Abstract
Cranial neural crest cells (NCCs) play an intimate role in craniofacial development. Multiple signaling cascades participate in patterning cranial NCCs, some of which are regulated by endothelin-A receptor (Ednra) signaling. Ednra−/− embryos die at birth from severe craniofacial defects resulting from disruption of neural crest cell patterning and differentiation. These defects include homeotic transformation of lower jaw structures into upper jaw-like structures, suggesting that some cephalic NCCs alter their “identity” in the absence of Ednra signaling. To elucidate the temporal necessity for Ednra signaling in vivo, we undertook two strategies. We first used a conditional knockout strategy in which mice containing a conditionally targeted Ednra allele (Ednrafl) were bred with mice from the Hand2-Cre and Wnt1-Cre transgenic mouse strains, two strains in which Cre expression occurs at different time periods within cranial NCCs. In our second approach, we used an Ednra-specific antagonist to treat wild type pregnant mice between embryonic days E8.0 and E10.0, a time frame encompassing the early migration and proliferation of cranial NCCs. The combined results suggest that Ednra function is crucial for NCC development between E8.25 and E9.0, a time period encompassing the arrival of NCCs in the arches and/or early post-migratory patterning. After this time period, Ednra signaling is dispensable. Interestingly, middle ear structures are enlarged and malformed in a majority of Ednrafl/fl;Wnt1-Cre embryos, instead resembling structures found in extinct predecessors of mammals. These observations suggest that the advent of Ednra signaling in cranial NCCs may have been a crucial event in the evolution of the mammalian middle ear ossicles.
Keywords: Transgenic mice, Endothelin, loxP/Cre, Craniofacial development, Neural crest cell, Ednra antagonist
Introduction
Cranial neural crest cells (NCCs) are an essential component of the vertebrate craniofacial skeleton (Chai and Maxson, 2006; Muelemans and Bronner-Fraser, 2007). These cells originate along the neural fold of the posterior midbrain and hindbrain and migrate into the pharyngeal arches, transient structures on the ventral embryo surface (Couly et al., 1993; Fraser et al., 1990; Le Douarin et al., 1993; Lumsden et al., 1991). Once there, NCCs differentiate into the bone, cartilage and connective tissue of the face and neck (Couly et al., 1993, 1996; Kontges and Lumsden, 1996; Lumsden et al., 1991; Noden, 1983; Noden, 1988). Most facial structures are derived from the first pharyngeal arch, which is classically subdivided into mandibular and maxillary prominences.
During pharyngeal arch morphogenesis, signals from surrounding tissues provide crucial patterning cues to NCCs, initiating signaling cascades that eventually lead to the development of the vertebrate jaw (Chai and Maxson, 2006). In the mouse, development of the bone, cartilage and connective tissue of the lower jaw is regulated in part by endothelin-1 (Edn1), which is expressed by the ectoderm, core paraxial mesoderm and pharyngeal pouch endoderm of the pharyngeal arches (Clouthier et al., 1998; Maemura et al., 1996; Yanagisawa et al., 1998a). Edn1 induces signaling from its cognate receptor, the endothelin-A receptor (Ednra), on NCCs (Clouthier et al., 1998; Yanagisawa et al., 1998a). This in turn initiates the expression of a transcription factor cascade (Clouthier et al., 2000; Ozeki et al., 2004; Ruest et al., 2004). Mice containing a targeted inactivation of the genes encoding Ednra, Edn1, or endothelin converting enzyme-1 (Ece1, the enzyme that cleaves Edn1 from an inactive to active form) are born with both craniofacial and cardiovascular malformations and die shortly after birth (Clouthier et al., 1998; Kurihara et al., 1994; Ozeki et al., 2004; Ruest et al., 2004; Yanagisawa et al., 1998a). These defects, which include the homeotic transformation of lower jaw structures into upper jaw-like structures, arise in part from mis-patterning of NCCs due to expansion of maxillary-like gene expression into the distal mandibular arch and a loss of normal mandibular arch-specific gene expression (Ozeki et al., 2004; Ruest et al., 2004). This action appears to be mediated in part by Dlx5 and Dlx6, two genes whose expression in the pharyngeal arches are largely dependent on Ednra signaling and whose combined loss also results in loss of mandibular identity (Beverdam et al., 2002; Depew et al., 2002). Defects in anterior arch derivatives and homeotic transformation of anterior arch dermal bones into more posterior-like bones are also observed in edn1-mutant zebrafish and in embryos with morpholino mediated partial reduction in Edn1 levels (Kimmel et al., 2003; Miller and Kimmel, 2001; Miller et al., 2000). In addition, mutations in either the gene encoding the furin protease that cleaves preproendothelin-1 into big endothelin or the gene encoding phospholipase C β3, the down-stream intracellular mediator of endothelin signaling, lead to anterior arch defects (Walker et al., 2006, 2007). In addition to zebrafish, blocking Ednra signaling in both chick and rat produces craniofacial defects (Kempf et al., 1998; Spence et al., 1999). These changes may in part be due to changes in Mef2c function, though the exact relationship of Ednra signaling and Mef2c function is not clear (Miller et al., 2007; Verzi et al., 2007). It thus appears that the function of Edn1/Ednra signaling in pattering NCCs within the mandibular pharyngeal arch is conserved among vertebrates. In contrast, while endothelin signaling is conserved among vertebrates, it is missing in the more basal organisms (Muelemans and Bronner-Fraser, 2007).
While Ednra signaling is required for NCC development, its timing and action are less clear. In the mouse, Ednra expression is first observed on NCCs shortly after they leave the neural folds (Sato et al. 2008) and data not shown). Expression of Edn1 on the arch ectoderm and endoderm occurs around the same time period. In E9.5 Ednra mutant chimeric embryos, in which Ednra−/− cells are mixed with wild type cells (referred to as ), Ednra−/− cells are excluded from the distal mandibular and second arches (Clouthier et al., 2003), indicating potential roles for Ednra signaling in NCC migration within the pharyngeal arches. Further, mouse embryos cultured in the presence of the Ednra-specific antagonist BQ-123 between E8.5 to E9.5 exhibit disrupted gene expression in the mandibular arch at E10.5 (Fukuhara et al., 2004), suggesting a potential role for Ednra in migratory and early post-migratory patterning. More recently, migration of NCCs to the pharyngeal arches of Ednra−/− embryos has been shown to be grossly normal, supporting a role for Ednra in initiating pattering events (Abe et al., 2007). Consistent with this is the finding that injection of human EDN1 protein into the arches of edn1 mutant zebrafish after most NCC migration is complete can rescue the mutant phenotype (Miller et al., 2000). Ednra signaling is, however, dispensable after these early events, as conditional inactivation of the Ednra gene after E10.5 in the cells within the mandibular arch that will eventually form the mandible does not result in lower jaw defects (Ruest et al., 2005).
While it is often difficult to define the window of gene function during development, such information is crucial in elucidating potential downstream signals and pathways that may be active in a specific signaling network. In addition, the timing of gene action can directly determine the developmental processes in which a gene product is involved. To investigate the timing of Ednra function and hence its role in NCC patterning during craniofacial development, we have taken two approaches. First, we have used a mouse strain carrying a conditional mutation in the Ednra gene (Kedzierski et al., 2003), and crossed these mice with two Cre transgenic mouse lines in which Cre expression in NCCs occurs between E8.25 and E9.5. In a second approach, we have used an Ednra-specific antagonist (Padilla et al., 2006; Sprogar et al., 2007; Wu et al., 2001) to knockdown Ednra signaling at specific time periods of NCC development. Our results indicate that Ednra signaling is required for NCC development in a very narrow temporal window spanning approximately 18 h. In addition, detailed analysis of aberrant craniofacial structures observed following reduction in Ednra signaling in both strains suggest a role for Ednra signaling in the evolution of the mammalian hearing apparatus.
Materials and methods
Mice
Ednraflox/flox (Ednrafl/fl) mice have been previously described (Kedzierski et al., 2003). Briefly, loxP sites were introduced by homologous recombination into the 3′ region of the gene, flanking the last three exons of the gene. Wnt1-Cre (Danielian et al., 1998) (a gift from Andrew McMahon through Henry Sucov) and Hand2-Cre (previously referred to as dHAND-Cre (Ruest et al., 2003) transgenic mouse strains have been previously described.
Breeding and genotyping
Ednrafl/fl mice were first bred with Wnt1-Cre and Hand2-Cre transgenic mice to generate Ednrafl/+;Wnt1-Cre and Ednrafl/+;Hand2-Cre animals. Ednrafl/+;Wnt1-Cre and Ednrafl/+;Hand2-Cre female mice were then bred with Ednrafl/fl male mice to generate Ednrafl/fl;Wnt1-Cre and Ednrafl/fl;Hand2-Cre embryos. Mice and embryos were genotyped by PCR analysis using genomic DNA prepared from tail biopsies or amniotic sacs. Ednrafl genotyping was performed using the primers 5′-ACACAACCATGGTGTCGA-3′ and 5′-CGGTTCTTATCCATCTCATC-3′. Reactions were visualized on a 1.5% agarose gel. PCR resulted in two bands in Ednrafl/+ samples (~420 bp and ~380 bp), one band in Ednrafl/fl (~420 bp) and one in Ednra+/+ samples (~380 bp). Cre genotyping was performed as previously described (Danielian et al., 1998) using the Cre primers 5′-GGACATGTTCAGGGATCGCCAGGCG-3′ and 5′-GCATAACCAGTGAAACAGCATTGCTG-3′.
Recombination PCR
E8.5 to E10.5 Ednrafl/fl;Wnt1-Cre or Ednrafl/fl;Hand2-Cre embryos were collected in PBS and mandibular arches dissected, placed in Eppendorf tubes and digested with proteinase K. Genomic DNA was extracted and used for recombination PCR using the primers 5′ ACACAACCATGTTGTCGAGGTCGA 3′ and 5′ GAGAACCTACAACTGGGGACACAAACAC 3′. This PCR reaction only amplifies the recombined Ednra allele, producing a 1.2 kb band (Kedzierski et al., 2003). Reaction products were visualized on a 1% agarose gel.
Skeleton staining
Skeleton staining was performed as previously described by McLeod (1980). Briefly, E18.5 embryos were eviscerated and skinned, taking care not to disrupt structures of the lower jaw. After fixation in 95% ethanol, embryos were incubated two days in acetone followed by staining for 3–5 days at 37 °C in 70% ethanol/5% glacial acetic acid containing 0.015% alcian blue (stock solution of 0.3% in 70% ethanol) and 0.005% alizarin red (stock solution of 0.1% in 95% ethanol). After staining, embryos were cleared by successive immersions in 1% potassium hydroxide in 20%, 50% and 80% glycerol. Stained skeletons were stored in 25% glycerol/75% ethanol. Skeletons were analyzed and photographed using an Olympus SZX12 stereomicroscope fitted with a DP11 digital camera.
