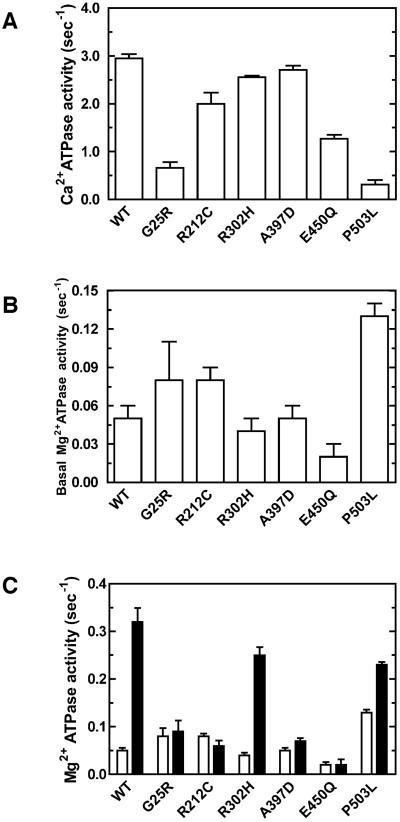
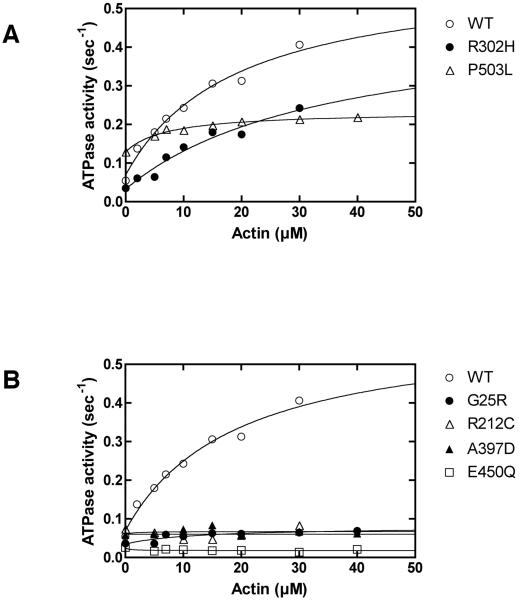
To evaluate the enzymatic function of the wild-type and mutant myosin VIIa, Ca2+-ATPase and Mg2+-ATPase activity of HuM7AIQ2 were measured (Fig. 2A, B). While the missense mutations changed the Ca2+ and Mg2+-ATPase activity of HuM7AIQ2, none of the mutations abolished the ATPase activities, suggesting that the mutations tested do not eliminate the ATP binding to the active site. Fig. 2C shows the actin activation of the Mg2+-ATPase activities of HuM7AIQ2 in the presence of 30 μM actin (see also Table I). In the absence of actin, basal Mg2+-ATPase activity of the wild-type was 0.05 ± 0.01 s−1. The ATPase activity of the wild-type was significantly activated by actin. The Mg2+-ATPase activity of G25R, R212C, A397D and E450Q mutants were not activated by actin, although their basal Mg2+-ATPase activities were not largely different from that of the wild-type. The basal Mg2+-ATPase activity of the P503L mutant was two-fold higher than that of wild type, which was further activated about twice by actin. On the other hand, the ATPase activity of R302H mutant was well activated by actin without the change in the basal ATPase activity. To further analyze the effect of mutation on the motor function, the ATPase activity of HuM7AIQ2 was measured as a function of actin concentration (Fig. 3A and 3B). The ATPase activities of the wild-type HuM7AIQ2 at various actin concentrations were fitted well with a Michaelis – Menten equation to yield Vmax and KATPase of 0.45 ± 0.05 s−1 and 12.8 ± 2.9 μM, respectively. The ATPase activity of R302H mutant was well activated by actin to yield Vmax and KATPase of 0.34± 0.05 s−1 and 15.2 ± 4.8 μM (Fig. 3A and Table I). The obtained Vmax value was almost identical to that of the wild-type. The results suggest that the USH1 phenotype of R302H mutation may not be due to the alteration of the motor function. On the other hand, the high basal activity of P503L mutant was further increased with actin to yield Vmax of 0.23 ± 0.02 s−1 (Fig. 3A and Table I). Interestingly, KATPase value was significantly reduced by this mutation. For G25R, R212C, A397D and E450Q mutations, there was no indication of the increase in the ATPase activity at wide rage of actin concentrations (Fig. 3B and Table I).
Figure 2. Ca2+ATPase and the actin-activated Mg2+ATPase activity of USH1B mutants.
