Abstract
Nuclear pore complexes have recently been shown to play roles in gene activation, however their potential involvement in metazoan transcription remains unclear. Here we show that the nucleoporins Sec13, Nup98 and Nup88, as well as a group of FG-repeat nucleoporins, bind to the Drosophila genome at functionally distinct loci that often do not represent NE contact sites. While Nup88 localizes to silent loci, Sec13, Nup98 and a subset of FG-repeat nucleoporins bind to developmentally regulated genes undergoing transcription induction. Strikingly, RNAi-mediated knockdown of intranuclear Sec13 and Nup98 specifically inhibits transcription of their target genes and prevents efficient reactivation of transcription after heat shock, suggesting an essential role of NPC components in regulating complex gene expression programs of multicellular organisms.
INTRODUCTION
In order to accommodate transport between the nucleoplasm and the cytoplasm, the nuclear envelope (NE) is fenestrated by nuclear pore complexes (NPCs), large multiprotein channels consisting of multiple copies of ~ 30 different nucleoporins (Nups) (Alber et al., 2007a; Hetzer et al., 2005; Wente, 2000). Nups can be classified into two categories: (i) scaffold Nups, which mainly consist of the large Nup107/160 and Nup93/205 complexes (Debler et al., 2008) and (ii) peripheral Nups. The latter extend from the membrane-embedded scaffold either into the pore channels, or as filaments into the cytoplasm or the nucleoplasm (Alber et al., 2007b; Beck et al., 2004; Brohawn et al., 2009). While the scaffold is thought to provide structural integrity to the highly curved pore membrane, the peripheral Nups, many of which contain phenylalanine-glycine (FG)-repeats, are responsible for establishing the permeability barrier (D’Angelo et al., 2009) and mediating nuclear trafficking (Weis, 2002).
In addition to their role as transport channels, NPCs have been implicated in chromatin organization and gene regulation (Akhtar and Gasser, 2007; Capelson and Hetzer, 2009). Studies in yeast revealed that Nups can associate with promoters of active genes (Schmid et al., 2006) and that the expression of inducible genes is increased by interactions with nuclear pores (Taddei et al., 2006). Furthermore, a genome-wide analysis in S. cerevisiae demonstrated that a subset of Nups can occupy regions of highly transcribed genes (Casolari et al., 2004). Additionally, Nups have been shown to function as chromatin boundaries in S. cerevisiae (Dilworth et al., 2005; Ishii et al., 2002). Boundary activity involves protection from nearby activating or repressive signals and constitutes another plausible function for NPCs in the organization of the genome into discrete chromatin domains. As further evidence for the role of the NPC in regulation of active chromatin, Nups have been found to participate in X-chromosome transcriptional hyper-activation in dosage compensation of Drosophila melanogaster (Mendjan et al., 2006).
Interestingly, the only genome-wide study of Nup-chromatin association in animal cells revealed a correlation between the binding sites of Nups and regions enriched in repressive histone modifications (Brown et al., 2008), which exhibited characteristics of sequences known to associate with the nuclear periphery in human cells (Guelen et al., 2008). The observed discrepancy between yeast and human data suggests that the genome-binding pattern of the NPC may be quite different or more complex in metazoa. Furthermore, many of the peripheral Nups in mammalian cells have been shown to be mobile and to move dynamically on and off the pore (Rabut et al., 2004). Therefore, it seems possible that Nup-chromatin interactions could occur at distant sites from the NE, a notion that has some experimental support from the observation of intranuclear Nups (i.e. not associated with the NE) in mammalian cells (Enninga et al., 2003; Griffis et al., 2002). However, the functional role of Nup-chromatin interactions and whether they occur exclusively at the nuclear periphery remain unresolved.
Given the functional implications of yeast Nups in gene regulation, we wanted to test if NPC components play a role in gene expression of multicellular organisms. Here we demonstrate that different Nups bind to distinct regions of the Drosophila genome and that many of these interactions can occur at off-pore locations. More significantly, we show that a subset of NE-independent NPC proteins play an essential role in the induction of transcription of its target genes during Drosophila development, suggesting a direct function for Nups in the regulation of gene expression in metazoa.
RESULTS
A subset of Nups binds to specific sites of the Drosophila genome
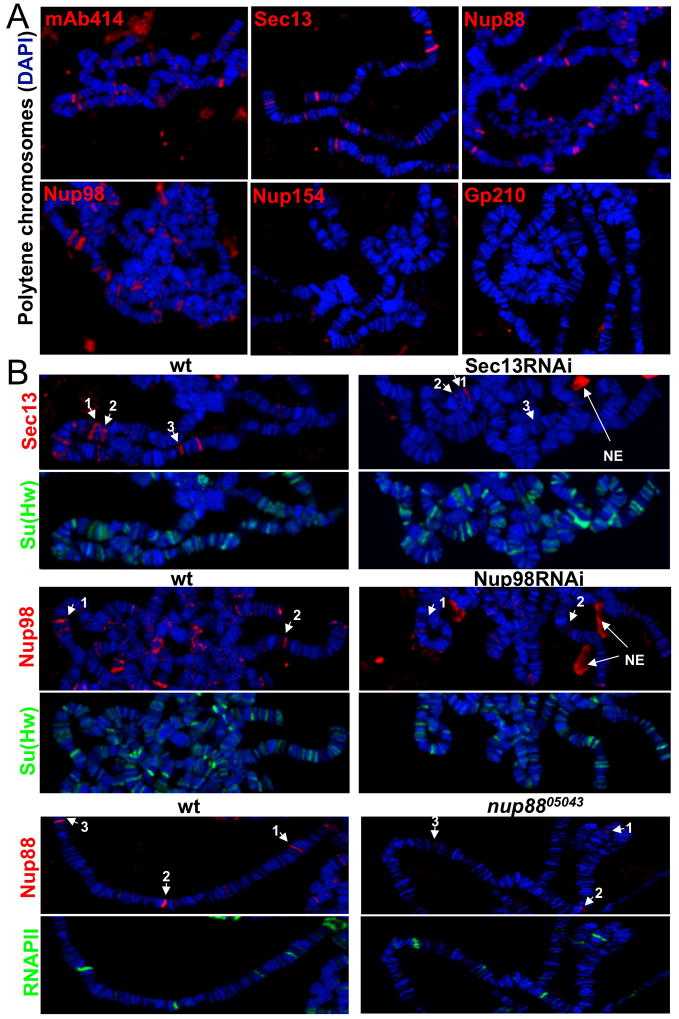
To study the potential role of Nups in metazoan gene regulation, we performed indirect immunofluorescence on polytene chromosome spreads using the antibody mAb414, which is a widely used marker of NE-associated NPCs and has been shown to react with the FG-repeat domain of Nup62, Nup153, Nup214 and Nup358 (Davis and Blobel, 1987). Strikingly, mAb414 stained dozens of specific sites on the chromosomes (Figure 1A). The staining pattern was highly reproducible among chromosomes of the same animal, but varied among larvae of different developmental stages (see below). To determine whether additional Nups might exhibit chromatin-binding behavior, we used specific antibodies against representative components of the major sub-complexes of the NPC (Figure S2A), including Sec13, a component of the stable Nup107/160 sub-complex (Siniossoglou et al., 1996; Vasu et al., 2001) and Nup154, a member of the stable Nup93/205 sub-complex (Gigliotti et al., 1998; Hawryluk-Gara et al., 2005). In addition, antibodies against the cytoplasmic filament component Nup88 (Roth et al., 2003), the nucleoplasmic pore component Nup98 and the transmembrane nucleoporin gp210 (Greber et al., 1990) were analyzed. As expected, all antibodies were able to stain nuclear pores at the NE in cells of fly imaginal discs (Figure S1A). On chromosomes, we detected specific binding patterns with anti-Sec13, anti-Nup88 and anti-Nup98 antibodies (Figure 1A), suggesting that various pore components are targeted to particular genomic sites. In contrast, the scaffold Nup154 and the transmembrane gp210 did not associate with chromatin.