Histology
For histological analysis, E18.5 embryos were fixed in 10% neutral buffered formalin. After dehydration in graded ethanols and xylene, embryos were embedded in paraffin and sectioned at 7–9 µm. Sections were counterstained with hematoxylin and eosin and then mounted with a coverslip using DPX mounting medium (BDH). Sections were examined and photographed with an E600 Nikon microscope equipped with a SPOT-RT digital camera.
In situ hybridization
Whole mount in situ hybridization analysis was performed as previously described using digoxigenin (DIG)-labeled antisense cRNA riboprobes against Hand1, Hand2, Dlx3, Dlx5 and Dlx6 (Ruest et al., 2003). Embryos were photographed as described above.
Malleus measurement
Mallei were dissected and photographed using a Nikon SMZ1500 microscope fitted with a Nikon DXM1200 digital camera. Captured images were measured using calibrated digital calipers included with Metamorph Imaging software (Universal Imaging Corporation). The length of each malleus was measured from the top of the malleus to the midpoint between the anterior process and manubrium. Malleus width was measured across the malleus at a level just below the point of fusion with Meckel's cartilage.
Antagonist treatment
Wild type 129S6 female mice were mated with 129S6 males, with noon of the date that the copulatory plug was found counted as E0.5. Plugged females were weighed daily to assess weight gain before and after antagonist or control treatment. At noon, midnight or both of the designated day of treatment (E8.0 to E10.5), females were given either 100 µl water (as a control) or 100 mg/kg body weight TBC3214 in 100 µl water by gavage. TBC3214 is an Ednra-specific non-peptidic antagonist that has good oral bioavailability (approximately 25% in rats), high potency (IC(50)=40 pM), and high Ednra/Ednrb selectivity (400,000-fold) (Wu et al., 2001). In addition, the half-life is relatively short (t(1/2)=>N4 h; (Wu et al., 2001)) and is well tolerated by mice, with no obvious signs of toxic affects in treated females (data not shown). This included normal weight gain after treatment compared with water-treated females (data now shown). Further, the rate of embryo resorption and average embryo weight was similar between antagonist-treated and water-treated females (Supplementary Fig. 1). Embryos from water- and TBC3214-treated females were collected at either E10.5 (whole mount in situ hybridization) or E18.5 (skeleton staining) and processed as described above.
Data analysis for antagonist study
A minimum of 3 litters (average of 4.7 litters) and a minimum of 20 embryos (average of 35.3 embryos)were analyzed for skeletal defects for each treatment modality with the antagonist. Embryos were individually scored for structural defect of each analyzed structure and data compiled as a percentage of embryos having a given structural malformation per litter. These percentages were then averaged for each structure and treatment modality, and standard deviation calculated.
Results
Differential timing of Ednra recombination in Ednrafl/fl;Wnt1-Cre and Ednrafl/fl;Hand2-Cre embryos
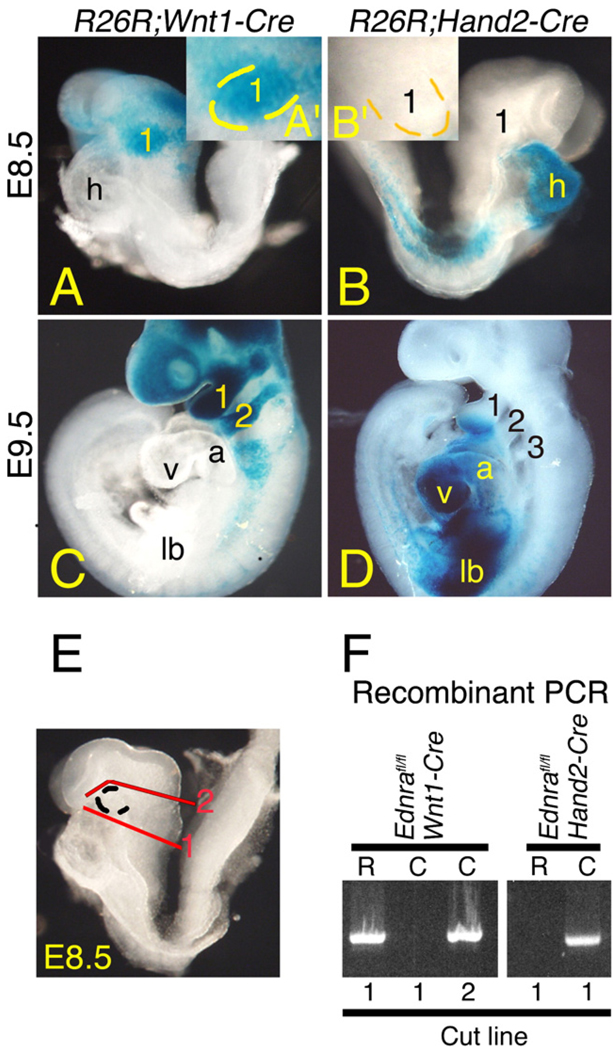
To examine the timing of Ednra signaling during cephalic NCC development, we took advantage of a conditional mutant mouse strain in which loxP sites flank the last three exons of the Ednra gene (we use the abbreviation “fl”) (Kedzierski et al., 2003). This conditional allele was inactivated using two specific Cre transgenic mouse strains in which Cre expression occurs at different time points within cephalic NCCs: Wnt1-Cre (Chai et al., 2000; Danielian et al., 1998) and Hand2-Cre (Ruest et al., 2003). To compare the early expression of Cre in these two strains, each was bred into the Cre reporter strain R26R (Soriano, 1999). In E8.5 R26R;Wnt1-Cre embryos, β-gal actosidase (β-gal) staining was observed in NCCs migrating towards and in the first arch by E8.5 (Fig. 1A), with the first and second arches extensively labeled by E9.5 (Fig. 1C). In contrast, β-gal staining in E8.5 R26R;Hand2-Cre embryos was confined to the heart and a portion of the lateral plate mesoderm (Fig. 1B), with staining in the pharyngeal arches not observed until E9.5 (Fig. 1D).
Fig. 1.
Timing of Cre expression and Ednra gene recombination in Wnt1-Cre and Hand2-Cre embryos. A–D. Lateral view of whole mount β-galactosidase (β-gal) staining in E8.5 (A, B) and E9.5 (C, D) R26R;Wnt1-Cre (A, C) and R26R;Hand2-Cre (B, D) embryos. A, B. In a 8.25–E8.5 R26R;Wnt1-Cre embryo, β-gal-labeled cells are observed between the midbrain/hindbrain and first pharyngeal arch (1; see also inset, A′). In an E8.5 R26R;Hand2-Cre embryo, β-gal labeled cells are observed in the heart (h) and lateral plate mesoderm but not arch 1 (see also inset, B′). C, D. At E9.5, labeled cells are observed in pharyngeal arches 1–3 of a R26R;Wnt1-Cre embryo (C) and arches 1 and 2 of a R26R;Hand2-Cre embryo (D). E, F. PCR that specifically detects recombination of the Ednrafl allele was performed on DNA isolated from the rostral (R) and caudal (C) halves of E8.5 Ednrafl/fl;Wnt1-Cre and Ednrafl/fl;Hand2-Cre embryos bisected along the plane denoted by the line marked “1” (E) (see Materials and methods for details). Recombination of the Ednrafl allele correlates with sites of β-gal staining in R26R;Wnt1-Cre and R26R;Hand2-Cre embryos. Recombination in Ednrafl/fl;Wnt1-Cre embryos is observed in the posterior half (including the first arch) when the cut line is moved rostral to the first arch (line 2 in E). a, atrium; lb, limb bud; v, ventricle.
We next crossed these strains into the Ednrafl background to inactivate the conditional Ednra allele. To first determine if the timing of Ednra gene recombination in the first arch of E8.5 Ednrafl/fl;Wnt1-Cre and Ednrafl/fl;Hand2-Cre embryos correlated with β-gal staining in R26R;Wnt1-Cre and R26R;Hand2-Cre embryos shown above, embryos were subdivided into rostral (first arch and head; “R”) and caudal (the remainder of the embryo; “C”) portions (red line #1 in Fig. 1E), with DNA isolated from each half. Using PCR analysis designed to specifically detect the recombined Ednra allele, recombination in Ednrafl/fl;Wnt1-Cre embryos was confined to the rostral portion of the embryo, while recombination in Ednrafl/fl;Hand2-Cre embryos was confined to the caudal (C) segment (Fig. 1F). To further confirm that the recombination in E8.5 Ednrafl/fl;Wnt1-Cre was occurring in cells moving toward the first arch, different Ednrafl/fl;Wnt1-Cre embryos were subdivided into rostral (all tissue above the first arch) and caudal (the remainder of the embryo) portions (red line #2 in Fig. 1E). In these samples, recombination was now observed in the caudal portion (Fig. 1F), confirming that the Ednra conditional allele was inactivated in these cells.