A. Ca2+ATPase activity. Ca2+ATPase activity was measured in the buffer containing 600 mM KCl, 30 mM Tris-HCl (pH 8.5), 10 mM MgCl2, 2 mM phosphoenol pyruvate, 20 unit/ml pyruvate kinase, 0.2 mg/ml calmodulin, 20 nM HuM7AIQ2 and 2 mM ATP at 25 °C. B. Basal Ma2+-ATPase activity of USH1B mutants. The ATPase activity was measured in the buffer containing 50 mM KCl, 20 mM MOPS-KOH (pH 7.5), 2 mM MgCl2, 1 mM EGTA, 2 mM phosphoenol pyruvate, 20 unit/ml pyruvate kinase, 0.2 mg/ml calmodulin, 20 nM HuM7AIQ2 and 2 mM MgATP at 25 °C. C. The actin-activated Mg2+ATPase activity in the presence of 30 μM actin. Open bars, in the absence of actin; closed bars, in the presence of 30 μM actin. The error bars indicate S.D. for n=3 from three independent preparations.
Table I.
Steady-state and the ADP release rate constants of human myosin VIIa.
| Ca2+ ATPase (s−1) |
Actin-activated Mg2+ATPase |
ADP release rate from actomyosin (s−1) |
||||
|---|---|---|---|---|---|---|
| v0 (s−1) | 30μM actin (s−1) | Vmax (s−1) | KATPase (μM) | |||
| Wild-type | 2.95 ± 0.09 | 0.05 ± 0.01 | 0.37 ± 0.04 | 0.45 ± 0.05 | 12.8 ± 2.9 | 0.56 ± 0.05 |
| G25R | 0.66 ± 0.12* | 0.08 ± 0.04 | 0.09 ± 0.04* | N.D. | N.D. | 0.23 ± 0.08* |
| R212C | 2.00 ± 0.23* | 0.07 ± 0.01 | 0.07 ± 0.02* | N.D. | N.D. | 0.58 ± 0.12 |
| R302H | 2.56 ± 0.03* | 0.05 ± 0.02 | 0.25 ± 0.02 | 0.34 ± 0.05 | 15.2 ± 4.8 | 0.67 ± 0.14 |
| A397D | 2.71 ± 0.09 | 0.05 ± 0.01 | 0.07 ± 0.01* | N.D. | N.D. | 0.43 ± 0.12* |
| E450Q | 1.27 ± 0.08* | 0.04 ± 0.01 | 0.02 ± 0.01* | N.D. | N.D. | 0.72 ± 0.20 |
| P503L | 0.31 ± 0.10* | 0.13 ± 0.01* | 0.23 ± 0.01* | 0.23 ± 0.02* | 3.3 ± 1.6* | 0.79 ± 0.04* |
All date are presented as mean value ± S.D. (n=3 from three independent preparations of each mutant)
P< 0.01 versus wild-type. N.D., Not Detected.
Figure 3. The actin-dependence of the actin-activated Mg2+-ATPase activity of USH1B mutants.
A. The actin-activated Mg2+-ATPase activity of USH1B mutants showing the actin dependent activation. B. The actin-activated Mg2+-ATPase activity of USH1B mutants showing little actin activation. Assay conditions are described in MATERIALS AND METHODS.
Rate of ADP release from acto-myosin
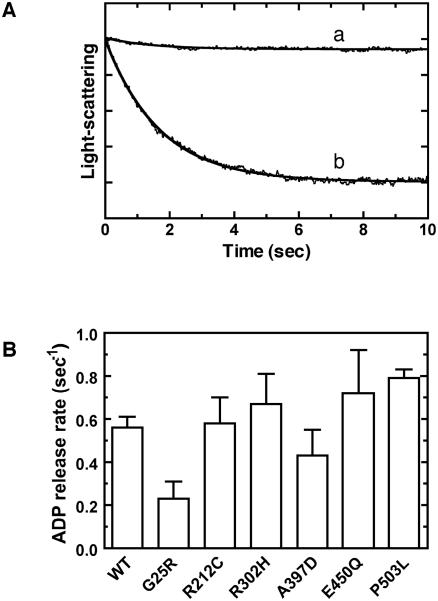
The previous kinetic study of Drosophila myosin VIIa showed that the step of ADP release is the rate-limiting step in acto-myosin VIIa ATPase cycle, indicating that Drosophila myosin VIIa is classified to be a high duty ratio motor which spends the majority of the ATPase cycle in the strongly actin-binding state (29). Since it is thought that the high duty ratio is required for the processive movement of myosin, myosin VIIa can be a processive motor if it forms a dimer (30). To assess the effect of USH1B mutations on the high duty ratio of myosin VIIa, we measured the rate of ADP release from acto-HuM7AIQ2 (Fig. 4 and Table I).
Figure 4. ADP dissociation from acto-myosin VIIa USH1B mutants.
A. Time course of light-scattering change after mixing acto-HuM7AIQ2/ADP with Mg2+-ATP. Measurements were performed in the buffer containing 50 mM KCl, 20 mM MOPS-KOH (pH 7.5), 2 mM MgCl2, 1 mM EGTA, 0.2 mg/ml calmodulin at 25 °C. Acto-HuM7AIQ2 (0.5 μM HuM7AIQ2 + 0.5 μM actin) in the presence of 0.1 mM Mg2+-ADP was mixed with 2 mM Mg2+-ATP. The intensity of light-scattering was monitored at 420 nm. The solid line (b) is the best fit to single exponential kinetics with kobs of 0.56 s−1. The line (a) represents the control in the absence of ATP. B. ADP dissociation rates of USH1B mutants. The ADP dissociation rates were measured as described in A for each USH1B mutant. The error bars represent the S.D. from 3 independent experiments.
