Abstract
Infection by herpes simplex virus type 1 (HSV-1) can cause clinical symptoms in the peripheral and central nervous system. Recurrent ocular shedding can lead to corneal scarring and vision loss making HSV-1 a leading cause of corneal blindness due to an infectious agent. The primary site of HSV-1 latency is sensory neurons within trigeminal ganglia. Periodically, reactivation from latency occurs resulting in virus transmission and recurrent disease. During latency, the latency-associated transcript (LAT) is abundantly expressed. LAT expression is important for the latency-reactivation cycle in animal models, in part, because it inhibits apoptosis, viral gene expression, and productive infection. A novel transcript within LAT coding sequences (AL3) and small nonprotein coding RNAs are also expressed in trigeminal ganglia of latently infected mice. In this review, an update of viral factors that are expressed during latency and their potential roles in regulating the latency-reactivation cycle is discussed.
1. Introduction
At least 90% of the population are infected with herpes simplex virus type 1 (HSV-1), and infection can cause a variety of disorders [1, 2]. Recurrent ocular HSV-1 is the leading cause of infectious corneal blindness in industrialized nations [3]. In a murine model, ocular infection appears to induce autoimmune disorders leading to corneal antigen destruction and stromal keratitis [4]. HSV-1 infections also cause gastrointestinal disorders, esophageal disorders, and approximately 25% of all genital herpes infections [5, 6].
HSV-1 is the most commonly identified cause of acute, sporadic viral encephalitis in the U.S. accounting for 10%–20% of all cases [7]. It is estimated that there are approximately 2,000 new cases per year in the U.S. HSV-1 and HSV-2 can cause acute necrotizing encephalitis in infants, children, and adults. Encephalitis due to HSV-2 in newborn infants is a widespread disease in the brain and commonly involves a variety of other organs in the body including skin, eyes, and lungs [8].
Herpes simplex virus- induced encephalitis (HSE) is characterized by severe destruction of temporal and frontal lobe structures, including limbic mesocortices, amygdala, and hippocampus. Without antiviral therapy, the mortality rate is as high as 70%, but even after antiviral therapy 20% of these patients die. Despite early treatment, chronic progressive tissue damage in magnetic resonance imaging can be found up to 6 months following the onset of symptoms. Approximately 2/3 of the HSE cases occur because of reactivation from latency [9], which explains why there is high morbidity and long-term complications despite antiviral treatment [10–12].
HSE is often associated with necrotic cell death resulting from virus replication and inflammatory changes secondary to virus-induced immune response [13]. However, there is not a perfect correlation between virus burden in the brain and the severity of histological changes and neurological symptoms. Furthermore, a small number of HSE patients are negative for HSV-1 DNA early in the course of infection suggesting that factors other than virus replication are involved in pathogenesis. The finding that mice lacking toll like receptor 2 (TLR2) are less susceptible to HSV-1-induced encephalitis than wild type mice or mice lacking TLR4 implies that TLR-2-mediated cytokine responses are detrimental to the host [14]. In addition, two children with HSE lack the intracellular protein UNC-93B and have impaired interferon responses [15]. It appears that the ability of the host to appropriately respond to HSV-1 infections in the brain is crucial for preventing HSE.
2. The Latency-Reactivation Cycle
2.1. The Latency-Reactivation Cycle Has 3 Distinct Steps
Despite a vigorous immune response during acute infection, HSV-1 establishes latency in ganglionic sensory neurons, typically trigeminal ganglia (TG) or sacral dorsal root ganglia [16, 17]. Although TG is a primary site of latency following ocular, oral, or intranasal infection [18–20], latent HSV-1 can also be detected in human adult nodose ganglia and the vagus nerve [5, 6]. Up to 40% of sensory neurons can be latently infected [21–25]. HSV-1 genomic DNA has also been detected in the central nervous system of a significant percentage of humans [18, 26, 27].
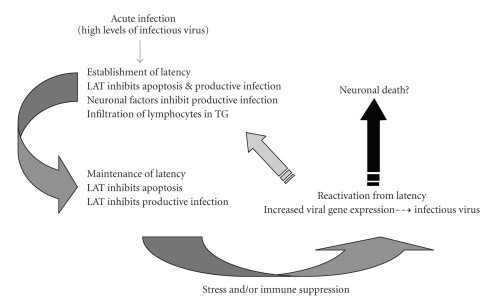
The steps of the latency-reactivation cycle have been operationally divided into three major steps: establishment, maintenance, and reactivation (Figure 1). Establishment of latency includes entry of the viral genome into a sensory neuron and acute infection. Viral gene expression is then extinguished, with the exception of the latency-associated transcript (LAT). For further details regarding viral gene expression during acute infection and establishment of latency, see Section 2.2.
Figure 1.
Steps in the latency-reactivation cycle of HSV-1. For details, see the text.