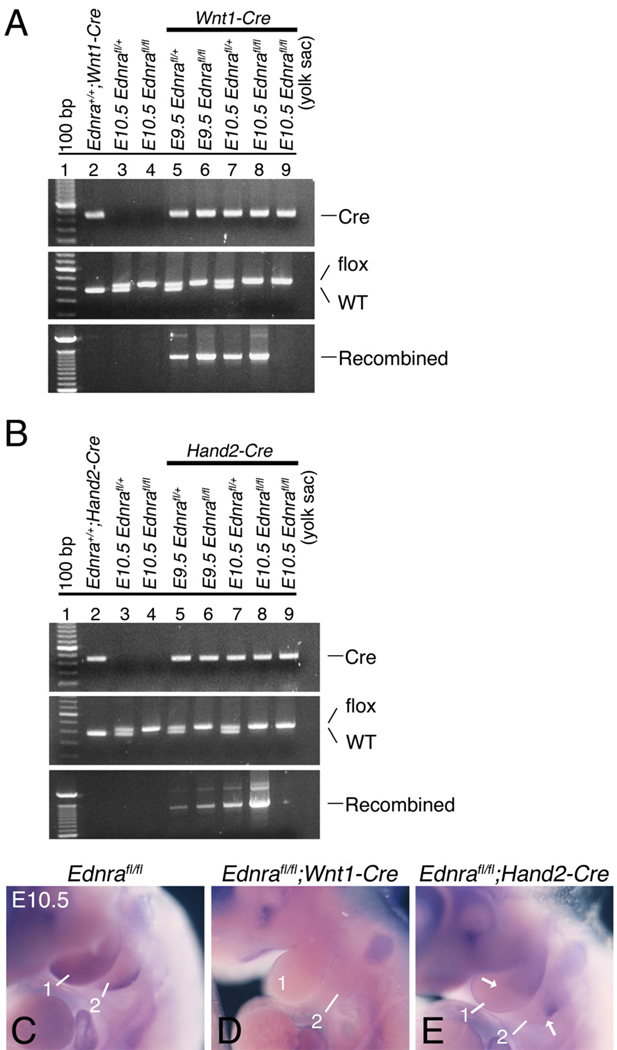
Since recombination of the Ednrafl gene was not observed in E8.5 Ednrafl/fl;Hand2-Cre embryos, we examined recombination within mandibular arch DNA in both E9.5 and E10.5 Ednrafl/fl;Wnt1-Cre and Ednrafl/fl;Hand2-Cre embryos. Recombination was observed in embryos that contained at least one Ednrafl allele and the Wnt1-Cre transgene (Fig. 2A, lanes 5–8). Recombination was not detected when using mandibular arch DNA extracted from embryos carrying either the Ednrafl allele or the Wnt1-Cre transgene alone (Fig. 2A, lanes 2–4). Further, no recombination was observed in yolk sac DNA from Ednrafl/fl;Wnt1-Cre embryos (Fig. 2A, lane 9), a site where the transgene is not expressed. Recombination of the conditional Ednra allele within mandibular arch DNA of Ednrafl/fl;Hand2-Cre embryos was also detectable at E9.5 and E10.5 (Fig. 2B, lanes 5–8). As observed in Ednrafl/fl;Wnt1-Cre embryos, Ednrafl gene recombination was not observed in either Hand2-Cre or Ednrafl/fl embryos (Fig. 2B, lanes 2–4). Recombination was also not observed in yolk sac DNA of Ednrafl/fl;Hand2-Cre embryos (Fig. 2B, lane 9). Due to the specific Cre strains used in this study, the Ednra flox allele was not recombined in the pharyngeal arch ectodermal, pouch endodermal and core paraxial mesodermal cells. This accounts for the continued presence of the flox Ednra allele band in our PCR assays.
Fig. 2.
Recombination of the Ednra conditional allele in the mandibular arch of E9.5 and E10.5 Ednrafl/fl;Wnt1-Cre and Ednrafl/fl;Hand2-Cre embryos. A, B. Agarose gels showing PCR-generated bands corresponding to presence of a Cre transgene (Cre), Ednrafl (flox) or Ednra wild type alleles and a recombined Ednrafl allele. DNA was isolated from the mandibular arch of E9.5 and E10.5 Ednrafl/+;Wnt1-Cre, Ednrafl/fl;Wnt1-Cre, Ednrafl/+;Hand2-Cre and Ednrafl/fl;Hand2-Cre embryos. Tail DNA from an adult Ednra+/+;Wnt1-Cre mouse and yolk sac DNA from E10.5 Ednrafl/fl;Wnt1-Cre and Ednrafl/fl;Hand2-Cre embryos were used as negative controls. Recombination of the Ednrafl allele is only observed in the presence of the Cre transgene. The presence of the flox band in all samples reflects the presence of cells in which the Wnt1-Cre or Hand2-Cre transgenes are not expressed. C–E. In situ hybridization analysis of conditional inactivation of the Ednra allele using a DIG-labeled probe that only detects the recombined allele. In E10.5 control embryos, Ednra expression can be seen throughout the mandibular (1) and second (2) arches, with highest expression in the distocaudal aspects (C). In Ednrafl/fl;Wnt1-Cre embryos, recombination of the conditional Ednra allele appears to have occurred throughout the pharyngeal arches (D). In Ednrafl/fl;Hand2-Cre embryos, conditional Ednra recombination is only observed in the distal half of the mandibular and second arches, corresponding to the area of Hand2 expression.
To further verify recombination in vivo, we performed in situ hybridization analysis using an Ednra riboprobe that specifically detected the region of the Ednra mRNA deleted following conditional gene recombination of the Ednra allele (Kedzierski et al., 2003). In E10.5 control embryos, expression of Ednra was observed throughout the mandibular and second arches, with expression highest distally and caudally (Fig. 2C). In E10.5 Ednrafl/fl;Wnt1-Cre embryos, Ednra expression was absent from the mandibular and second pharyngeal arches (Fig 2D), consistent with pattern of gene recombination obtained when using the Wnt1-Cre strain (Chai et al., 2000). In contrast, arch Ednra expression in Ednrafl/fl;Hand2-Cre embryos was only absent in distal half of the first and second arches (Fig. 2E), consistent with pattern of gene recombination observed when using the Hand2-Cre strain (Ruest et al., 2003). These findings show that Ednra gene inactivation has occurred using both Cre strains and also illustrates the spatial specificity of Ednra gene inactivation when using both Cre strains.
Craniofacial defects in E18.5 Ednrafl/fl;Wnt1-Cre embryos
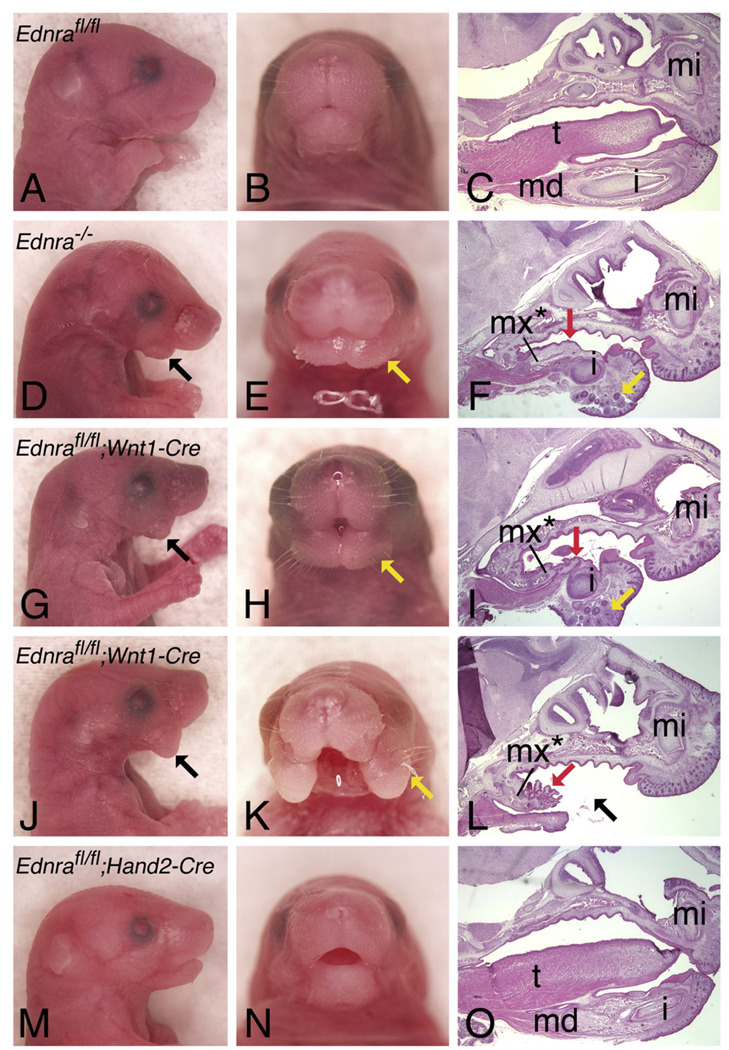
To determine the effect of Ednra gene recombination in Ednrafl/fl;Wnt1-Cre and Ednrafl/fl;Hand2-Cre embryos, we performed gross analysis of near-term embryos. E18.5 Ednrafl/fl;Wnt1-Cre embryos had defects similar to those observed in Ednra−/− embryos (Figs. 3D–F), including a shortened lower jaw(Fig. 3G) that contained mystacial vibrissae (yellow arrows in Fig. 3H) and a midline mandibular cleft (Fig. 3H). Analysis of sagittal sections stained with hematoxylin and eosin further illustrated tongue hypoplasia (Fig. 3I) and the presence of rugae, raised epithelial ridges normally confined to the palate of the mouth, on the floor of the mouth (Fig. 3I). Lower incisors were present but primarily set in mesenchyme (Fig. 3I). In addition, 24% (8/33) of Ednrafl/fl;Wnt1-Cre embryos had a more severe phenotype, in which the two mandibular arch halves appeared as large flaps of soft tissue that failed to fuse (Figs. 3J–L). This more severe phenotype correlated with specific Wnt1-Cre animals, suggesting the presence or absence of modifier alleles in these complex genetic crosses. In contrast, the lower jaw of E18.5 Ednrafl/fl;Hand2-Cre embryos appeared normal (Figs. 3M–O). Since Hand2 daughter cells are found throughout the lower jaw of near-term mouse embryos (Ruest et al., 2003) and most ventral cartilages are absent in the hand2 zebrafish mutant hands off (Miller et al., 2003; Yelon et al., 2000), we believe that an absence of a phenotype in Ednrafl/fl;Hand2-Cre embryos reflects inactivation of the Ednra gene after Ednra signaling has accomplished its required function.
Fig. 3.
Gross analysis of E18.5 Ednrafl/fl;Cre embryos. Lateral (A, D, G, J,M) and frontal (B, E, H, K, N) views and sagittal sections (C, F, I, L, O) through the head of wild type (A–C), Ednra−/− (D-F), Ednrafl/fl;Wnt1-Cre (G–L) and Ednrafl/fl;Hand2-Cre (M–O) embryos. Sagittal sections were stained with hematoxylin and eosin. Defects in Ednrafl/fl;Wnt1-Cre embryo are almost identical to those observed in Ednra−/− embryos, including a midline cleft in the lower jaw in addition to the presence of mystacial vibrissae (yellow arrows), normally only observed on the snout. Red arrows (F, I, L) point to the rugae present on the oral surface of the lower jaw, resembling rugae present on the roof of the oral cavity. Some Ednrafl/fl;Wnt1-Cre embryos have a more severe phenotype (J–L), characterized by an absence of tissue in the distal lower jaw (black arrow). No defects are observed in Ednrafl/fl;Hand2-Cre embryo (M–O). i, incisor; md, mandible; t, tongue.