The rate of ADP release from acto-myosin was measured by monitoring the light-scattering change after mixing acto-HuM7AIQ2/ADP with 2 mM MgATP. In the absence of ADP, acto-HuM7AIQ2 dissociated rapidly upon addition of ATP, but the dissociation rate was markedly decreased in the presence of ADP. This is because the dissociation of ADP from acto-HuM7AIQ2 is slower than the ATP induced dissociation of acto-HuM7AIQ2 and limits the entire process. Conversely, the obtained dissociation rate of acto-HuM7AIQ2 determined by measuring the change in light-scattering represents the ADP release rate from acto-HuM7AIQ2. As shown in Fig. 4A, the decrease in the light-scattering upon dissociation of acto-HuM7AIQ2/ADP followed single exponential kinetics. The rate of ADP release from acto-HuM7AIQ2/ADP determined by this method was 0.56 ± 0.05 s−1 for the wild-type (Fig. 4A). This value was comparable with the Vmax of the actin-activated Mg2+-ATPase activity, suggesting that the rate of ADP release is the rate-limiting step in acto-human myosin VIIa ATPase cycle. This result suggests that human myosin VIIa is a high duty ratio motor like Drosophila myosin VIIa. The USH1B mutantions examined did not largely affect the rate of ADP release from acto-HuM7AIQ2 (Fig. 4B), except for G25R mutant, the rate constant of which was a half of the wild-type. Since the USH1B mutations, except for R302H, significantly reduced the Mg2+-ATPase rate in the presence of actin, the result indicates that the duty ratio of the mutants is largely reduced from that of the wild-type. Therefore, the results suggest the USH1B mutants, except for R302H, hamper not only the over-all ATP hydrolysis cycle but also the duty ratio of the cycle.
Actin cosedimentation assay
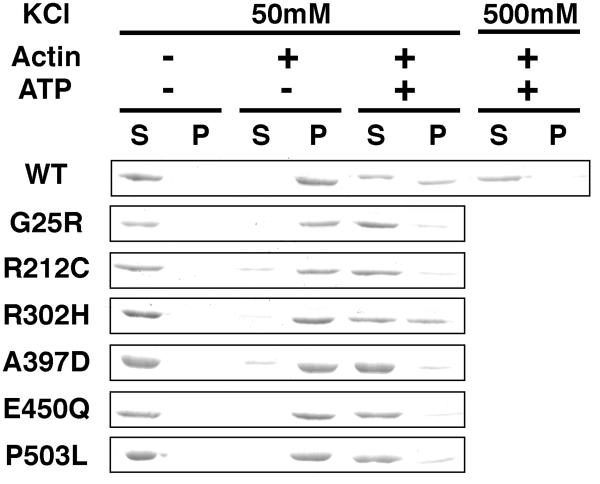
To assess the actin binding affinity of the USH1B mutants, actin cosedimentation assays were performed (Fig. 5). Unlike the non-processive conventional myosin II, the processive high duty ratio myosins such as myosin Va have a high affinity for actin, even in the presence of ATP (41, 42). Consistent with the high duty ratio nature of human myosin VIIa, the HuM7AIQ2 wild-type showed approximately 50% of the protein cosedimented with actin in the presence of ATP in low salt condition. On the other hand, all HuM7AIQ2 mutants cosedimented with actin in the absence of ATP, but not in the presence of ATP, suggesting that all mutations did not hamper the ATP-dependent actin binding property of myosin VIIa. However, all mutants, except for R302H, showed that 85-90% of proteins were dissociated from actin in the presence of ATP, indicating that the USH1B mutations reduce the affinity for actin in the presence of ATP. This result supports the notion that the USH1B mutations reduce the duty ratio of myosin VIIa.
Figure 5. Actin cosedimentation assay.
Actin cosedimentation assays were performed in the buffer containing 50 mM KCl, 20 mM MOPS-KOH (pH 7.5), 2 mM MgCl2, 1 mM EGTA, 0.2 mg/ml calmodulin. HuM7AIQ2 (0.5 μM) were mixed with 1 mg/ml F-actin in the presence or absence of ATP regeneration system (2 mM phosphoenol pyruvate, 20 unit/ml pyruvate kinase) and 2 mM MgATP, and incubated for 10 min at room temperature. Then, the reaction mixtures were centrifuged at 300,000 X g for 10 min. The supernatants and dissolved pellets were subjected to the densitometry analysis of SDS-PAGE. S, supernatant; P, pellet.