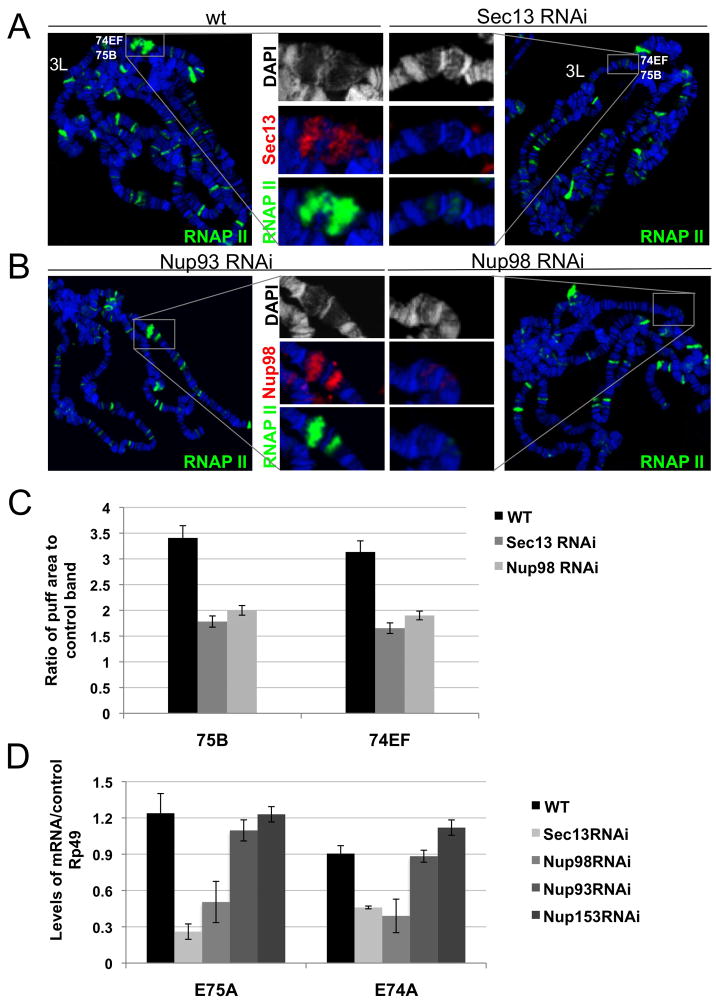
Figure 1. A subset of nuclear pore components binds specific sites in the Drosophila genome.
(A) Polytene chromosome spreads were stained with mAb414, anti-Sec13, anti-Nup88, anti-Nup98, anti-Nup154 and anti-gp210 as indicated; chromosomes are stained with DAPI (blue) here and throughout the paper, unless otherwise indicated. (B) Polytene chromosomes of third instar larvae of wandering LB stage from wt and Sec13 RNAi (top), Nup98 RNAi (middle) or nup8805043 homozygotes (bottom) were stained with anti-Sec13 and anti-Su(Hw), anti-Nup98 and a nti-Su(Hw), anti-Nup88 and anti-RNAP II, respectively. White arrows with numbers denote the same genomic locations for a particular Nup between chromosomes of wt and mutant larvae in a given panel. See also Figure S1.
To confirm the specificity of Nup-chromatin interactions, we knocked down Sec13 and Nup98 using inducible UAS-RNAi transgenic lines directed against the sec13 and nup98 gene transcripts (Dietzl et al., 2007), which can be activated in salivary glands by a GAL4 gene under the control of an Sgs3 promoter (Cherbas et al., 2003). The binding of both proteins at specific sites was reduced on chromosomes of larvae, in which Sec13 or Nup98 have been depleted by RNAi. In contrast, Suppressor of Hairy-wing (Su(Hw)), an unrelated chromatin-binding protein was not affected in these animals (Figure 1B). The extent of the RNAi-induced knockdown of expression and the specificity of employed antibodies were further evidenced by a decrease in the total levels of Sec13 and Nup98 mRNA and protein in the salivary glands of the knock-down larvae relative to wild-type (wt) (Figure S1B-D). Similarly, in a hypomorphic mutant of nup88, generated by the insertion of a P-element in the 5′ UTR of the nup88 gene and characterized previously (Roth et al., 2003), the staining of Nup88 was diminished at the specific binding sites found in wt animals (Figure 1B). Together, these results provide cell biological evidence that various NPC components are present at specific chromatin sites in differentiating animal cells.
NPC components can bind chromatin in an off-pore manner inside the nucleus
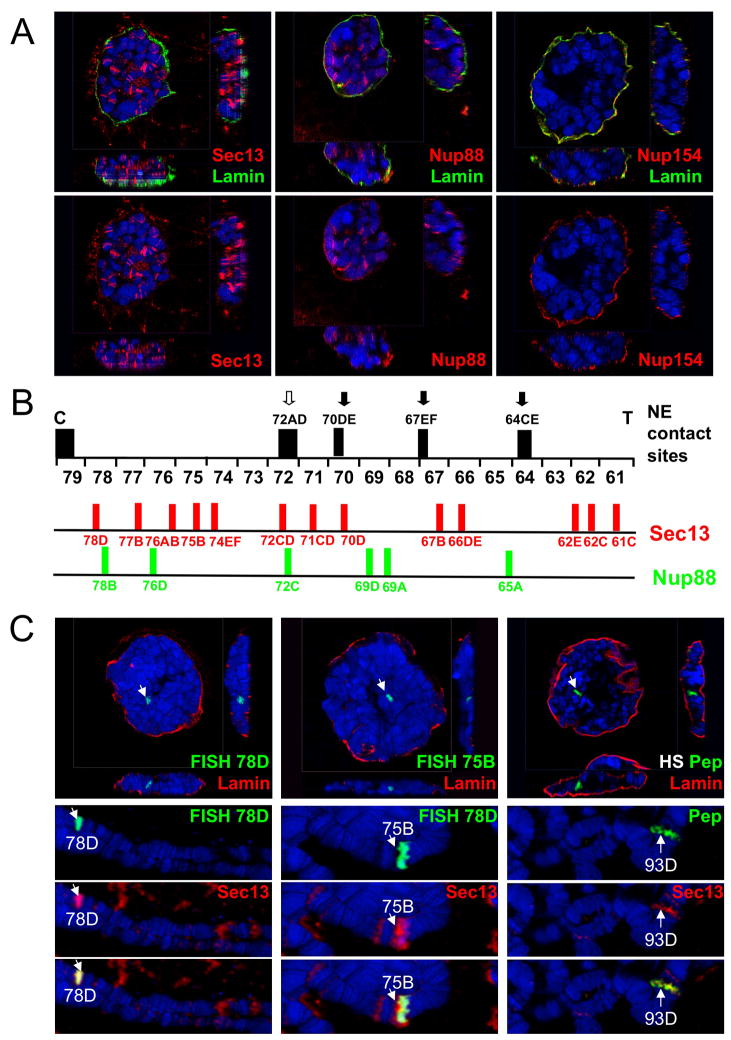
Detection of genome-wide binding of various NPC components raised the question if these contacts all occur at the NE. To analyze the subnuclear localization of Nup-chromatin interactions, we performed indirect immunofluorescence analysis on intact salivary gland nuclei using specific Nup antibodies in combination with an anti-Lamin antibody to mark the nuclear periphery (Gruenbaum et al., 2003). The nuclei were analyzed by confocal microscopy and three-dimensional reconstructions were generated from multiple z-stacks. Consistent with our findings on chromosome spreads, the stable component Nup154 exclusively stained the NE but was not detectable at chromosome sites (Figure 2A). In striking contrast, many of the chromatin-bound Sec13 and Nup88 loci did not colocalize with Lamin and thus the NE, suggesting that these sites are occupied by an intranuclear pool of NPC components (Figure 2A).