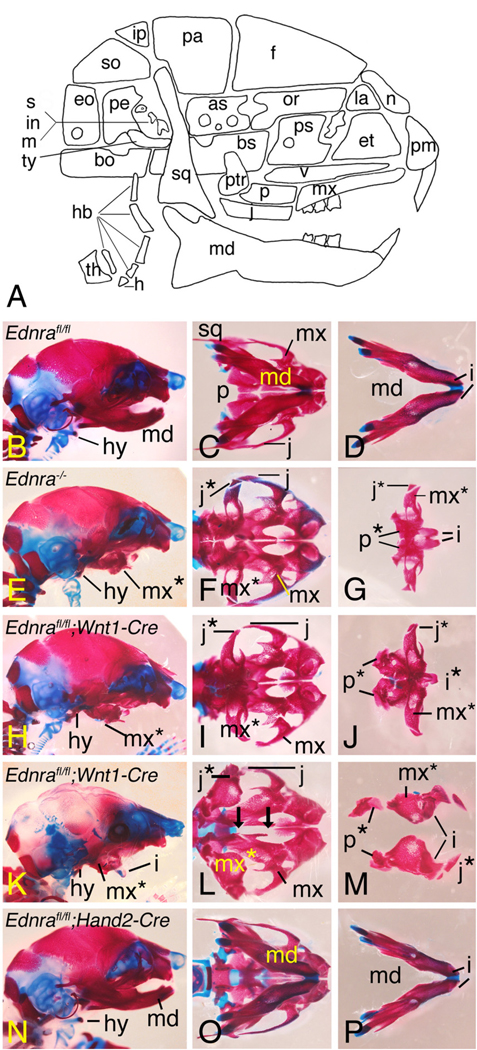
To examine skeletal structure in the lower jaw in more detail, E18.5 Ednrafl/fl;Wnt1-Cre embryos were stained with alizarin red/alcian blue to examine bone and cartilage, respectively. As observed in Ednra−/− embryos (Figs. 4E–G), the mandible in Ednrafl/fl;Wnt1-Cre embryos was shortened and flattened (Fig. 4H). On ventral view, the mandible in these embryos resembled a small maxilla (Fig. 4I). The mandible attached to the jugal bone of the zygomatic arch via a bone that appeared to be a duplicated jugal bone (Figs. 4I, J). Duplications of the palatine, pterygoid and lamina obturans bones and the ala temporalis cartilage were also present (Figs. 4I, J and data not shown). Most of Meckel's cartilage was also missing, though the symphysis was present, surrounded by a small amount of membranous bone (data not shown). As described above, the incisors were set in mesenchyme rather than bone. In Ednrafl/fl;Wnt1-Cre embryos with the more severe phenotype (Figs. 4K–M), the two pieces of tissue that extended down from the maxilla were composed almost completely of soft tissue, with each containing an incisor (Figs. 4L, M). In 5/6 stained skulls examined, incisor polarity was changed, with each incisor projecting downwards (Fig. 4K). In contrast to these changes, defects were not observed in the lower jaw structure of E18.5 Ednrafl/fl;Hand2-Cre (Figs. 4N–P).
Fig. 4.
Skeletal analysis of E18.5 Ednra conditional knockout embryos. Lateral (B, E. H, K, N) and ventral (C, D, F, G, I, J, L, M, O, P) views of the skull and mandible of Ednrafl/fl (wild type) (B–D), Ednra−/− (E–G), Ednrafl/fl;Wnt1-Cre (H–M), Ednrafl/fl;Hand2-Cre (N–P) embryos. A. A schematic drawing showing the bones of the mouse skull (abbreviations given below). B–D. Skull and mandible (md) structure of a Ednrafl/fl (wild type) embryo. E–G. In Ednra−/− embryos, the mandible is flattened and resembles a maxilla (referred to as a pseudo-maxilla (mx*)) (E, F), though overall size is smaller. Duplications of the jugal bones (j*) and palatine bones (p*) are also observed. H–J. Similar craniofacial defects are observed in Ednrafl/fl;Wnt1-Cre embryos. In some mutant embryos, the lower incisors are malformed and fused at their basis (i* in I), a phenomenon not observed in Ednra−/− embryos. K–M. In some Ednrafl/fl;Wnt1-Cre embryos, a severe lower jaw phenotype is present, characterized by a cleft lower jaw (K, L). The angle of photography in L makes it appear that the jugal bone in L projects more distally than the incisors, though this is not actually the case. The two black arrows in K indicate a gap between the palatine bones, resulting in a cleft palate. N–P. Defects are not observed in Ednrafl/fl;Hand2-Cre embryos. as, alisphenoid; bo, basioccipital; bs, basisphenoid; eo, exoccipital; et, ethamoid; f, frontal; h, hyoid; hb, hypobranchials; i, incisor; in, incus; ip, interparietal; j, jugal; la, lacrimal; m, malleus; mx, maxilla; n; nasal; or, orbitosphenoid; p, palatine; pa, parietal; pe, petrosal; pm, premaxilla; ps, presphenoid; ptr, pterygoid; s, stapes; so, supraoccipital; sq, squamosal/temporal; th, thyroid cartilage; ty, tympanic; v, vomer.
Because incomplete gene inactivation resulting in mosaicism is a common problem with conditional mouse knockouts (Kwan, 2002; Nagy, 2000), we also examined the lower jaw structure of E18.5 Ednrafl/−;Wnt1-Cre embryos. In these embryos, one Ednra allele contains the conditional mutation (fl) and one contains the conventional mutation (null, here referred to as “−”). In these embryos, changes in jaw structure were identical to that of Ednrafl/fl;Wnt1-Cre embryos (Supplementary Figs. 2A–C), indicating that the extent of gene recombination was similar between Ednrafl/fl and Ednrafl/− embryos. Similarly, defects were also not observed in E18.5 Ednrafl/−;Hand2-Cre embryos (data not shown), indicating that the absence of a phenotype was not due to insufficient gene inactivation. We confirmed this hypothesis by breeding the Hand2-Cre line with the Smoothened conditional knockout line to generate Smofl/fl;Hand2-Cre embryos. In these embryos, severe mandibular defects similar to those observed in Smofl/fl;Wnt1-Cre embryos were observed (data not shown), illustrating that the Hand2-Cre line is capable of inactivating conditional alleles. It is important to note that while the absence of a phenotype in Ednrafl/fl;Hand2-Cre and Ednrafl/−;Hand2-Cre embryos could reflect an absence of Ednra, Edn1 or Ece1 in the arch environment, numerous studies have shown that Ednra, Edn1 and Ece1 are all expressed within the arch environment through at least E11.5 (Clouthier et al., 1998; Fukuhara et al., 2004; Kurihara et al., 1994; Maemura et al., 1996; Ozeki et al., 2004; Yanagisawa et al., 1998a,b). We therefore believe that our findings reflect a temporal requirement for Ednra signaling rather than reflect a limited window of endothelin family member expression in the arch.
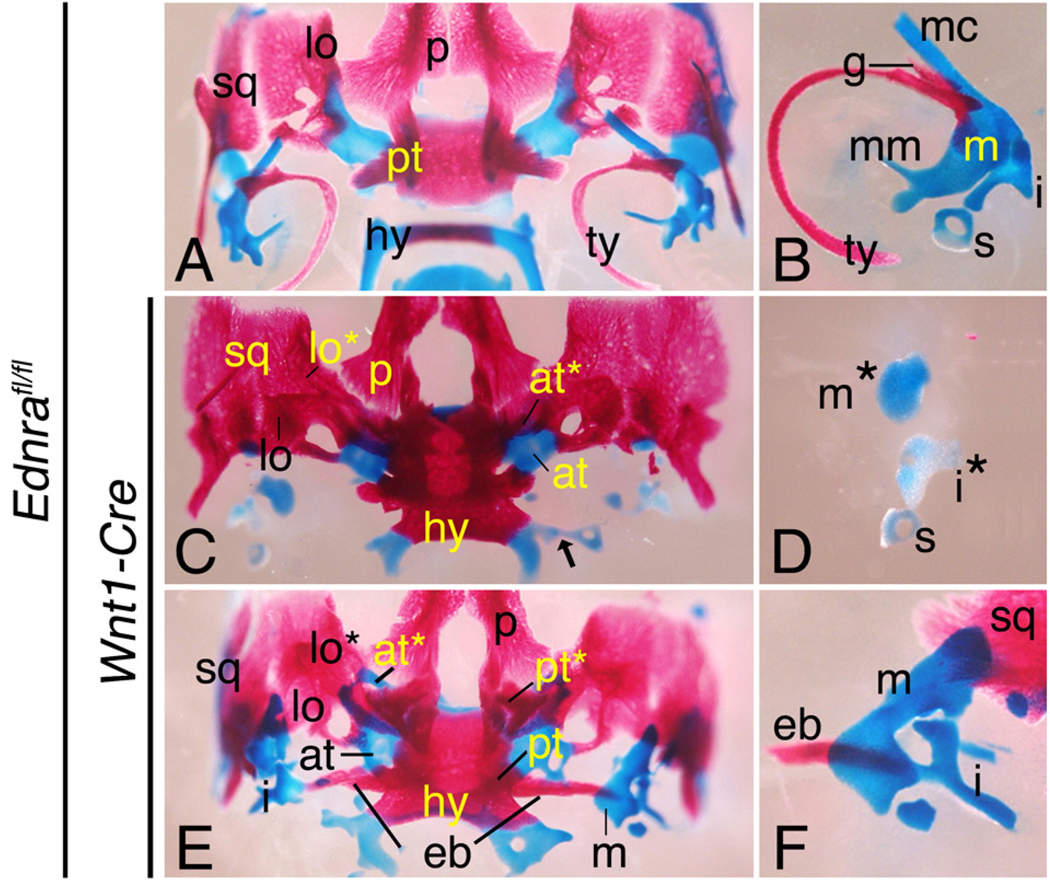
Differential changes in middle ear ossicles in Ednrafl/fl;Wnt1-Cre embryos
Defects were also present in middle ear structures of Ednrafl/fl;Wnt1-Cre embryos, though the extent of malformation was more variable. In Ednra−/− embryos, the malleus and incus are not detectable, though small nodules of undefined cartilage are present (Clouthier et al., 1998). By gross analysis, identical changes were observed in 25% (12/48) of mallei from E18.5 Ednrafl/fl;Wnt1-Cre embryos (Figs. 5C, D). In contrast, 75% of mallei examined from Ednrafl/fl;Wnt1-Cre embryos were enlarged in one or both sides of the skull (Figs. 5E, F), suggesting a threshold affect of Ednra signaling on malleus development. We randomly chose 14 of these enlarged mallei for measurement using Metamorph imaging software. We also measured 8 mallei from wild type embryos and 7 mallei from Ednrafl/fl;Wnt1-Cre embryos in which the mallei resembled that of Ednra−/− embryos. Of the enlarged mallei, we found that 8/14 were similar in length (641.09 µm (±49. 9 µm) to that of wild type embryos (676.68 µm (±62. 4 µm), while 6/14 mallei were longer (1082.04 µm (±194. 3 µm) than wild type embryos. In addition, 12/14 of these mallei were wider (558.57 µm (±150. 6 µm) than wild type mallei (313.30 µm (±20.54 µm). The incus in Ednrafl/fl;Wnt1-Cre embryos was also malformed, contained several processes and often articulated with the pterygoid in the skull base via an ectopic bone (Figs. 5E, F). This bone was present in at least one side of the skull base in 14/24 Ednrafl/fl;Wnt1-Cre embryos examined, with the severity of the lower jaw phenotype not impacting the presence or absence of the ectopic bone (data not shown). It should be noted that the incus defects are more severe than those observed for the palatoquadrate in suc/et1 mutant zebrafish, likely reflecting the impact of the loss of the malleus on incus shape in mouse embryos (Clouthier et al., 2003). The stapes of Ednrafl/fl;Wnt1-Cre embryos appeared normal, though the stapes on at least one side of the skull was attached to the greater horn of the hyoid in 16/24 Ednrafl/fl;Wnt1-Cre embryos examined (Fig. 5C), a defect observed at the same frequency in Ednra−/− embryos (data not not Shown)This would be considered an intermediate pharyngeal arch phenotype, where the most distal derivative of the second arch fuses to the most proximal derivative.