Actin-translocating activity
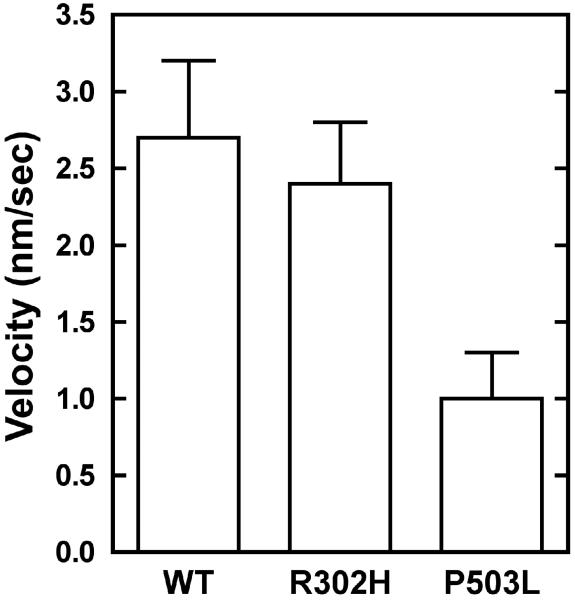
To directly determine the mechanical activity of USH1B mutants, the actin-translocating activity was measured by observing the movement of the rhodamine-labeled actin filaments under the fluorescence microscope. The actin-translocating velocities of the wild-type and USH1B mutant myosin VIIa measured in an in vitro motility assay are shown in Fig. 6. The velocity of the wild-type was 2.7 ± 0.5 nm/sec. This value is much slower than any other myosin so far reported. P503L mutant, which showed high basal ATPase activity, was able to translocate actin filaments, however, the velocity was significantly slower than the wild-type. On the other hand, the velocity of R302H mutant was similar to the wild-type, consistent with the actin-activated ATPase activity that showed little difference from the wild-type.
Figure 6. Actin-translocating velocity of USH1B mutants in in vitro motility assay.
The movement of the fluorescent-labeled actin filament was observed in 25 mM KCl, 20 mM MOPS-KOH (pH 7.5), 2 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 36 μg/ml catalase, 4.5 mg/ml glucose, 216 μg/ml glucose oxidase, 0.2 mg/ml calmodulin, and 2 mM MgATP at 25 °C. The error bars indicate S.D. for n=30-40 from three independent preparations.
DISCUSSION
While it is known that myosin VIIa is the responsible gene for USH1B, the effect of USH1B mutations on the function of myosin VIIa at the molecular level is unknown. In the present study, we succeeded in isolating human myosin VIIa and clarified, for the first time, how the USH1B mutation hampers the function of myosin VIIa at the molecular level. While the USH1B mutations are distributed in the entire region of the myosin VIIa gene, the majority of the missense mutations are located in the motor domain suggesting that the USH1B phenotype is due to the malfunction of myosin VIIa motor activity. Therefore, we focused our efforts on the missense mutations in the motor domain of myosin VIIa, and examined the effects of these mutations on the motor activity.
One of possible effects of mutation is that the mutation may affect the proper folding and stability of the protein. However, the yields of all USH1B mutant proteins examined in this study were virtually the same as that of the wild-type, suggesting that all mutations examined do not affect the proper folding of myosin VIIa.
In all the functional assays performed in this study, R302H mutation showed little effect on the motor activity of myosin VIIa. It was originally described (43) that the R302H mutation undergoes difficulty in trying to correlate phenotype/genotype associations because some of the myosin VIIa cDNA clones, isolated from different cDNA libraries, had a histidine at position 302 (26). Furthermore, Arg302 is not conserved among members of the myosin superfamily, suggesting that Arg302 is not critical for the authentic function of myosin VIIa. Therefore, we concluded that the R302H mutation is not directly responsible for USH1B phenotype.
All other missense mutations tested in this study showed severe dysfunction of the myosin VIIa motor function. The Mg2+-ATPase activities of G25R, R212C, A397D and E450Q mutants were not activated by actin at all, although they had the basal Mg2+-ATPase activity that is similar to the wild-type. These results suggest that these mutations do not hamper the ATP binding and the following ATP hydrolysis, but do impair the transduction pathway between the actin-binding interface and the ATPase active site.
Since these mutants bind to actin in the absence of ATP and dissociate from actin in the presence of ATP, it is anticipated that these mutants can produce the weak actin binding intermediate of myosin VIIa upon ATP binding. We anticipate that the product release of these mutants predominantly takes place without the actin re-binding pathway, thus, no apparent actin activation of the ATPase activity. It is unlikely that these mutants carry the cargo molecules because the majority of the molecules are in the actin-dissociated form in the presence of ATP. On the other hand, P503L mutation showed a two-fold larger basal Mg2+-ATPase activity than wild-type, which was further activated twice by actin. A significant increase in the basal ATPase activity may cause an inefficient energy usage in cells. The ADP release rate of this mutant was higher than the wild-type, suggesting that the duty ratio of this mutant is less than 30%. Therefore, it is less likely that this mutant myosin VIIa can function as a cargo transporter.