Figure 2. Nup-chromatin interactions occur independently of the NE.
(A) Fixed intact polytene nuclei were stained with anti-Lamin and anti-Sec13, anti-Nup88 or anti-Nup154 antibodies and analyzed by confocal microscopy. 3D reconstructions were generated from multiple z-stacks. (B) NE contact sites, which correspond to regions of high probability of contacting NE, derived from (Hochstrasser et al., 1986), were plotted against mapped binding sites of Sec13 and Nup88 along chromosome 3L. 61–79 correspond to 23 cytological divisions of 3L, each of which is further subdivided into 6 subdivisions A–F; C and T mark the centromere and telomere, respectively. NE contacts sites are marked by black (intercalary heterochromatin) or un-filled (not intercalary heterochromatin) arrows and approximate cytological locations. For Sec13, all observed binding sites on 3L during wandering third instar larval development are shown. (C) FISH probes generated from BAC clones spanning 78D and 75B were hybridized to either fixed intact polytene nuclei, followed by an anti-Lamin staining, or to chromosomes from the sister salivary glands, followed by anti-Sec13 staining; alternatively (right panel), fixed intact nuclei and chromosomes from the sister glands of heat-shocked (HS) larvae were stained with anti-Pep, which predominantly stains 93D after HS, and anti-Lamin or anti-Sec13 antibodies. Top panels show 3D reconstructions from z-stacks.
To obtain additional evidence for the NE-independent nature of Nup binding sites, we mapped Sec13 and Nup88 loci and compared them to the previously described NE contact sites of polytene chromosomes, which were computed as nuclear surface contact frequencies from sectioned salivary gland nuclei (Hochstrasser et al., 1986) (Figure 2B). A detailed analysis of chromosome 3L revealed that multiple Sec13 and Nup88 sites do not correspond to NE contact sites. In addition, we mapped the 3D localization of two prominent Sec13 binding sites, 78D and 75B, by fluorescent in situ hybridization (FISH) in intact polytene nuclei. These genomic sites, which co-localize with Sec13 on chromosomes were consistently detected away from the NE in the nucleoplasm (Figure 2C). These findings further support the notion of an intranuclear pool of Nups interacting with specific genomic sites away from the NE-embedded nuclear pores.
NPC components bind to functionally distinct regions of the genome
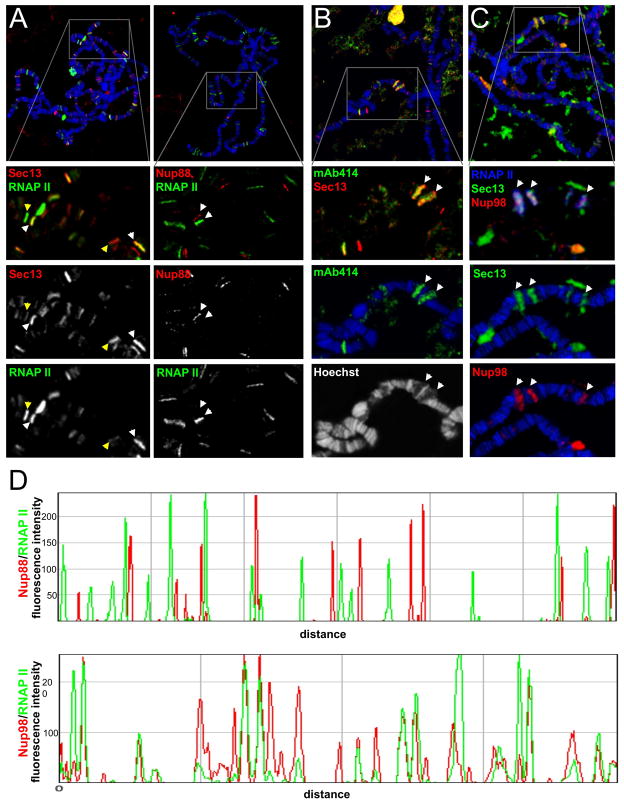
To test if NPCs may be involved in transcriptional activation, we directly compared Nup-chromatin interactions to the localization pattern of active, hyperphosphorylated form of RNA polymerase II (RNAP II) (Palancade and Bensaude, 2003; Patturajan et al., 1998). In support of the reported duality of NPC behavior in chromatin organization, binding of Sec13, Nup98 and mAb414-positive Nups were found to correlate with sites of active transcription, whereas Nup88 was found at loci that did not co-localize with active RNAPII (Figure 3A–D).
Figure 3. Nucleoporins Sec13, Nup98 and FG-repeat Nups are recruited to sites of active transcription.
(A) Chromosome spreads were co-stained with either anti-Sec13 (left) or anti-Nup88 (right) and RNAP II (MARA3 antibody that recognizes phosphorylated RNAP II). Proportional or non-proportional binding behavior of Sec13 is indicated with white and yellow arrows, respectively. Nup88 is found at sites devoid of RNAP II. (B) Chromosomes were co-stained with anti-Sec13 and mAb414, which shows recruitment of FG Nups to a subset of Sec13-positive sites, particularly at highly transcribing puffs. (C) Chromosomes were triple-stained with anti-Nup98, anti-Sec13 and anti-RNAP II (farred, color-coded as blue), showing co-localization of all 3 at multiple sites. (D) Co-localization analysis of a typical chromosomal arm, co-stained with either Nup88 and RNAP II (top plot) or with Nup98 and RNAP II (bottom plot), demonstrating lack of co-localization or co-localization, respectively. See also Figure S2.
Interestingly, while nearly all Sec13 sites co-localized with RNAP II, we noticed that the levels of these two proteins were often inversely proportional. For instance, a fraction of loci exhibited strong RNAP II but relatively weak Sec13 staining, while reciprocally a subset of strong Sec13 sites overlapped with a modest signal from RNAP II (Figure 3A, yellow arrows). Furthermore, Nup98 was found to be recruited to the majority of Sec13 target loci (Figure 3C, white arrows), while the mAb414-positive FG nucleoporins were present at a subset of Sec13 sites and appeared to be particularly enriched at highly decondensed regions called puffs (Figure 3B, white arrows), suggesting that their recruitment is associated with high levels of transcription. This idea is also supported by images of the highly spread puffs (Figure S2B), which show the binding of FG Nups and Sec13 at overlapping but separable regions of a transcribing unit, suggesting that they may have distinct functions in gene expression.