Fig. 5.
Aberrant bone development in E18.5 Ednra conditional knockout embryos. Ventral (A, C, E) and lateral (B, D, F) views of the skull and middle ear ossicles of control (Ednrafl/fl)(A, B), and Ednrafl/fl;Wnt1-Cre (C–F) embryos. A, B. In Ednrafl/fl embryos, normal middle ear structures are observed, including the malleus (m), incus (i) and stapes (s) and the tympanic (ty) and gonial (g) bones. C, D. In some Ednrafl/fl;Wnt1-Cre embryos, most middle ear structures are absent, though a few cartilaginous bodies are present (m*, abberant malleus; i*, abberant incus). The hyoid (h) is also pulled ventro-rostrally and fused with the basisphenoid, matching the Ednra−/− phenotype. The stapes is also often attached to the greater horns of the hyoid (hy) (black arrow in panel C). E, F. In most Ednrafl/fl;Wnt1-Cre embryos, the malleus and incus are enlarged. The malleus is malformed and resembles an articular. The incus is also malformed and often fused with an ectopic bone that also articulates with the pterygoid (pt). at, ala temporalis; at*, duplicated ala temporalis; lo, lamina obturans; lo*, duplicated lamina obturans; p, palatine bone, pt*, duplicated pterygoid bone; sq, squamosal bone.
Mandibular arch gene expression in Ednrafl/fl;Wnt1-Cre embryos
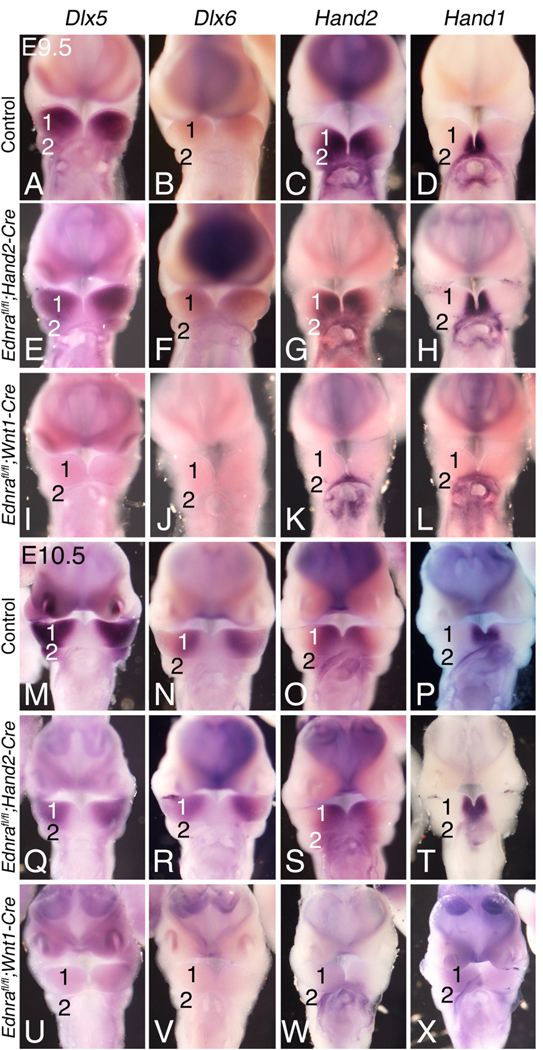
Targeted inactivation of either Edn1 or Ednra in the mouse disrupts expression of multiple genes in the pharyngeal arches (Clouthier et al., 1998, 2000; Fukuhara et al., 2004; Kitano et al., 1998; Ozeki et al., 2004; Ruest et al., 2004). To examine whether similar gene expression patterns were disrupted in conditional knockout embryos, we performed whole mount in situ hybridization analysis on E9.5 and E10.5 embryos, focusing on gene expression changes within the mandibular arch. Expression of Dlx5, Dlx6, Hand2 and Hand1 was similar between E9.5 wild type (Figs. 6A–D) and Ednrafl/fl;Hand2-Cre (Figs. 6E–H) embryos, while expression of all four genes was disrupted in E9.5 Ednrafl/fl;Wnt1-Cre embryos (Figs. 6I–L). Like at E9.5, gene expression was similar between E10.5 wild type (Figs. 6M, N) and Ednrafl/fl;Hand2-Cre embryos (Figs. 6Q–T), while expression of all four genes was almost completely absent in E10.5 Ednrafl/fl;Wnt1-Cre embryos (Figs. 6U–X). Loss of expression in Ednrafl/fl;Wnt1-Cre embryos but not in Ednrafl/fl;Hand2-Cre embryos argues that Ednra signaling is required for initiation but not maintenance of genes involved in lower jaw development.
Fig. 6.
Gene expression analysis in Ednra conditional knockout embryos. Ventral view of E9.5 (A–L) and E10.5 (M–X) control (A–D,M–P), Ednrafl/fl;Hand2-Cre (E–H, Q–T) and Ednrafl/fl;Wnt1-Cre (I–L, U–X) embryos following whole mount in situ hybridization analysis of Dlx5 (A, E, I, M, Q, U), Dlx6 (B, F, J, N, R, V), Hand2 (C, G, K, O, S, W) and Hand1 (D, H, L, P, T, X) expression. The heart has been removed to aid in visualization of the pharyngeal arches. A–D. Gene expression in E9.5 control embryos. E–L. At E9.5, expression of all four genes is normal in Ednrafl/fl;Hand2-Cre embryos (E, H), but almost completely absent in Ednrafl/fl;Wnt1-Cre embryos (I, L). M–P. Gene expression in E10.5 control embryos. Q–X. At E10.5, all four transcription factors show normal expression in Ednrafl/fl;Hand2-Cre embryos (Q, P), but are again almost completely absent in Ednrafl/fl;Wnt1-Cre embryos (U–X). 1, mandibular arch; 2, pharyngeal arch 2.
Craniofacial defects in wild type embryos following Ednra antagonism in utero
To better define the exact timing of Ednra action, we took a second approach, in which Ednra signaling was blocked for short periods during development using an Ednra-specific antagonist (Padilla et al., 2006; Sprogar et al., 2007). The antagonist, TBC3214 (Encysive Pharmaceuticals), is an orally-bioavailable non-peptidic antagonist with a sub-nanomolar IC50 (Wu et al., 2001). Because of this and its short half life (~4 h), it is an ideal compound by which to elucidate the timing of Ednra function in vivo. Wild type pregnant female mice were given the antagonist by gavage between E8.0 and E10.0, a time period covering both migration of cranial NCCs to the arches, and their subsequent patterning, proliferation and initial differentiation. The antagonist was given at either one time point or two time points separated by 12 h. This latter dosing was performed to ensure that with the short half-life of the antagonist, adequate receptor blocking was achieved.