Taken together, the present results suggest that USH1B mutations cause either the complete loss of the motor activity of myosin VIIa or the severe reduction of the duty ratio of myosin VIIa. It was shown that Drosophila myosin VIIa is a high duty ratio motor (29) and can move procesively when it forms dimer (30). To date, it is unclear whether human myosin VIIa can form dimer and move processively. On the other hand, it has been suggested that mammalian myosin VIIa serves as a transporter in the pericuticular necklace of the inner ear hair cells (21, 23) and photoreceptor cells (24, 25), therefore, it is plausible that human myosin VIIa can move processively, thus functioning as a cargo transporter. This also implies that the processive nature of myosin VIIa is critical for the physiological function of myosin VIIa in the inner ear and retina. The present results are consistent with earlier cell biological findings, and further supports the idea that myosin VIIa functions as a cargo transporter in the cells.
The present results also provide important information for understanding the relationship between the structure and function of myosin VIIa molecule. Of interest is the mutation at Gly25 that is located near the amino terminus of the molecule. Some of the other myosin classes do not contain this region, and the others showed no sequence homology at this region. Nevertheless, G25R mutation completely abolished the actin-translocating activity and the actin activated-ATPase activity. It is plausible that the unique N-terminal domain of myosin VIIa is responsible for the myosin VIIa specific properties of the motor activity, such as a fast actin-attached ATP hydrolysis rate (29). The importance of the N-terminal region for the myosin motor activity was recently reported for Dictyostelium myosin II, in which the deletion of the N-terminal region of approximately 80 residues reduced the ATPase and motile activities and the affinities for ADP and actin (13). Since the primary structure of myosin VIIa at this region is not conserved among myosin family members, it is difficult to predict the structural changes of myosin VIIa at this region by G25R mutation. Further structural studies are required to clarify the mechanism of G25R induced inhibition of myosin VIIa motor activity.
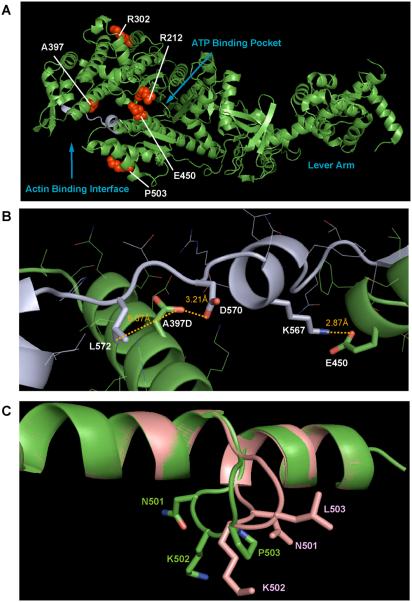
Ala397 is highly conserved among all myosins and lies on the surface of the upper 50 kDa domain in the cleft of the myosin motor domain, which extends from the ATP-binding site to the actin-binding site (Fig. 7A). Since structural studies showed that the cleft half closes upon ATP binding, it is thought that the opening and closing of the cleft provide the physical link between the ATP- and actin-binding sites. Consistent with this notion, A397D did not change the basal Ma2+-ATPase activity, but abolished the actin-activated Mg2+-ATPase and motile activities, indicating that Ala397 is essential for the actin dependent process of the ATPase reaction. Based upon the 3D crystal structure of myosin Va, A397 is in close proximity to D570 and L572. The distance between the Oxygen atom of the substituted Asp side chain and the Oxygen atom of the Asp570 side chain is within 3 Å, and it is possible that the substitution of Ala397 to Asp may cause repulsive interaction. The substitution of Ala to Asp may result in steric hindrance with the bulky Leu side chain. Supporting this view, homology modeling suggested that the substitution of Ala397 to Asp pushes the side chain of Asp397 away from Asp570 to increase the distance between the side chain of Asp397 to Asp570 and Leu572 (Fig. 7B).
Figure 7. 3D model for the disruption of the motor activity of human myosin VIIa by the USH1B mutations.
A. Crystal structure of the motor domain and first IQ motif. Since the crystal structure of myosin VIIa is not known, the crystal structure of chicken myosin Va motor domain and first IQ motif (49) (1w7j.pdb) is used as a model. Arg212, Arg302, Ala397, and Glu450 are conserved between chicken myosin Va and human myosin VIIa. Pro503 in myosin VIIa is equivalent to Met503 in myosin Va. The side chains of these residues are shown in red (space-filling model). The strut loop region is shown in light blue. B. 3D model structure around the strut loop region. The effect of A397D and P503L mutations on the molecular level were analyzed with the modeling program Swiss-Model (http://swissmodel.expasy.org/) and Swiss-PDB-viewer software. Note that Asp is substituted for Ala at position 397. The strut loop region is shown in light blue. C. 3D model structure around P503 residue. The loop containing P503 of the wild type and P503L mutant are shown in green and pink, respectively.