Binding behavior of Nups during development
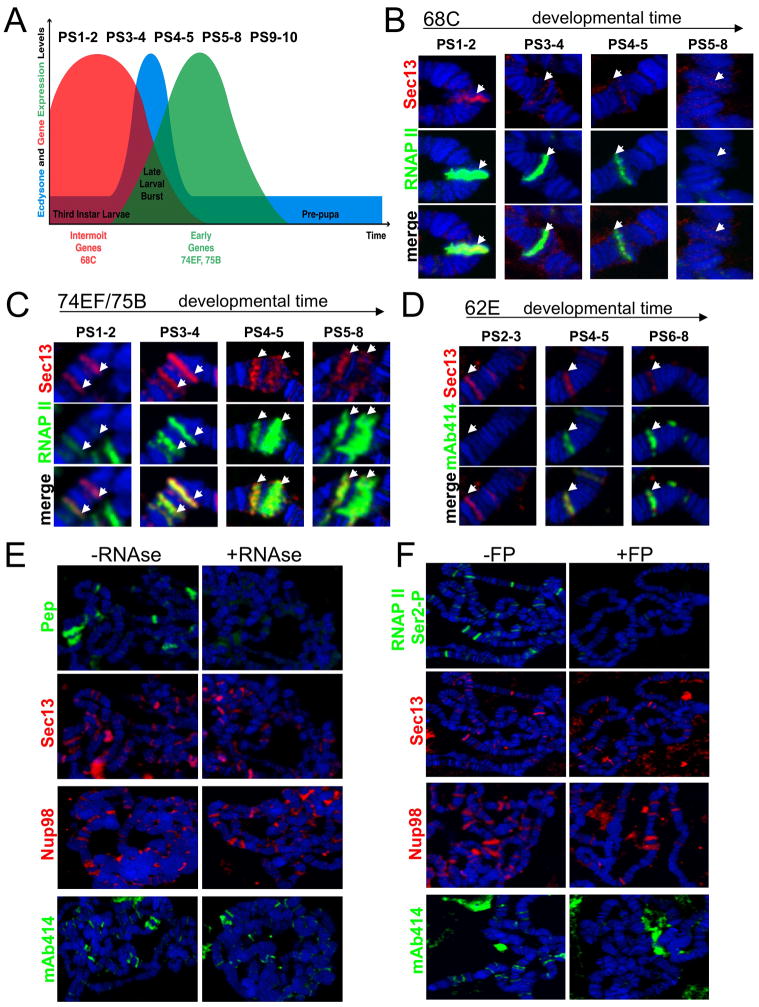
The inversely proportional levels of active RNAP II and Sec13/Nup98 at sites of co-localization suggested that these Nups either play a role in transcriptional initiation, or conversely, the termination of transcriptional activity. To discriminate between these two possibilities, we studied the well-characterized transcriptional program during the wandering third instar stage of larval development, which can be subdivided into morphologically distinguishable Puff Stages (PS) 1–10 (Ashburner, 1967) via the appearance and disappearance of puffs at specific genomic loci. Two main groups of genes were monitored during this ~10 hour period of development (Huet et al., 1993; Thummel, 1990); (i) the Intermolt genes, such as Sgs3 at position 68C, which are expressed first and become down-regulated in response to a pulse of the molting hormone ecdysone and (ii) the Early or ecdysone-induced genes, such as those at positions 74EF and 75B, which are turned on as a result of the same pulse (Figure 4A). Analysis of locus 68C, which undergoes termination of transcription, allowed us to detect Sec13 at the earliest stage (Figure 4B), but showed a loss of Sec13 binding as transcriptional activity diminished. In contrast, at gene loci 74EF, 75B and 76AB, whose expression was induced during this period, we detected bound Sec13 preceding transcription, followed by an enrichment in Sec13 staining as transcription was initiated (Figure 4C, S3A).
Figure 4. Recruitment of Sec13 and mAb414-reactive Nups to transcribing genes during development, which can depend on RNAP II CTD Ser2 phosphorylation.
(A) Schematic illustration of the gene expression program of third instar larval stage, showing the expression profiles of Intermolt genes (red), Early genes (green), and the relative levels of ecdysone hormone (blue), with representative Puff Stages (PS) shown above. (B–C) Polytene chromosomes from larvae of different developmental stages (PS) were stained with anti-Sec13 and RNAP II; (B) during down-regulation of an Intermolt locus 68C, Sec13 levels are lost as RNAP II levels are decreased; (C) Developmental binding of Sec13 during up-regulation of Early loci 74EF and 75B, demonstrating presence of Sec13 either preceding or coinciding with appearance of phosphorylated RNAP II. (D) Polytene chromosomes from larvae of different developmental stages (PS) were stained with mAb414 and Sec13. Locus 62E is shown. (E) Isolated chromosomes were treated with buffer +/−RNase A and stained with anti-Pep, anti-Sec13, anti-Nup98 or mAb414. (F) Salivary glands were treated with buffer −/+FP and stained with control H5 antibody (anti-phosphoSer2 CTD of RNAP II), anti-Sec13, anti-Nup98 or mAb414. See also Figure S3.
To directly test if Sec13 can be recruited to ecdysone-induced genes, recombinant V5-tagged dSec13 protein was injected into salivary glands isolated from early third instar larvae, which were subsequently cultured in the presence of ecdysone. This hormone treatment has been shown to effectively stimulate transcriptional induction of ecdysone-responsive genes in vitro (Ashburner, 1971). The binding of Sec13-V5 to the 74EF/75B puffs was assessed by staining with anti-V5 serum, which co-localized with anti-Sec13 signal at these loci only on chromosomes from glands injected with Sec13-V5, but not with a control V5 fusion protein (Figure S3D). Analysis of Nup98 recruitment to chromatin during development revealed a temporal pattern similar to Sec13 at the 74EF and 75B ecdysone-induced sites (data not shown). These results suggest that Sec13 and Nup98 bind genes that are undergoing activation and may play a role in the induction of transcription at target loci.
Interestingly, developmental analysis of the binding of mAb414-positive FG-repeat Nups demonstrated that their recruitment to and persistence at Sec13 target sites accompanies the appearance and enrichment of active RNAP II (Figures 4D and S3B-C). These findings imply that while Sec13 and Nup98 may bind before and possibly participate in transcriptional initiation, the FG-repeat Nups bind at a step after commitment to transcription has occurred during development.
Chromatin binding of FG-repeat Nups is sensitive to inhibition of phosphorylation of RNAP II at Ser2
Since recruitment of mAb414-Nups to transcribing genes suggested a potential role of these Nups in mRNA export, we tested if it is mediated by RNA. When polytene chromosomes were incubated with RNAse A, the presence of Pep, which binds to ecdysone-induced genes in an RNA-dependent manner (Hamann and Stratling, 1998), was strongly reduced (Figure 4E). Yet no significant changes in chromatin binding of Sec13, Nup98 or the mAb414-reactive FG-repeat Nups were observed between RNase-treated and untreated chromosomes, suggesting that interaction of nucleoporins with transcribing loci was not mediated by mRNA. Next, we asked if Sec13, Nup98 and FG-repeat Nups recruitment requires the process of transcription itself. We treated salivary glands with Flavopiridol (FP), an inhibitor of the kinase Positive Transcription Elongation Factor b (P-TEFb) that specifically phosphorylates Serine 2 of RNAP II CTD (Chao and Price, 2001). Inhibition of Serine 2 phosophorylation is readily observed on chromosomes from glands treated with FP (Figure 4F and (Ni et al., 2004)), and has been reported to result in inefficient transcriptional elongation and reduced recruitment of mRNA processing factors (Cho et al., 2001; Shim et al., 2002). Interestingly, while binding of Sec13 and Nup98 remained largely unaffected by FP treatment, chromatin association of FG-repeat Nups was dramatically abolished under the same conditions (Figure 4F). These results provide further evidence that Sec13 and Nup98 are recruited to active sites before transition into transcriptional elongation, while mAb414-positive Nups are recruited by a mechanism that appears to be dependent on Serine 2 phosphorylation of RNAP II or on events accompanying the switch into elongation.