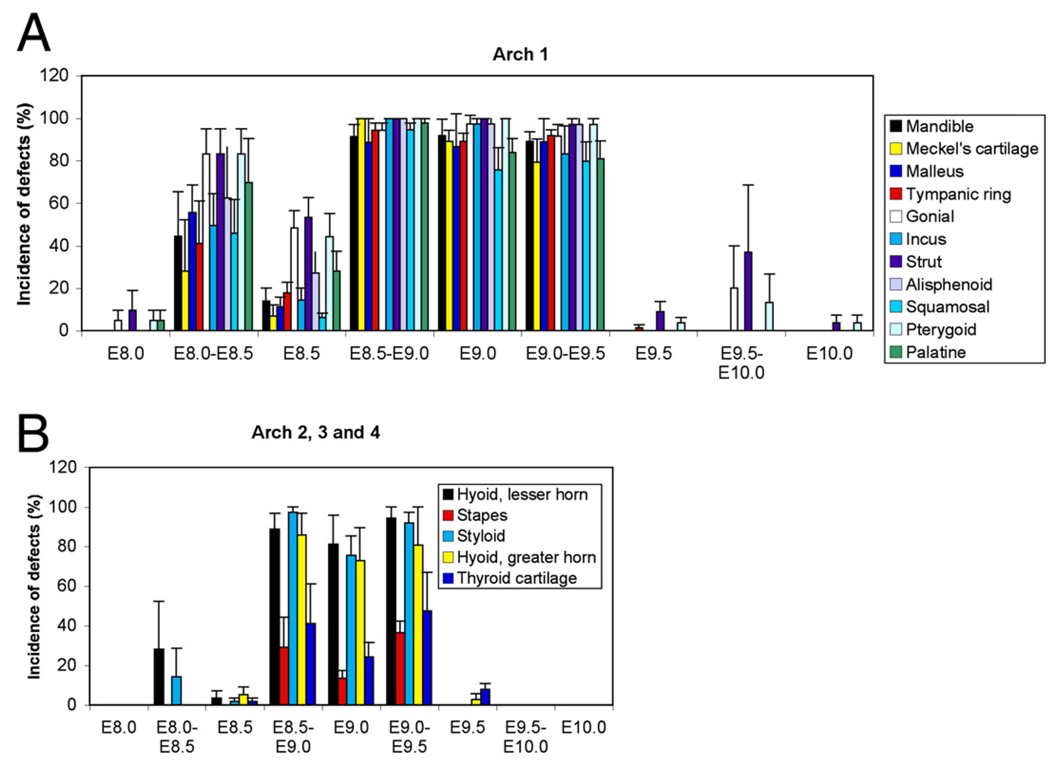
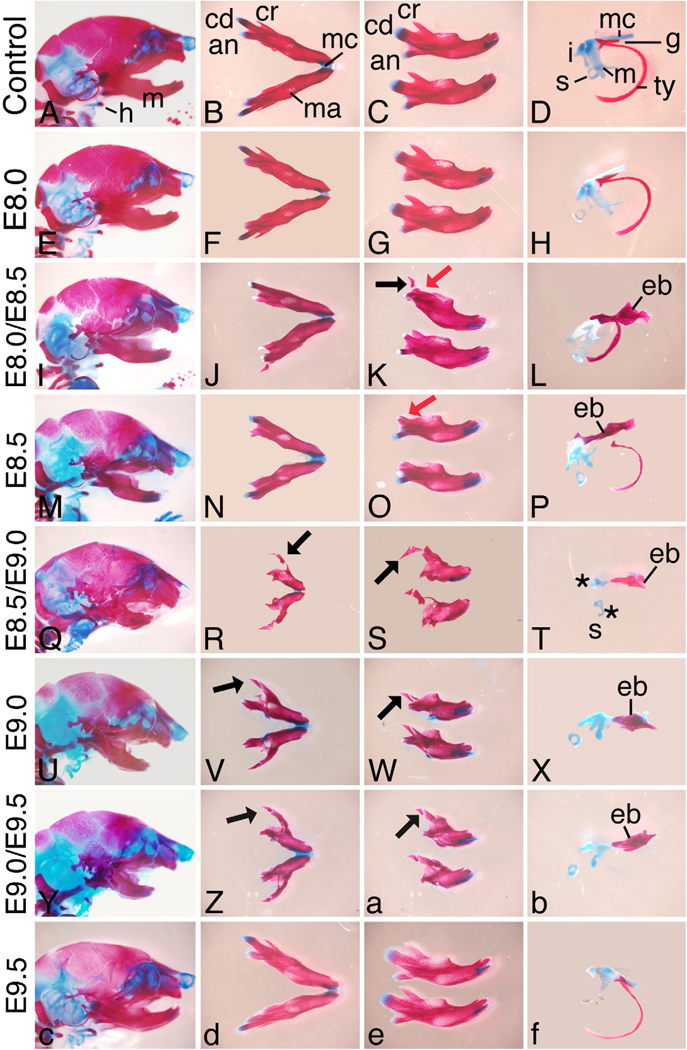
While Ednra signaling likely acts through IP3 intracellular signaling pathway (Ivey et al., 2003; Dettlaff-Swierez et al, 2005; Walker et al., 2007) that can be difficult to measure in vivo, we examined both bone and cartilage formation and gene expression to confirm receptor blockade and to determine the crucial timing of Ednra action. To first examine the affect of Ednra blockade on overall facial development, embryos from treated females were collected at E18.5 and stained with alizarin red and alcian blue. Skeletal structures derived from the pharyngeal archeswere then scored for defects, with any deviation from normal structure scored as a defect. Antagonist treatment at E8.0 resulted in minor changes in about 10% of embryos, with defects confined to the gonial, pterygoid and squamosal bones (Fig. 7A, 8E–H, Supplementary Fig. 3C, D. Supplementary Table 1). The changes in the pterygoid and gonial coincided with the presence of an ectopic bone in the skull base (Supplementary Table 1); this structure was similar to the one observed in the Ednrafl/fl;Wnt1-Cre embryos (Fig. 5E). In contrast, treatment at E8.0/E8.5 resulted in defects in all first arch-derived structures (Figs. 7A, 8I–L and Supplementary Fig. 3E, F. Supplementary Table 1). These included slight hypoplasia of many first arch derivatives and the presence of the ectopic bone (Fig. 8L). In addition, a duplication of the jugal bone was present (Fig. 8K). Defects were also present in arch 2 derivatives (Fig. 7B), while defects were not observed arch 3 and 4 derivatives. Treatment at E8.5 also resulted in defects similar to those observed with antagonist treatment at E8.0/E8.5, though the overall incidence was decreased alone (Figs. 7A, 8M–P, Supplementary Figs. 3G, H and Supplementary Table 1). Defects were also observed at a low incidence rate in arch 2, 3 and 4 derivatives (Fig. 7B).
Fig. 7.
Affect of temporal antagonist treatment on facial development. Incidence of defects per litter was analyzed for each enumerated structure and treatment modality. The error bars represent the standard deviation (see Materials and methods for a detailed description of data analysis). (A) Analysis of the incidence of defects in structures derived from the first pharyngeal arch. (B) Incidence of defects in structures derived form the second, third and fourth pharyngeal arch.
Fig. 8.
Analysis of craniofacial skeleton in E18.5 embryos. Lateral view of the skull (A, E,M, Q, U, Y and c), ventral (B, F, J, N, R, V, Z and d), intra-lateral (top) and contra-lateral (bottom) (C, G, K, O, S,W, a and e) views of the mandible bone and lateral view of middle ear structures (D, H, L, P, T, X, b and f) of embryos from females treated with either water at E8.5/E9.0 (A–D) or the antagonist at the listed time points (E–f). Antagonist treatment as early as E8.5/E9.0 begins to cause changes in the structure of the mandible bone and middle ear structures. The mandible becomes shortened, with loss of the coronoid (cr) process. In embryo treated between E8.5/E9.0 and E9.0/E9.5, a bone resembling the jugal bone of the zygomatic arch is observed in place of the coronoid process (black arrows in panels S,Wand a). While shortened, the mandible bone retains an obvious mandibular structure. Lower incisors are present at all treatment regimens. In the middle ear, an ectopic bone strut (eb) is obvious following treatment between E8.0/E8.5 to E9.0/E9.5. The malleus (m) and incus (i) become increasingly dysmorphic, with treatment at E8.5/E9.0 resulting in almost complete loss of both elements. an, angular process; cd, condylar process; g, gonial bone; mc, Meckel's cartilage; s, stapes; s*, malformed stapes; ty, tympanic ring bone.
Antagonist treatment at E8.5/E9.0, E9.0 and E9.0/9.5 all resulted in severe defects in first arch derivatives, with little variation observed in the type or incidence of defect within or between groups. Defects included significant mandibular hypoplasia, duplication of the jugal and alisphenoid bones, loss or severe malformation of the malleus, loss of the tympanic ring bone and Meckel's cartilage and the presence of the ectopic bones (Fig. 7A, Fig. 8Q–b and Supplementary Fig. 3I–P). Defects in structures derived from the more caudal arches included fusion of the hyoid bone to the pterygoid bones (following blockade at E8.5/E9.0 and E9.0), similar to the fusion and disruption of normal thyroid cartilage architecture (Fig. 7B and Supplementary Table 1). All observed defects were consistent with those observed in Ednra−/− and Ednrafl/fl;Wnt1-Cre embryos. One notable exception was the mandible, which while hypoplastic, never showed a complete transformation to a maxilla-like structure (as described above, a duplication of the jugal bone was observed).
In contrast to the changes observed with treatment between E8.5 and E9.0, treatment at E9.5 alone resulted in very few changes (Fig. 7A, B, Fig. 8c–f, Supplementary Figs. 3O, P), through the ectopic bone was still present in 10% of embryos (Supplementary Table 1). In addition, defects in arches 3 and 4 were still present at a low level (Supplementary Table 1). The only defects observed following treatment at E9.5/10.0 were defects in the gonial, pterygoid and the presence of the ectopic bone (possibly all due to presence of the ectopic bone) (Supplementary Figs. 3Q, R), though these defects were observed in up to 40% of embryos (Supplementary Table 1). Antagonist treatment at E10.0 alone resulted in low incidence of defects, limited to the pterygoid and ectopic bone (Supplementary Figs. 3S, T and Supplementary Table 1).
Changes in gene expression following Ednra antagonism
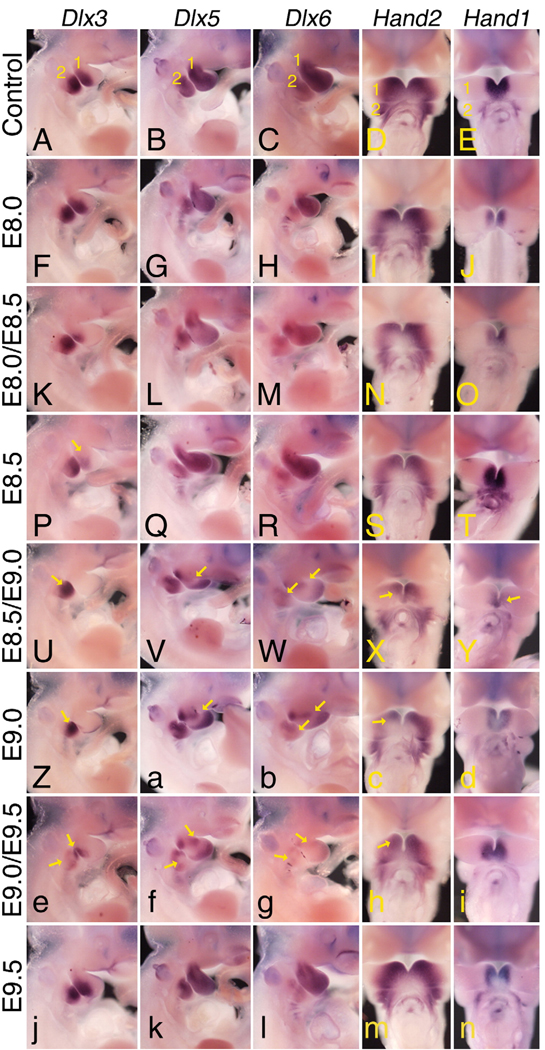
To examine how these changes in jaw and middle ear structures were reflected in changes at the molecular level, we examined the expression pattern of five genes whose expression is disrupted in Ednra−/− and Edn1−/− embryos. Antagonist treatment at E8.0 did not result in consistent changes in gene expression in Dlx3, Dlx5, Dlx6 and Hand2, though the Hand1 expression domain was shifted distally (Figs. 9A–E). After treatment at E8.0/E8.5, the expression of Dlx3 was weaker in the first arch; Hand1 expression was again shifted distally in the mandibular arch, with Dlx6 unchanged (Figs. 9K–O). Following antagonist treatment at E8.5 alone, only Dlx3 showed a clear change in the first arch (yellow arrow in Fig. 9P). However, following treatment at E8.5/E9.0 (Figs. 9U–Y) or E9.0 alone (Figs. 9Z–d), mandibular arch expression of all five genes was changed to some extent, with Dlx3 being virtually absent in the first arch at both time points (Figs. 9U, Z). Interestingly, Hand1 (Figs. 9Y, d) and Hand2 (Figs. 9X, c) expression along the rostral mandibular arch was never lost, which may explain the absence of distal mandibular defects. Antagonist treatment at E9.0/E9.5 resulted in disrupted expression of Dlx3, Dlx5 and Dlx6 in the first and second arches, Hand1 and Hand2 less severely affected (Figs. 9e–i). The expression of all five genes in arches one and two appeared unaffected by antagonist treatment at E9.5 (Figs. 9j–n). Together, these results illustrate that blocking Ednra signaling at discrete time periods affects the development of particular craniofacial structures by disrupting specific temporal signaling networks responsible for rostral–caudal patterning of the pharyngeal arches.