Pro503 is well conserved among the myosin superfamily which lies on the outer surface of the lower domain of myosin (Fig. 7A), and this region is thought to be one of the actin-binding sites (14). Interestingly, the P503L mutation increased the basal Mg2+-ATPase activity and decreased the actin activation and the actin gliding velocity, suggesting that the P503L mutation hampers the link between the actin binding site and the ATPase active site. Based upon homology modeling, it was predicted that P503L mutation alters the position of the loop A500-I506 (Fig. 7C). It is plausible that this conformational change partially mimic the actin bound conformation of myosin.
Arg212 and Glu450 are present in switch I and switch II/relay helix regions, respectively (Fig. 7A), which are conserved in all myosins and essential for the myosin motor function. Therefore, it is expected that these mutations would severely impair the motor function. Based upon the structural studies of myosin II, Arg212 is thought to produce a salt bridge with Glu442 in the switch II loop (44). This salt-bridge is important for the hydrolysis step, because the mutation of Arg212 diminish Pi-burst (45-47). The crystal structure of Dictyostelium myosin II reveals that the replacement of the basic residue at this position prevents the motor domain of myosin to form “closed” conformation that represents a conformational transition of myosin motor domain (48). The present study of myosin VIIa R212C mutant is consistent with these earlier studies of myosin II, and suggests that the replacement of the basic residue at the position 212 with cysteine is expected to abolish the salt bridge formation, thus hampering the hydrolysis of ATP and destabilizing the formation of “closed” conformation of the motor domain of myosin VIIa.
On the other hand, Glu450 is in close proximity to Lys567, which is in the “strut loop” lying in the position in the cleft connecting the upper and lower 50 kDa domain of the myosin motor domain. Val572 and Asp570 described above are on the same loop (Fig. 7B). The distance between the Oxygen atom of the Glu450 and the nitrogen atom of the Lys567 is within 3 Å, which may produce salt bridge. It is plausible that E450Q mutation disrupts the salt-bridge, thus changing the position of the “strut loop” and hampering the ATP hydrolysis cycle of myosin VIIa.
The strut loop is one of the three loops connecting the upper and lower 50K subdomains. The importance of this loop for the myosin motor activity was reported for Dictyostelium myosin II, in which the insertion or deletion of the residues in this loop abolished strong binding to actin, although the basal ATPase activities were normal (15). The present results are consistent with the previous results of Dictyostelium myosin II, and support a notion that A397 or E450 mutation may induce local structural changes at strut loop, thus causing a loss of motor function.
In summary, this is the first report that has clarified the effects of USH1B mutations on the function of the responsible gene product, human myosin VIIa. The USH1B mutations severely hamper the motor function of myosin VIIa. The results indicate that the impairment of the motor function of myosin VIIa is responsible for USH phenotype in humans.
The abbreviations used are
- USH1B
Usher syndrome type IB
- CaM
calmodulin
- PBS
Phosphate buffer saline
- MOPS
4-morpholinepropanesulfonic acid
Footnotes
This work was supported by National Institute of Health Grants DC006103, AR 048526, AR 048898 and AR 41653 (to M.I.).
REFERENCES
- 1.Kaplan J, Gerber S, Bonneau D, Rozet JM, Delrieu O, Briard ML, Dollfus H, Ghazi I, Dufier JL, Frezal J, et al. A gene for Usher syndrome type I (USH1A) maps to chromosome 14q. Genomics. 1992;14:979–987. doi: 10.1016/s0888-7543(05)80120-x. [DOI] [PubMed] [Google Scholar]
- 2.Smith RJ, Lee EC, Kimberling WJ, Daiger SP, Pelias MZ, Keats BJ, Jay M, Bird A, Reardon W, Guest M, et al. Localization of two genes for Usher syndrome type I to chromosome 11. Genomics. 1992;14:995–1002. doi: 10.1016/s0888-7543(05)80122-3. [DOI] [PubMed] [Google Scholar]
- 3.Wayne S, Der Kaloustian VM, Schloss M, Polomeno R, Scott DA, Hejtmancik JF, Sheffield VC, Smith RJ. Localization of the Usher syndrome type ID gene (Ush1D) to chromosome 10. Hum Mol Genet. 1996;5:1689–1692. doi: 10.1093/hmg/5.10.1689. [DOI] [PubMed] [Google Scholar]
- 4.Kimberling WJ, Moller CG, Davenport S, Priluck IA, Beighton PH, Greenberg J, Reardon W, Weston MD, Kenyon JB, Grunkemeyer JA, et al. Linkage of Usher syndrome type I gene (USH1B) to the long arm of chromosome 11. Genomics. 1992;14:988–994. doi: 10.1016/s0888-7543(05)80121-1. [DOI] [PubMed] [Google Scholar]