Nucleoporins Sec13 and Nup98 play an essential role in transcription of developmentally induced genes
To test if Sec13 and Nup98 are required for the induction of transcription at their target loci, we analyzed gene expression in the Sec13 knock-down larvae, using an inducible Sec13 RNAi transgenic line (Figure 1B). When compared to wt levels at PS 3–8 stages, Sec13 RNAi larvae exhibited a pronounced reduction in transcriptional activity of the early genes 74EF and 75B, to which Sec13 is normally recruited during development (Figure 5A). This reduction was apparent by both decreased RNAP II staining and the average puff areas of 74EF and 75B, normalized to a non-puffing control band (Figure 5A,C). Similar functional results were obtained for the Nup98 RNAi knock-down, as evidenced by the reduction of RNAP II and by the quantification of puffing in the knockdown population, while in contrast, RNAi against the scaffold component Nup93 had no effect (Figure 5B, C). Consistently, the mRNA levels of E74A and E75A genes (produced from 74EF puff and 75B puff, respectively (Huet et al., 1993)) in salivary glands are reduced when Sec13 or Nup98 levels are decreased by RNAi compared to controls (Figures 5D and S4B,C).
Figure 5. Sec13 and Nup98 play a functional role in transcriptional regulation.
(A) Levels of phosphorylated RNAP II and of chromatin decondensation (puffing) of Early genes at 74EF and 75B (inset) are reduced in Sec13 RNAi knock-down larvae as compared to wt; and (B) in Nup98 RNAi knock-down larvae relative to Nup93 RNAi knock-down larvae. (C) Measured puff areas at 75B and 74EF were significantly reduced in Sec13 RNAi animals relative to wt, when normalized to the control non-puffing band. (D) RT-PCR analysis using primers for the Early gene transcripts E75A (75B) and E74A (74EF) in Sec13 RNAi and Nup98 RNAi relative to wt, Nup93 RNAi and Nup153 RNAi larvae. Ratios of mRNA levels of Early gene transcripts to control gene rp49 are shown. See also Figure S4.
Importantly, the overall genome-wide levels of RNAP II appeared to be unaffected on chromosomes of Nup RNAi tissues, arguing against a more general transport or nuclear defect. Furthermore, we did not observe a reduction of Sec13 at the NE-located pores in the Sec13 RNAi tissues, which is consistent with its reported stability at the NE in non-dividing cells (D’Angelo et al., 2009), as opposed to pronounced reduction of Sec13 at a genomic locus (Figure S4A).
Sec13 and Nup98 are required for transcriptional re-activation of genes repressed by heat shock
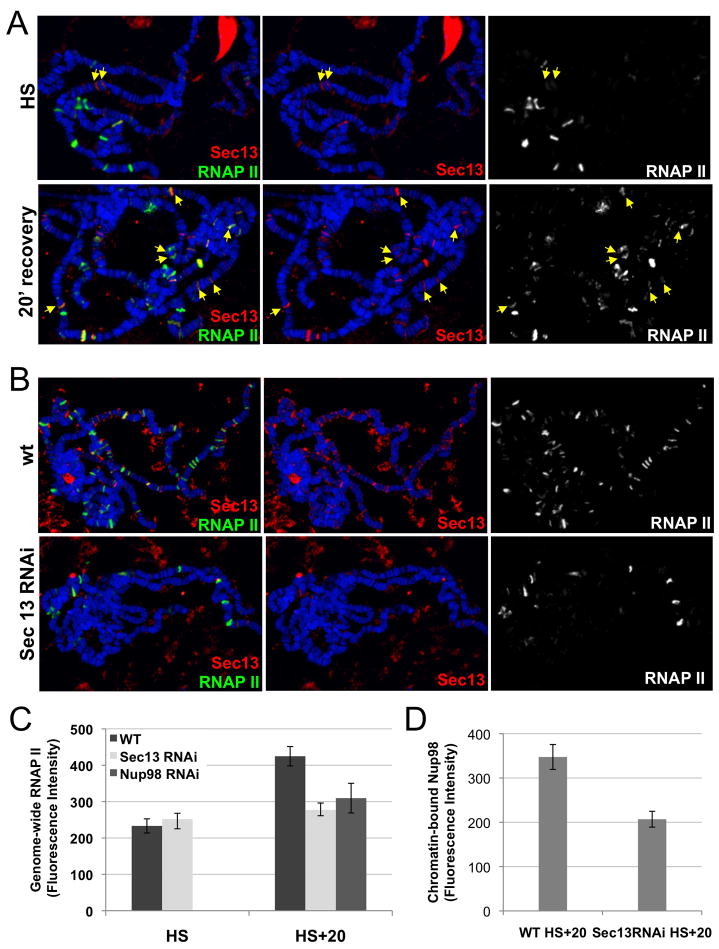
Next we analyzed the potential recruitment and effect of Sec13 and Nup98 during transcriptional behavior induced by heat shock (HS), when transcription is globally repressed, except at the 9 induced HS loci (Mirault et al., 1978; Spradling et al., 1975). We observed significant recruitment of Sec13 to only 2 of the 9 induced HS loci, 93D and 67B (Figure 6A, top panel), arguing against general involvement of Sec13 in heat shock-activated transcription. A similar pattern was observed for Nup98, which was most enriched at the 93D site during HS (Figure S5C). In contrast, Sec13 was strongly recruited to a large number of genes undergoing transcriptional re-activation during a 20 minutes recovery after HS (Figures 6A and S5AB). Strikingly, Sec13 RNAi larvae exhibited a compromised transcriptional recovery after HS, as evidenced by a reduction in new transcription sites beyond the 9 HS loci and by a lack of change in genome-wide RNAP II normally observed in the wt response (Figure 6B, C). The post-heat shock recruitment and transcriptional phenotype of Nup98 were found to closely parallel those of Sec13 (Figures S5C and 6C). Importantly, Nup98 was not observed at recovering sites in Sec13RNAi larvae compared to wt (Figures 6D and S5C). This dependence suggests that Sec13 may be an initial NPC component recruited to active or soon-to-be active genomic sites and may in turn recruit Nup98. In summary, these results reveal chromatin-bound NPC components as key players in the regulation of developmental and stress-induced transcription programs in metazoa.
Figure 6. Nucleoporins Sec13 and Nup98 are involved in re-activation of transcription after heat shock-induced repression.
(A) Chromosomes were stained with RNAPII and Sec13 right after heat shock (HS) or after 20 min recovery (HS + recovery). Yellow arrows point to sites of bound Sec13 that do not correspond to the 9 known heat shock loci (marked by RNAPII at the HS time point). (B) Chromosomes of wt or Sec13 RNAi larvae were stained with RNAP II and Sec13 at the HS + recovery time point. RNAP II accumulation at the recovering transcription sites, observed in wt, is not seen in the Sec13 RNAi larvae. (C) Genome-wide levels of active RNAP II were compared between HS and HS + recovery time points for wt, Sec13 RNAi and Nup98 RNAi larvae. (D) Genome-wide levels of Nup98 recruited during heat shock recovery are shown to be reduced in Sec13 RNAi larvae relative to wt. See also Figure S5.