Fig. 9.
Whole mount in situ hybridization analysis following antagonist treatment. Lateral (Dlx3, Dlx5 and Dlx6) and ventral (Hand2 and Hand1) views of control (A–E) and TBC3214-treated (F–n) embryos hybridized with DIG-labeled cRNA riboprobes against Dlx3 (A, F, K, P, U, Z, e and j), Dlx5 (B, G, L, Q, V, a, f and k) and Dlx6 (C, H, M, R,W, b, g and l), Hand2 (D, I, N, S, X, c, h and m) and Hand1 (E, J, O, T, Y, d, i and n). Each of the genes examined shows a distinct sensitivity to antagonist treatment. All three Dlx genes first show loss of expression in the first arch after antagonist treatment at E8.0/E8.5, with loss in the second arch appearing after treatment at E9.0/E9.5. By E9.5, expression is fully restored. Note that disrupted expression of both Dlx5 and Dlx6 is more evident at E8.5 than at E8.0/E8.5, reflecting the differences observed in skeletal defects at these time periods. Hand2 and Hand1 expression show a narrower temporal sensitivity to antagonist treatment, with disrupted expression primarily observed between E8.5 and E9.0.
Discussion
Here we have shown that Ednra signaling is crucial for early cranial NCC development. Using conditional inactivation of the Ednra gene in cranial NCCs at both E8.5 and E9.5, we found that only inactivation at E8.5 resulted in craniofacial defects, with these defects resembling those of Ednra−/− embryos (Clouthier et al., 1998; Ruest et al., 2004). Inactivation of Ednra at E9.5 did not disrupt normal jaw development. We further refined this timing by blocking Ednra signaling in pregnant wild type female mice to show that Ednra signaling is required between E8.25 and E9.0. Again, blocking receptor activity after E9.0 resulted in very few changes in skull structure. In both cases, an ectopic bone formed between the gonial bone and pterygoid bone. These findings support a model in which Ednra signaling is required for both early establishment of a proximal distal patterning program and induction of a gene expression profile necessary to execute this program. Further, we propose that increasing Ednra signaling levels in the mandibular arch may have contributed to the evolution of the vertebrate middle ear.
Timing and function of Ednra signaling
In this current study, we have shown that Ednra signaling is first required by NCCs between E8.0 and E8.5, with this requirement continuing between E9.0 and E9.5. This time period is earlier than previously described using short term in vitro culture in the presence of another Ednra antagonist (Fukuhara et al., 2004), with the difference likely reflecting the potency of the antagonist for its receptor (IC50: 40 pM) and increased sensitivity of using an orally bioavailable antagonist coupled with analysis of embryos at E18.5. Initial Ednra activity between E8.0 and E8.5 could suggest that Ednra signaling is either required for NCC movement/targeting within the arches to specific arch sub-domains or required for immediate patterning of post-migratory NCCs. Recent evidence would favor the latter explanation. Ednra signaling is not required for mouse cranial NCCs to reach the pharyngeal arches (Abe et al., 2007; Clouthier et al., 2003), even though Ednra expression is observed in migrating NCCs by E8.25 (Sato et al., 2008) and is not required by NCC-derived cells in the arches after E10.5 (Ruest et al., 2005). Similarly, Ednra1;Ednra2 double morphant zebrafish embryos also do not have defects in NCC migration (Nair et al., 2007). In addition, crude injection of human EDN1 into the mandibular arch of suc;edn1−/− zebrafish after most migration is complete leads to a rescue of the suc;edn1−/− phenotype (Miller et al., 2000). Together, these findings have led to the hypothesis that Ednra signaling establishes or maintains patterning along the arch dorsal–ventral axis (Nair et al., 2007). If this hypothesis is correct, our current findings indicate that this patterning process is initiated very early in arch development, likely immediately before or after NCCs reach their destination. We cannot determine whether the early sensitivity to Ednra blockade (the E8.0/E8.5 window) represents a specific developmental event, since receptor blockade at E8.0/E8.5 produces defects always observed at later receptor blockade time points.
It is plausible that, like in the zebrafish, early signaling establishes the D-V pattern and that later Ednra signaling reinforces this developmental program (Nair et al., 2007). However, directly assessing the similarity in temporal signaling between mouse and zebrafish is difficult due to the presence of two ednra genes in zebrafish, one of which is found in the arch ectoderm and may positively regulate edn1 expression (Nair et al., 2007). In the mouse, there is only one known Ednra gene, with its expression in the arches confined to the NCC-derived mesenchyme (Clouthier et al., 1998; Yanagisawa et al., 1998a; Yanagisawa, 1994). Our present finding that the phenotype of E18.5 Ednrafl/fl;Wnt1-Cre embryos is identical to that of E18.5 Ednra−/− embryos confirms this fact.
Several aspects of arch development that appear to be shared between both mouse and zebrafish are that Ednra-specific D-V patterning along the arch axis occurs over a rather long period (~18 h in the mouse) and that disrupting Ednra signaling at any point during this time disrupts lower jaw development. Further, an exquisite sensitivity to the level of Ednra signaling exists in NCCs. In Ednra1 morphant zebrafish embryos, distal lower jaw development is normal while the intermediate domain is disrupted (Nair et al., 2007). In Ednra1; Ednra2 double morphants, both distal and intermediate domains are disrupted. Thus, the overall level of Ednra signaling is crucial in directing different aspects of mandibular arch patterning. This view is supported by our current finding that while most defects observed in Ednra−/− embryos are observed following Ednra antagonism between E8.5 and E9.0, complete homeotic transformation of the mandible into a maxilla is not observed following any of the one- or two-dose antagonist treatment regimens. This indicates that the length of antagonist administration was not sufficient to produce this specific homeotic event and further highlights that the specification of NCCs towards a specific skeletal lineage is a broad developmental event requiring changes in the expression of numerous genes over an extended development window.
A second hypothesis for Ednra action has been proposed in which Ednra signaling regulates segregation of NCCs to the distal mandibular arch (Walker et al., 2006). In suc;edn1−/− embryos, the mandibular arch does not appear to properly elongate, though this defect is not present in older mutant embryos. This outgrowth defect has led to the proposal that absence of Edn1 leads to an initial failure of arch elongation (Walker et al., 2006, 2007). Inhibiting elongation would disrupt a proposed Edn1 gradient within the mandibular arch that is hypothesized to contribute to D-V patterning (Kimmel et al., 2003, 2007). These changes would subsequently result in a failure to segregate and pattern intermediate and ventral populations of NCCs within the arch. While we cannot rule out a role for Ednra signaling in mandibular arch outgrowth in the mouse, delayed elongation of this arch is not readily apparent (data not shown). Further, a role for Ednra signaling in NCC segregation would have to be restricted to a subset of NCCs within the mandibular arch. In Ednra−/− embryos, global changes in gene expression within the intermediate and distal regions of the pharyngeal arches are not observed (Clouthier et al., 1998, 2000; Ruest et al., 2004). While it is possible that D-V patterning in the mandibular arch of mice and zebrafish occurs through different mechanisms, we would propose that NCCs of different midbrain/hindbrain regions have differential requirements for Ednra signaling. Chimera analysis in mice has shown that all first arch structures that have a cell autonomous requirement for Ednra signaling contain a midbrain NCC contribution (Clouthier et al., 2003), based on homology with chick NCC fate maps (Kontges and Lumsden, 1996). Perhaps it is these cells in the first arch that are most sensitive to Ednra signaling for D-V patterning and/or segregation. Conditional inactivation of the Ednra gene in a hindbrain or rhombomere-specific manner will be required to address this question.
Changes in malleus shape and the importance of endothelin signaling
Varying Edn1 levels are believed to play a role in the shape and size of second arch elements in zebrafish, possibly reflecting a gradient of Edn1 within the arch. These elements, the opercle and branchiostegal rays, are part of the gill breathing apparatus. Following partial downregulation of edn1 using edn1 morpholinos, an increase in the size of the opercle was observed (referred to as the opercle-gain phenotype (Kimmel et al., 2003). The opercle-gain phenotype is hypothesized to reflect a change in an Edn1 gradient within the arch, leading to expansion of the future opercle (Kimmel et al., 2003). Here we have shown that the malleus, a first arch derivative, is larger than normal in 75% of in Ednrafl/fl;Wnt1-Cre embryos. While many aspects of vertebrate auditory system evolution are controversial, it is clear that changes in ossicle size have occurred throughout evolution, resulting in varying frequency reception (Allin, 1975). It is tempting to speculate that increasing Ednra signaling during mammalian evolution (through expansion of an Edn1 gradient or other mechanisms are not well understood) has been key to the development of multiple structures within the pharyngeal arches, including the gill apparatus and middle ear. To address this possibility, it will be important to determine if the appearance of the endothelin gene in the vertebrate lineage corresponds to anatomical changes within the middle ear.