- 5.Chaib H, Kaplan J, Gerber S, Vincent C, Ayadi H, Slim R, Munnich A, Weissenbach J, Petit C. A newly identified locus for Usher syndrome type I, USH1E, maps to chromosome 21q21. Hum Mol Genet. 1997;6:27–31. doi: 10.1093/hmg/6.1.27. [DOI] [PubMed] [Google Scholar]
- 6.Wayne S, Lowry RB, McLeod DR, Knaus R, Farr C, Smith RJH. Localization of the Usher syndrome type If gene (Ush1F) to chromosome 10. Am J Hum Genet. 1997;61:A300. [Google Scholar]
- 7.Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Ondek B, Williams DS. Mutant myosin VIIa causes defective melanosome distribution in the RPE of shaker-1 mice. Nat Genet. 1998;19:117–118. doi: 10.1038/470. [DOI] [PubMed] [Google Scholar]
- 9.Ernest S, Rauch GJ, Haffter P, Geisler R, Petit C, Nicolson T. Mariner is defective in myosin VIIA: a zebrafish model for human hereditary deafness. Hum Mol Genet. 2000;9:2189–2196. doi: 10.1093/hmg/9.14.2189. [DOI] [PubMed] [Google Scholar]
- 10.Todi SV, Franke JD, Kiehart DP, Eberl DF. Myosin VIIA defects, which underlie the Usher 1B syndrome in humans, lead to deafness in Drosophila. Curr Biol. 2005;15:862–868. doi: 10.1016/j.cub.2005.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rayment I. The structural basis of the myosin ATPase activity. J Biol Chem. 1996;271:15850–15853. doi: 10.1074/jbc.271.27.15850. [DOI] [PubMed] [Google Scholar]
- 12.Spudich JA. The myosin swinging cross-bridge model. Nat Rev Mol Cell Biol. 2001;2:387–392. doi: 10.1038/35073086. [DOI] [PubMed] [Google Scholar]
- 13.Fujita-Becker S, Tsiavaliaris G, Ohkura R, Shimada T, Manstein DJ, Sutoh K. Functional characterization of the amino-terminal region of myosin-2. J Biol Chem. 2006 doi: 10.1074/jbc.M605171200. [DOI] [PubMed] [Google Scholar]
- 14.Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki N, Ohkura R, Sutoh K. Insertion or deletion of a single residue in the strut sequence of Dictyostelium myosin II abolishes strong binding to actin. J Biol Chem. 2000;275:38705–38709. doi: 10.1074/jbc.M001966200. [DOI] [PubMed] [Google Scholar]
- 16.Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 17.Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci U S A. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasson T, Heintzelman MB, Santos-Sacchi J, Corey DP, Mooseker MS. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci U S A. 1995;92:9815–9819. doi: 10.1073/pnas.92.21.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahly I, El-Amraoui A, Abitbol M, Petit C, Dufier JL. Expression of myosin VIIA during mouse embryogenesis. Anat Embryol (Berl) 1997;196:159–170. doi: 10.1007/s004290050088. [DOI] [PubMed] [Google Scholar]
- 21.Hasson T, Gillespie PG, Garcia JA, MacDonald RB, Zhao Y, Yee AG, Mooseker MS, Corey DP. Unconventional myosins in inner-ear sensory epithelia. J Cell Biol. 1997;137:1287–1307. doi: 10.1083/jcb.137.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Self T, Mahony M, Fleming J, Walsh J, Brown SD, Steel KP. Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development. 1998;125:557–566. doi: 10.1242/dev.125.4.557. [DOI] [PubMed] [Google Scholar]
- 23.Richardson GP, Forge A, Kros CJ, Fleming J, Brown SD, Steel KP. Myosin VIIA is required for aminoglycoside accumulation in cochlear hair cells. J Neurosci. 1997;17:9506–9519. doi: 10.1523/JNEUROSCI.17-24-09506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Vansant G, Udovichenko IP, Wolfrum U, Williams DS. Myosin VIIa, the product of the Usher 1B syndrome gene, is concentrated in the connecting cilia of photoreceptor cells. Cell Motil Cytoskeleton. 1997;37:240–252. doi: 10.1002/(SICI)1097-0169(1997)37:3<240::AID-CM6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Udovichenko IP, Brown SD, Steel KP, Williams DS. Myosin VIIa participates in opsin transport through the photoreceptor cilium. J Neurosci. 1999;19:6267–6274. doi: 10.1523/JNEUROSCI.19-15-06267.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen ZY, Hasson T, Kelley PM, Schwender BJ, Schwartz MF, Ramakrishnan M, Kimberling WJ, Mooseker MS, Corey DP. Molecular cloning and domain structure of human myosin-VIIa, the gene product defective in Usher syndrome 1B. Genomics. 1996;36:440–448. doi: 10.1006/geno.1996.0489. [DOI] [PubMed] [Google Scholar]