Nups are associated with active genes in S2 cells
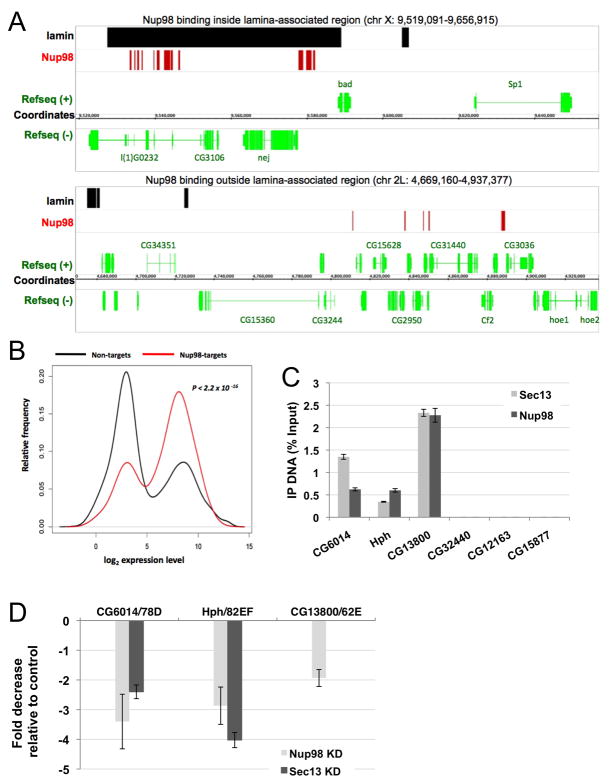
To determine whether the binding to active genes and transcriptional regulation by Nups occur in other cells types, we analyzed chromatin binding of Nup98 in Drosophila S2 culture cells. ChIP-on-chip assays were performed with antibodies against Nup98 and a control IgG on Drosophila whole genome arrays. Analysis of the promoter-associated binding sites identified 841 genes in the fly genome that were specifically immunoprecipitated by the Nup98 antiserum, exhibiting no specific linkage to the X chromosome (Figure S7A, B and Supplemental Table S1). To further determine how chromatin contact sites of Nups correlate with the nuclear periphery, Nup98 target loci were compared to the previously characterized genome-wide binding sites of Lamin (Pickersgill et al., 2006). Similarly to our observations in polytene nuclei (Figure 2B), Nup98 binding sites were found preferentially (~98%) outside of reported Lamin sites (Figure 7A). To examine the subnuclear position of Nup98 target genes in more detail, we determined the relative position of the FISH signal relative to the NE. In contrast to Lamin-associated genes, which preferentially localize near the NE, Nup98 target genes exhibited a random distribution (Figure S6A), further validating the notion that nuclear pore components can bind to chromatin inside the nucleoplasm.
Figure 7. Nucleoporins bind and play a functional role in expression of target genes in tissue culture cells.
(A) A representative example of Nup98 target sites obtained from ChIP-on-chip analysis, plotted relative to Lamin binding sites and annotated genes. (B) Distribution of Nup98 target and non-target genes relative to expression levels. (C) ChIP analysis of candidate target genes of Sec13 and Nup98 (CG6014/78D, Hph/82EF, CG13800/62E) and of control neighboring genes in Drosophila S2 cells, showing enrichment of target genes in fractions immunoprecipitated by Nup antisera (blue and red bars, respectively) as percentage of input DNA, with levels immunoprecipitated by the normal rabbit IgG subtracted. (D) Fold decrease in mRNA levels of Nup target genes in S2 cells treated with dsRNA against Sec13 or Nup98, relative to S2 cells treated with control dsRNA, as detected by microarray analysis. See also Figures S6 and S7.
To determine whether Nups may be associated with active genes in S2 cells, Nup98-bound genes were compared to genome-wide expression, obtained by hybridizing cellular mRNA to Affymetrix Drosophila 2.0 gene arrays. We observed that Nup98 target genes exhibit high expression levels when compared to non-target genes (Figure 7B), demonstrating that Nup98 is preferentially targeted to active genes. Furthermore, the analysis of genome-wide mRNA levels in cells in which Nup98 was knocked down with dsRNA (Figure S6C and Supplemental Table S2), revealed a significant reduction in expression of Nup98 target genes compared to non-target genes (Figure S6B), suggesting that Nup98 is required for normal expression of its target genes in S2 cells.
To analyze the functional role of chromatin-bound Nups more directly, we explored the effect of Nup98 and Sec13 on expression of specific target genes. We first selected potential target genes that were also bound by Nup98 and Sec13 on polytene chromosomes. Three candidate loci identified in this manner, CG6014 at 78D, CG13800 at 62E and Hph at 82EF, were successfully immunoprecipitated by both Nup98 and Sec13 antisera from S2 cells, while control neighboring genes CG32440, CG12163 and CG15877 were not (Figures 7C and S6E). Furthermore, the same 3 target genes were selectively immunoprecipitated using a Myc antibody from cells transfected with a Nup98-Myc fusion construct, but not from mock-transfected cells (Figure S6D). According to expression analysis, transcripts from all three genes were strongly reduced by decreased levels of Nup98, and two of them were also reduced in cells treated with dsRNA against Sec13, as assessed from the Sec13 knock-down microarray (Figure 7D and Supplemental Table S2). Altogether, these results demonstrate binding of Nup98 and Sec13 to specific genes and support the uncovered role of Nup98 and Sec13 in transcriptional regulation of their target genes.
DISCUSSION
We provide evidence for an essential role of chromatin-associated nucleoporins in gene expression of a multicellular organism that seems to be critical for establishing developmental transcription programs and transcriptional response to cellular stress. Surprisingly, the role of Nups in gene regulation is carried out to a great extent by an intranuclear, NE-independent pool of NPC components. This has major implications for our understanding of nuclear organization and its proposed roles in gene regulation. Our data suggest that in metazoa there is no strict requirement of genes to move to peripheral pores, because Nups have the capacity to interact with genomic sites independently of the NE. It remains to be determined whether these intranuclear chromatin-binding Nups shuttle between genomic sites and the nuclear periphery, which would be consistent with the dynamic behavior of some pore components (Rabut et al., 2004). The former possibility is supported by previously reported observations that the dynamic shuttling of mammalian Nup98 and Nup153 is dependent on active transcription by RNAP II and I (Griffis et al., 2002; Griffis et al., 2004). It is tempting to speculate that the mobility of NPC components may establish a mechanism of communication between sites of production of mRNA and sites of its final exit.
Alternatively, the observed role of Nups in transcription could represent NPC-independent sub-complexes inside the nucleus and thus reflect NPC-independent functions of Nups, similar to those of the Nup107/160 complex in kinetochore function in mitosis (Belgareh et al., 2001; Loiodice et al., 2004). Importantly, the role of Sec13 at transcription sites is not linked to its role in secretion since the knockdown of COPII components did not affect transcription (not shown). Intranuclear Nups have been described in different cell types, however their role remained enigmatic. Recent reports of tissue-specific expression of certain Nups in post-mitotic cells, in which NPC assembly is generally absent (D’Angelo et al., 2009), might reflect cell type specific differences in Nup-chromatin interactions.
Given their early recruitment that either precedes or coincides with transcriptional initiation, Sec13 and Nup98 appear to be instrumental in establishing a transcriptionally active state of a particular subset of genes. This novel role of Sec13 and Nup98 in transcriptional induction agrees with a previous study in yeast, which reported an interaction between Nups and promoters of genes during transcriptional initiation and proposed that transient contact with pore proteins may be a general feature of gene activation (Schmid et al., 2006). On the other hand, other binding sites of NPC identified by the ChIP approach (Casolari et al., 2005; Casolari et al., 2004) were preferentially located at the 3′ ends of active genes in yeast. This function is likely reflected in the differential recruitment and transcription elongation dependency of FG-repeat Nups, which seem to be involved in later events of transcription.