Supplementary Material
Acknowledgments
The authors would like to thank James E.T. Smith, Bo Mason, Tinisha Taylor, Akiko Abe and Katherine Kuhn for their technical assistance, Makoto Abe, Joy Richman, Giovanni Levi, Guillermo Rougier and Jixiang Ding for their helpful discussions and Eugenia C. Olesnicky Killian for the suggestions, discussions and critical review of the manuscript. The authors would like to specifically thank Tom Brock of Encysive Pharmaceuticals for generously providing TBC3214 for this study. This work was supported in part by grants from the Kentucky Excellence in Education Research Trust Fund, the American Heart Association Ohio Valley Affiliate (0160184B and 355379B) and the NIH (DE14181) to D.E.C. D.E.C. was a recipient of a Career Development Award from the NIH/NIDCR (DE14765). L.B.R. was a recipient of a Research Fellowship from the Heart and Stroke Foundation of Canada.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2009.01.005.
References
- Abe M, Ruest L-B, Clouthier DE. Fate of cranial neural crest cells during craniofacial development in endothelin-A receptor deficient mice. Int. J. Dev. Biol. 2007;51:97–105. doi: 10.1387/ijdb.062237ma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allin EF. Evolution of the mammalian middle ear. J. Morphol. 1975;147:403–438. doi: 10.1002/jmor.1051470404. [DOI] [PubMed] [Google Scholar]
- Beverdam A, Merlo GR, Paleari L, Mantero S, Genova F, Barbieri O, Janvier P, Levi G. Jaw transformation with gain of symmetry after Dlx5/Dlx 6 inactivation: mirror of the past. Genesis. 2002;34:221–227. doi: 10.1002/gene.10156. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson JRE. Recent advances in craniofacial morphogenesis. Dev. Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Han J, Rositch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, Kumada M, Hammer RE, Yanagisawa M. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev. Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Hammer RE, Richardson JA, Yanagisawa M. Cell-autonomous and nonautonomous actions of endothelin-A receptor signaling in craniofacial and cardiovascular development. Dev. Biol. 2003;261:506–519. doi: 10.1016/s0012-1606(03)00128-3. [DOI] [PubMed] [Google Scholar]
- Couly GF, Coltey PM, Le Douarin NM. The triple origin of skull in higher vertebrates: a study in chick-quail chimeras. Development. 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- Couly GF, Grapin-Botton A, Coltey P, Le Douarin NM. The regeneration of the cephalic neural crest, a problem revisited: the regenerating cells originate from the contralateral or from the anterior and posterior neural fold. Development. 1996;122:3393–3407. doi: 10.1242/dev.122.11.3393. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Current Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Dettlaff-Swierez DA, Wettschureck N, Moers A, Huber K, Offermanns S. Characteristic defects in neural crest cell-specific Gαq/Gα11- and Gα12/Gα13-deficient mice. Dev. Biol. 2005;282:174–182. doi: 10.1016/j.ydbio.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- Fraser SE, Keynes RJ, Lumsden AGS. Segmentation in the chick embryo hindbrain is defined by cell lineage restriction. Nature. 1990;344:431–435. doi: 10.1038/344431a0. [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Kurihara Y, Arima Y, Yamada N, Kurihara H. Temporal requirement of signaling cascade involving endothelin-1/endothelin receptor type A in branchial arch development. Mech. Dev. 2004;121:1223–1233. doi: 10.1016/j.mod.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Ivey K, Tyson B, Ukidwe P, McFadden DG, Levi G, Olson EN, Srivastava D, Wilkie TM. Gαq and Gα11 proteins mediate endothelin-1 signaling in neural crest-derived pharyngeal arch mesenchyme. Dev. Biol. 2003;255:230–237. doi: 10.1016/s0012-1606(02)00097-0. [DOI] [PubMed] [Google Scholar]
- Kedzierski RM, Grayburn PA, Kisanuki YY, Williams CS, Hammer RE, Richardson JA, Schneider MD, Yanagisawa M. Cardiomyocyte-specific endothelin a receptor knockout mice have normal cardiac function and an unaltered hypertrophic response to angiotensin II and isoproterenol. Mol. Cell. Biol. 2003;23:8226–8232. doi: 10.1128/MCB.23.22.8226-8232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf H, Linares C, Corvol P, Gasc JM. Pharmacological inactivation of the endothelin type A receptor in the early chick embryo: a model of mispatterning of the branchial arch derivatives. Development. 1998;125:4931–4941. doi: 10.1242/dev.125.24.4931. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ullmann B, Walker M, Miller CT, Crump JG. Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development. 2003;130:1339–1351. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Walker MB, Miller CT. Morphing the hyomandibular skeleton in development and evolution. J. Exp. Zool. (Mol. Dev. Evol.) 2007;308B:609–624. doi: 10.1002/jez.b.21155. [DOI] [PubMed] [Google Scholar]
- Kitano Y, Kurihara H, Kurihara Y, Maemura K, Ryo Y, Yazaki Y, Harii K. Gene expression of bone matrix proteins and endothelin receptors in endothelin-1-deficient mice revealed by in situ hybridization. J. Bone Miner. Res. 1998;13:237–244. doi: 10.1359/jbmr.1998.13.2.237. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, Oda H, Kuwaki T, Cao W-H, Kamada N, Jishage K, Ouchi Y, Azuma S, Toyoda Y, Ishikawa T, Kumada M, Yazaki Y. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- Kwan K-M. Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis. 2002;32:49–62. doi: 10.1002/gene.10068. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Ziller C, Couly GF. Patterning of neural crest derivatives in the avian embryo: in vivo and in vitro studies. Dev. Biol. 1993;159:24–49. doi: 10.1006/dbio.1993.1219. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Sprawson N, Graham A. Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development. 1991;113:1281–1291. doi: 10.1242/dev.113.4.1281. [DOI] [PubMed] [Google Scholar]
- Maemura K, Kurihara H, Kurihara Y, Oda H, Ishikawa T, Copeland NG, Gilbert DJ, Jenkins NA, Yazaki Y. Sequence analysis, chromosomal location, and developmental expression of the mouse preproendothelin-1 gene. Genomics. 1996;31:177–184. doi: 10.1006/geno.1996.0029. [DOI] [PubMed] [Google Scholar]
- McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- Miller CT, Kimmel CB. Morpholino phenocopies of endothelin 1 (sucker) and other anterior arch class mutations. Genesis. 2001;30:186–187. doi: 10.1002/gene.1061. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K-H, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3838. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DYR, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003;130:1353–1365. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, Kimmel CB. mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish. Dev. Biol. 2007;308:144–157. doi: 10.1016/j.ydbio.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muelemans D, Bronner-Fraser M. Insights from amphioxus into the evolution of vertebrate cartilage. PLoS One. 2007;2:e787. doi: 10.1371/journal.pone.0000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Nair S, Li W, Cornell R, Schilling TF. Requirements for endothelin type-A receptors and endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebrafish. Development. 2007;134:335–345. doi: 10.1242/dev.02704. [DOI] [PubMed] [Google Scholar]
- Noden DM. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev. Biol. 1983;96:144–165. doi: 10.1016/0012-1606(83)90318-4. [DOI] [PubMed] [Google Scholar]
- Noden DM. Interactions and fates of avian craniofacial mesenchyme. Development. 1988;103:121–140. doi: 10.1242/dev.103.Supplement.121. [DOI] [PubMed] [Google Scholar]
- Ozeki H, Kurihara Y, Tonami K, Watatani K, Kurihara H. Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech. Dev. 2004;121:387–395. doi: 10.1016/j.mod.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Padilla DJ, Epp TS, McDonough P, Marlin DJ, Erickson HH, Poole DC. Effects of a specific endothelin-1A antagonist on exercise-induced pulmonary haemorrhage (EIPH) in thoroughbred horses. Equine Vet. J. 2006 Suppl. 36:198–203. doi: 10.1111/j.2042-3306.2006.tb05539.x. [DOI] [PubMed] [Google Scholar]
- Ruest L-B, Dager M, Yanagisawa H, Charité J, Hammer RE, Olson EN, Yanagisawa M, Clouthier DE. dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev. Biol. 2003;257:263–277. doi: 10.1016/s0012-1606(03)00068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest L-B, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development. 2004;131:4413–4423. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest L-B, Kedzierski R, Yanagisawa M, Clouthier DE. Deletion of the endothelin-A receptor gene within the developing mandible. Cell Tissue Res. 2005;319:447–454. doi: 10.1007/s00441-004-0988-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Kawamura Y, Asai R, Amano T, Uchijima Y, Dettlaff-Swiercz DA, Offermanns S, Kurihara Y, Kurihara H. Recombinase-mediated cassette exchange reveals the selective use of Gq/G11-dependent and -independent endothelin 1-endothelin type A receptor signaling in pharyngeal arch development. Development. 2008;135:755–765. doi: 10.1242/dev.012708. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Spence S, Anderson C, Cukierski M, Patrick D. Teratogenic effects of the endothelin receptor antagonist L-753,037 in the rat. Reprod. Toxicol. 1999;13:15–29. doi: 10.1016/s0890-6238(98)00064-1. [DOI] [PubMed] [Google Scholar]
- Sprogar S, Volk J, Drevensek M, Drevensek G. The effects of TBC3214, a selective endothelin ETA receptor antagonist, on orthodontic tooth movements in rats. Eur. J. Orthod. 2007;29:605–608. doi: 10.1093/ejo/cjm064. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Agarwar P, Brown C, McCulley DJ, Schwarz JJ, Black BL. The transcription factor MEF2C is required for craniofacial development. Dev. Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Talbot JC, Stock DW, Kimmel CB. Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev. Biol. 2006;295:194–205. doi: 10.1016/j.ydbio.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Swartz ME, Eberhart JK, Kimmel CB. phospholipase C, beta 3 is required for Endothelin1 regulation of pharyngeal arch patterning in zebrafish. Dev. Biol. 2007;304:194–207. doi: 10.1016/j.ydbio.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Decker ER, Blok N, Li J, Bourgoyne AR, Bui H, Keller KM, Knowles V, Li W, Stavros FD, Holland GW, Brock TA, Dixon RAF. Acyl substitution at the ortho position of anilides enhances oral bioavailability of thiophene sulfonamides: TBC2314, and ETA selective endothelin antagonist. J. Med. Chem. 2001;44:1211–1216. doi: 10.1021/jm000349x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M. The endothelin system: a new target for therapeutic intervention. Circulation. 1994;89:1320–1322. doi: 10.1161/01.cir.89.3.1320. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, de Wit D, Emoto N, Hammer RE. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998a:825–836. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Hammer RE, Richardson JA, Williams SC, Clouthier DE, Yanagisawa M. Role of endothelin-1/endothelin-A receptor-mediated signaling pathway in the aortic arch patterning in mice. J. Clin. Invest. 1998b;102:22–33. doi: 10.1172/JCI2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DY. The bHLH transcription factor hand2 plays parallel roles in zebrafish and pectoral fin development. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.