- 27.De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 28.Inoue A, Ikebe M. Characterization of the motor activity of mammalian myosin VIIA. J Biol Chem. 2003;278:5478–5487. doi: 10.1074/jbc.M210489200. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe S, Ikebe R, Ikebe M. Drosophila myosin VIIA is a high duty ratio motor with a unique kinetic mechanism. J Biol Chem. 2006;281:7151–7160. doi: 10.1074/jbc.M511592200. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, Kovacs M, Sakamoto T, Zhang F, Kiehart DP, Sellers JR. Dimerized Drosophila myosin VIIa: a processive motor. Proc Natl Acad Sci U S A. 2006;103:5746–5751. doi: 10.1073/pnas.0509935103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 32.Ikebe M, Kambara T, Stafford WF, Sata M, Katayama E, Ikebe R. A hinge at the central helix of the regulatory light chain of myosin is critical for phosphorylation-dependent regulation of smooth muscle myosin motor activity. J Biol Chem. 1998;273:17702–17707. doi: 10.1074/jbc.273.28.17702. [DOI] [PubMed] [Google Scholar]
- 33.Reynard AM, Hass LF, Jacobsen DD, Boyer PD. The correlation of reaction kinetics and substrate binding with the mechanism of pyruvate kinase. J Biol Chem. 1961;236:2277–2283. [PubMed] [Google Scholar]
- 34.Watanabe S, Mabuchi K, Ikebe R, Ikebe M. Mechanoenzymatic characterization of human myosin Vb. Biochemistry. 2006;45:2729–2738. doi: 10.1021/bi051682b. [DOI] [PubMed] [Google Scholar]
- 35.Wang F, Thirumurugan K, Stafford WF, Hammer JA, 3rd, Knight PJ, Sellers JR. Regulated conformation of myosin V. J Biol Chem. 2004;279:2333–2336. doi: 10.1074/jbc.C300488200. [DOI] [PubMed] [Google Scholar]
- 36.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: regulation by calcium, calmodulin, and the tail domain. J Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li XD, Mabuchi K, Ikebe R, Ikebe M. Ca2+-induced activation of ATPase activity of myosin Va is accompanied with a large conformational change. Biochem Biophys Res Commun. 2004;315:538–545. doi: 10.1016/j.bbrc.2004.01.084. [DOI] [PubMed] [Google Scholar]
- 38.Park H, Ramamurthy B, Travaglia M, Safer D, Chen LQ, Franzini-Armstrong C, Selvin PR, Sweeney HL. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol Cell. 2006;21:331–336. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 39.Todorov PT, Hardisty RE, Brown SD. Myosin VIIA is specifically associated with calmodulin and microtubule-associated protein-2B (MAP-2B) Biochem J. 2001;354:267–274. doi: 10.1042/0264-6021:3540267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Udovichenko IP, Gibbs D, Williams DS. Actin-based motor properties of native myosin VIIa. J Cell Sci. 2002;115:445–450. doi: 10.1242/jcs.115.2.445. [DOI] [PubMed] [Google Scholar]
- 41.Nascimento AA, Cheney RE, Tauhata SB, Larson RE, Mooseker MS. Enzymatic characterization and functional domain mapping of brain myosin-V. J Biol Chem. 1996;271:17561–17569. doi: 10.1074/jbc.271.29.17561. [DOI] [PubMed] [Google Scholar]
- 42.Tauhata SB, dos Santos DV, Taylor EW, Mooseker MS, Larson RE. High affinity binding of brain myosin-Va to F-actin induced by calcium in the presence of ATP. J Biol Chem. 2001;276:39812–39818. doi: 10.1074/jbc.M102583200. [DOI] [PubMed] [Google Scholar]
- 43.Weston MD, Kelley PM, Overbeck LD, Wagenaar M, Orten DJ, Hasson T, Chen ZY, Corey D, Mooseker M, Sumegi J, Cremers C, Moller C, Jacobson SG, Gorin MB, Kimberling WJ. Myosin VIIA mutation screening in 189 Usher syndrome type 1 patients. Am J Hum Genet. 1996;59:1074–1083. [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher AJ, Smith CA, Thoden JB, Smith R, Sutoh K, Holden HM, Rayment I. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 45.Onishi H, Ohki T, Mochizuki N, Morales MF. Early stages of energy transduction by myosin: roles of Arg in switch I, of Glu in switch II, and of the salt-bridge between them. Proc Natl Acad Sci U S A. 2002;99:15339–15344. doi: 10.1073/pnas.242604099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li XD, Rhodes TE, Ikebe R, Kambara T, White HD, Ikebe M. Effects of mutations in the gamma-phosphate binding site of myosin on its motor function. J Biol Chem. 1998;273:27404–27411. doi: 10.1074/jbc.273.42.27404. [DOI] [PubMed] [Google Scholar]
- 47.Furch M, Fujita-Becker S, Geeves MA, Holmes KC, Manstein DJ. Role of the salt-bridge between switch-1 and switch-2 of Dictyostelium myosin. J Mol Biol. 1999;290:797–809. doi: 10.1006/jmbi.1999.2921. [DOI] [PubMed] [Google Scholar]
- 48.Smith CA, Rayment I. X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 A resolution. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 49.Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. Embo J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]