In yeast, one explanation for the dual 5′ and 3′ end binding was suggested to be the presence of both gene ends at the NPC through interactions with different Nups, which would result in DNA looping. Such chromatin loops at the NPC were projected to serve as sites of assembly for transcription and mRNA processing machinery (Blobel, 1985; Menon et al., 2005) and/or to delimit active chromatin domains (Casolari et al., 2005). This role may be conserved by metazoan Nups at intranuclear active sites, where they can serve as a platform for co-regulated assembly of transcription machinery and mRNA export factors. The FG repeats, found in Nup98 and the mAb414-reactive Nups, are known to interact with mRNA export receptors, such as Mex67/TAP (Strasser et al., 2000), while the particular GLFG type of FG domain of Nup98 can associate with the CBP/p300 histone acetyltransferase (Kasper et al., 1999). Since Sec13 has been found to be very stable at the NPC (Rabut et al., 2004), it is unlikely that Sec13 itself shuttles but rather may exist in a separate intranuclear population. It is possible then that Sec13 either at the pore or at a transcribing locus inside the nucleus serves as the docking point for Nup98, which in turn brings about further recruitment of transcriptional regulators or mRNA export or other RNA processing factors. In this manner, the communication between intranuclear transcription sites and the NE-embedded nuclear pores, suggested above, may be carried out by a shuttling Nup98 component.
Furthermore, other reports revealed that Nups are able to interact with chromatin regions enriched in repressive histone modifications or in non-active genes (Brown et al., 2008; Casolari et al., 2004). In support of these observations, our findings demonstrate that other nucleoporins such as Nup88 may be targeted to regions in the genome where they may participate in alternative gene regulatory processes. Significantly, both Nup88 (Agudo et al., 2004) and Nup98 have been implicated in tumorigenesis. Particularly, genomic fusions of Nup98 to transcription regulators have been shown to be the underlying mutations behind multiple types of leukemia (Slape and Aplan, 2004). The uncovered association of Nup88 and Nup98 with chromatin opens new directions for understanding the roles of NPC components in cancer.
EXPERIMENTAL PROCEDURES
Fly Stocks and Staging
UAS-Sec13-RNAi transgenic line GD12035, UAS-Nup98/Nup96-RNAi line GD6897, UAS-Nup93-RNAi line GD7196, UAS-Nup153-RNAi line GD10636, UAS-Sec31-RNAi lines GD13867 were obtained from Vienna Drosophila RNAi Center (VDRC Stocks v50367, v31198, v16188, v47155, v35867 (1) and v35868 (2), respectively). Hypomorphic Nup88 allele mbo05043 has been described in Uv et al 2000. To facilitate developmental staging of larvae, we employed previously described blue food method (Andres and Thummel, 1994), which relies on the fact that larvae cease their food intake once wandering third instar larval stage is reached. In larvae raised on food with 0.02% Bromophenol Blue, Blue (B) larvae, which correspond to PS 1–3, Light Blue (LB) larvae, which correspond to PS 3–8, and Clear larvae, which correspond to PS 8–10, could be distinguished (PS stages as described in Ashburner 1967). To compare transcriptional behavior of wt and Sec13, Nup98, Nup93, Nup153 or Sec31 RNAi larvae, we selected LB wandering larvae.
Antibodies
Rabbit polyclonal antibodies were raised against purified recombinant protein of 6X histidine tagged-full length human Sec13 and affinity purified using GST-full length human Sec13 protein coupled to sepharose beads. It was used at 50μg/ml (1:20) for polytene immunofluorescence (IF) staining and 5μg/ml (1/200) for Western blotting analysis. Rabbit polyclonal antibodies were also raised against and affinity purified with recombinant GST-fusion of dNup98 amino acids 770–939. It was used at 50 μg/ml (1:20) for IF and at 10 μg/ml (1:100) in Western.
For Drosophila polytene IF, mAb414 (Covance MMS-120R) was used at 1:20–1:50, anti-dNup88 at 1:50, anti-dNup154 at 1:20, anti-Gp210 at 1:20, anti-RNAP II MARA3 (which recognizes either Ser5-P or Ser2-P of CTD, but not unphosphorylated CTD (Patturajan et al., 1998)) at 1:2, anti-RNAP II Ser2-P (H5 from Covance, MMS-129R) at 1:20, anti-Pep at 1:10 and anti-Su(Hw) at 1:500 (both a gift from Dr. E.P. Lei), anti-dLaminDm0 (Dr. P.A. Fisher) at 1:200, and anti-V5 (Abcam) at 1:100.
Immunofluorescence
Preparation of polytene chromosomes was done similarly to Gerasimova et al 1995. All blocking and antibody incubations were done in 2% BSA/0.1% Tween-20/PBS, overnight (O/N) at 4C for primary and 1 hr at RT for secondary. For 3D analysis of intact polytene nuclei, detached salivary glands were fixed in 8% acetic acid/2% PFA for 2–5 min, mounted on slides and incubated with primary antibodies O/N. For diploid cell stainings, imaginal discs, attached to larval cuticle, were fixed in 2% PFA for 20 min, washed in IF buffer (1% BSA, 0.1% TritonX, PBS) and incubated O/N. For 3D analysis, image acquisition was done with confocal sections of 100–200 nm and reconstructed with Leica confocal software. For quantification of puff size, average areas of 74EF and 75B puffs and of a band at 74A, as stained by DAPI, from chromosomes of 12 Sec13 RNAi, 13 Nup98 RNAi and 11 wt LB larvae were measured in Adobe Photoshop CS2. The average representative fluorescence intensity of Sec13 staining at the NE was quantified by measuring pixels in an area of fixed size, which corresponded to co-staining with Lamin Dm0, while Sec13 staining at the 74EF locus was quantified by measuring pixels in an area of 74EF locus. Genome-wide levels of RNAP II and Nup98 were measuring with integrated density over entire chromosome spreads at the HS or HS+20 time points (6–10 of either wt, Sec13 RNAi or Nup98 RNAi larvae for each time point). Co-localization analysis of red and green channel was performed using a macro that draws line profiles from a RGB image (Image J).
RNase A, Flavopiridol and Heat Shock treatments
For RNase treatment, salivary glands from larvae of LB stage were dissected in PBS and pre-treated in PBS/0.1% TritonX for 1.5 min. The two lobes of the same pair of salivary glands were separated, with one incubated in 0.7% NaCl (−RNase) and the other in 0.7% NaCl with 1 mg/ml RNase A (Ambion) (+RNase) for 8–20 min. For Flavopiridol (FP) treatment, the two lobes of salivary glands from LB stage were separated, one incubated in 50% Grace’s Medium (Sigma-Aldrich) (−FP), the other in 50% Grace’s Medium plus 800 nM FP (+FP) for 30–50 min. For heat shock, larvae were incubated for 30 min-1hr at 37C, recovered at RT for 0–20 min.
RT-PCR
Total RNA from 5–10 salivary gland pairs from LB stage larvae was isolated using Trizol reagent. Adjusted equal amounts of RNA from glands of different genotypes were reverse-transcribed using QuantiTect RT kit and used as templates for PCR (see Supplemental Table S3). Quantification was done on RT-PCR reactions from at least three independent RNA preparations for each genotype, using Image J.
FISH
Probes were generated from BACs RP98-24H03 for 78D, RP98-15B04 for 62E, RP, and RP98-26J11 for 75B (Children’s Hospital & Research Center Oakland) by digesting 2 micrograms of DNA with DNAse I (1:2000) for 90 min, purifying on Quagen PCR columns and labeling with FITC-labeling kit (ENZO Bioprobe Nick Translation Kit with Fluorescein-12-dUTP). Probes were applied to samples on poly-L-lysine coated slides at 80°C, incubated for 6 min, sealed with rubber cement and incubated O/N at 42C. Washes were done in buffers W1 (0.4X SSC, 0.3% NP40) and W2 (2X SSC, 0.1% NP40) at 73C for 2 min each, then 2X in 2X SSC for 15 min at RT, then in 1X PBS for 15 min at RT, followed by IF staining as described above.
Injections of salivary glands
Recombinant Drosophila Sec13-His-V5 and dTomato-His-V5 were purified using nickel beads and dialyzed into PBS to 2 mg/ml final concentration. Salivary glands from PS 1–2 or before-wandering larvae were mounted onto agarose drops on coverslips and injected with protein at ~0.05–0.5 microl/cell. The glands were incubated in Grace’s medium with 6 microM ecdysone (Sigma) for 1 hour, then fixed and stained as described above.
RNAi and microarray analysis in S2 cells
S2 cells were maintained in Schneider’s media (Invitrogen) with 10% FBS (Gibco). The RNAi treatment was carried out essentially as described in Worby et al 2001. dsRNA was generated from PCR templates (see Table 1 for primers) using the Megascript T7 Kit (Ambion). For each RNAi experiment, 15 microg of dsRNA was added to 1 ml of 106 cells/ml, incubated in serum-free media for 1 hr, then supplemented with 2 ml of regular media and incubated for 48 hrs. At 48 hrs, the RNAi treatment was repeated and final samples were harvested at 96 hrs. Total RNA was prepared using RNAeasy kit (Quagen) and labeled for microarray analysis by the Salk Functional Genomics Core Facility. The hybridizations were done on Affymetrix Drosophila 2.0 gene chips, and the raw data analyzed by Affymetrix software. Nup98 and Sec13 RNAi data sets were compared to dWhite data sets, with 1.5–2 fold cut-offs.
Chromatin Immunoprecipitation
S2 cultures were cross-linked with 1% formaldehyde solution for 10min, lysed in RIPA buffer and sonicated to generate DNA fragments <1kb. Chromatin was pre-cleared and immunoprecipitated using Nup98 antibody at 1:50 dilution and Sec13 antibody at 1:200 dilution. DNA was eluted, purified and subjected to PCR. For ChIP-on-chip analysis, the Nup98 antibody- or normal rabbit IgG- immunoprecipitated DNA in duplicates was labeled and co-hybridized with input DNA to 2.1M Drosophila whole genome arrays by NimbleGen Systems, Inc. ChIP peak data were generated by NimbleGen, and genes with peaks of scaled log2-ratio ≥1 and FDR score ≤ 0.05 were selected as Nup98 targets. Chromosome views were generated using Affymetrix Integrated Genome Browser. Analyses of Nup98 binding and expression correlation were performed using the R package for statistical computing.
Supplementary Material
Acknowledgments
We thank Tony Hunter and members of the Hetzer lab for critically reading the manuscript, Leanne Jones for sharing fly reagents and expertise and Andy Dillin for sharing injection and microscopy equipment. We also thank Elissa P. Lei, P.A. Fisher, S. Gigliotti and C. Samakovlis for generously providing antibodies. The project was supported by R01GM057438 from the NIHGMS. M.C. was supported by the Damon Runyon Cancer Research Foundation fellowship (DRG 1952-07).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agudo D, Gomez-Esquer F, Martinez-Arribas F, Nunez-Villar MJ, Pollan M, Schneider J. Nup88 mRNA overexpression is associated with high aggressiveness of breast cancer. Int J Cancer. 2004;109:717–720. doi: 10.1002/ijc.20034. [DOI] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 1994;44:565–573. doi: 10.1016/s0091-679x(08)60932-2. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. I. Autosomal puffing patterns in a laboratory stock of Drosophila melanogaster. Chromosoma. 1967;21:398–428. doi: 10.1007/BF00336950. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Induction of puffs in polytene chromosomes of in vitro cultured salivary glands of Drosophila melanogaster by ecdysone and echysone analogues. Nat New Biol. 1971;230:222–224. doi: 10.1038/newbio230222a0. [DOI] [PubMed] [Google Scholar]
- Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci U S A. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Hetzer MW. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development. 2003;130:271–284. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci U S A. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol. 2005;171:955–965. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enninga J, Levay A, Fontoura BM. Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol Cell Biol. 2003;23:7271–7284. doi: 10.1128/MCB.23.20.7271-7284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigliotti S, Callaini G, Andone S, Riparbelli MG, Pernas-Alonso R, Hoffmann G, Graziani F, Malva C. Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the nup155 vertebrate nucleoporin gene. J Cell Biol. 1998;142:1195–1207. doi: 10.1083/jcb.142.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber UF, Senior A, Gerace L. A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. Embo J. 1990;9:1495–1502. doi: 10.1002/j.1460-2075.1990.tb08267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell. 2004;15:1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Goldman RD, Meyuhas R, Mills E, Margalit A, Fridkin A, Dayani Y, Prokocimer M, Enosh A. The nuclear lamina and its functions in the nucleus. Int Rev Cytol. 2003;226:1–62. doi: 10.1016/s0074-7696(03)01001-5. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hamann S, Stratling WH. Specific binding of Drosophila nuclear protein PEP (protein on ecdysone puffs) to hsp70 DNA and RNA. Nucleic Acids Res. 1998;26:4108–4115. doi: 10.1093/nar/26.18.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell. 2005;16:2382–2394. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M, Walther TC, Mattaj IW. Pushing the Envelope: Structure, Function, and Dynamics of the Nuclear Periphery. Annu Rev Cell Dev Biol. 2005 doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber H, Sedat JW. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. J Cell Biol. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet F, Ruiz C, Richards G. Puffs and PCR: the in vivo dynamics of early gene expression during ecdysone responses in Drosophila. Development. 1993;118:613–627. doi: 10.1242/dev.118.2.613. [DOI] [PubMed] [Google Scholar]
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci U S A. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirault ME, Goldschmidt-Clermont M, Moran L, Arrigo AP, Tissieres A. The effect of heat shock on gene expression in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):819–827. doi: 10.1101/sqb.1978.042.01.082. [DOI] [PubMed] [Google Scholar]
- Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur J Biochem. 2003;270:3859–3870. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Roth P, Xylourgidis N, Sabri N, Uv A, Fornerod M, Samakovlis C. The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J Cell Biol. 2003;163:701–706. doi: 10.1083/jcb.200304046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16:2135–2146. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Slape C, Aplan PD. The role of NUP98 gene fusions in hematologic malignancy. Leuk Lymphoma. 2004;45:1341–1350. doi: 10.1080/10428190310001659325. [DOI] [PubMed] [Google Scholar]
- Spradling A, Penman S, Pardue ML. Analysis of drosophila mRNA by in situ hybridization: sequences transcribed in normal and heat shocked cultured cells. Cell. 1975;4:395–404. doi: 10.1016/0092-8674(75)90160-9. [DOI] [PubMed] [Google Scholar]
- Strasser K, Bassler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. The Journal of cell biology. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Thummel CS. Puffs and gene regulation--molecular insights into the Drosophila ecdysone regulatory hierarchy. Bioessays. 1990;12:561–568. doi: 10.1002/bies.950121202. [DOI] [PubMed] [Google Scholar]
- Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. Nucleocytoplasmic transport: cargo trafficking across the border. Curr Opin Cell Biol. 2002;14:328–335. doi: 10.1016/s0955-0674(02)00337-x. [DOI] [PubMed] [Google Scholar]
- Wente SR. Gatekeepers of the nucleus. Science. 2000;288:1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.