Maintenance of latency is a phase that lasts for the life of the host and is operationally defined as a period when infectious virus is not detected by standard virus isolation procedures. In general, abundant expression of viral genes that are required for productive infection does not occur. LAT is abundantly expressed during this stage of latency.
Reactivation from latency is initiated by external stimuli (stress and immunosuppression, e.g.) that stimulate viral gene expression. Abundant viral gene expression is detected in sensory neurons and infectious virus can be isolated from TG, eye swabs, and/or nasal swabs. It is not clear whether a neuron that undergoes reactivation and produces infections virus survives and resumes latency or is killed. For further discussion of factors that regulate reactivation from latency, see Sections 2.4–2.6. The ability of HSV-1 to reactivate from latency results in recurrent disease and virus transmission.
2.2. Viral Gene Expression during Productive Infection versus Latency
2.2.1. Viral Gene Expression during Productive Infection
Binding and entry of HSV-1 to cells are mediated by viral glycoproteins and cellular factors [28–30]. A cellular mediator of viral entry (HveA or HVEM) is primarily expressed in activated T cells and belongs to the tumor necrosis factor receptor family [31]. Entry of HSV-1 into epithelial and other nonlymphoid cells is mediated by an unrelated membrane glycoprotein that resembles the poliovirus receptor (HveB and HveC) [32]. HveC is active as an entry mediator for all herpesviruses examined to date, HSV-1, bovine herpesvirus 1 {BHV-1}, and pseudorabies virus, {PRV}. HveC is abundantly expressed in neurons and can block viral entry in several neuron-like cell lines [32]. After uncoating, the viral genome is present in the nucleus and viral gene expression ensues.
HSV gene expression is temporally regulated in three distinct phases: immediate early (IE), early (E), or late (L) [33]. IE transcription does not require protein synthesis and is stimulated by VP16 [34]. E gene expression is dependent on at least one IE protein, and generally E genes encode nonstructural proteins that play a role in viral DNA synthesis. L gene expression is maximal after viral DNA replication, requires IE protein production, and L proteins comprise the virion particle.
Five IE genes encode ICP0, ICP4, ICP22, ICP27, or ICP47. ICP4 [35–38] and ICP27 [39–41] are required for virus growth in tissue culture. In general, ICP4 represses IE gene expression [37, 42–46] and activates E or L gene expression by interacting with RNA polymerase II transcription factors [46, 47]. ICP27 redistributes small nuclear ribonucleoprotein complexes, interferes with splicing of IE transcripts, and promotes E and L poly A site selection [48–51]. ICP47 prevents transport of antigenic peptides into the endoplasmic reticulum [52] and is crucial for neurovirulence because it inhibits CD8+ T cell responses [53]. ICP0 can activate expression of all classes of viral genes, in part because it increases steady-state levels of mRNA [54].
ICP0 also binds several cellular proteins: (1) elongation factor 1α [55], (2) cyclin D3 [56], (3) an ubiquitin-specific protease [57, 58], and (4) PML [59–62]. Interactions between ICP0 and chromatin-remodeling enzymes activate viral transcription. For example, a histone deacetylase inhibitors enhance viral gene expression and productive infection [63, 64]. Secondly, ICP0 alters a complex that inhibits gene expression (REST/CoREST/histone deacytlase repressor complex) [65]. Finally, HSV-1 ICP0 interacts with HDAC2 [66] and blocks histone deacetylation to stimulate viral gene expression [65, 67]. Since ICP0 has recently been shown to remove histones from viral chromatin during productive infection [68], it is tempting to suggest that ICP0 has a similar function during reactivation from latency. These activities of ICP0 promote virus replication in differentiated cells [69].
2.2.2. Viral Gene Expression Is Extinguished after Infection of Sensory Neurons
Following infection of rodents, rabbits, or humans with HSV-1, productive infection is initiated in the mucosal epithelium. Virus particles or subparticles then enter sensory neurons and are transported intra-axonally to the sensory ganglia. Since HSV-1 infection typically occurs via the oral, ocular, or nasal route, the 5th cranial nerve, trigeminal ganglia (TG), is a primary site for latency [18, 19]. Extensive viral gene expression and replication occur within TG for approximately a week following infection of animal models that support HSV infection [70, 71]. Productive viral gene expression that occurs in TG appears to be different than what is seen in cultured cells [72]. Infectious virus can readily be detected in homogenates prepared from TG during acute infection. However, it is difficult to conclude whether this infectious virus is the result of productive infection in sensory neurons or the result of transport from peripheral sites of infection. Replication is not required for establishment of latency because mutants that cannot replicate will establish latency, but at a reduced level [73–81].
2.2.3. IE Promoters Are Differentially Regulated in Sensory Neurons Relative to Nonneuronal Cell Types
Several studies using transgenic mice that contain IE promoters linked to a reporter gene have concluded that IE promoters are differentially regulated by neuronal specific factors. For example, the HSV-1 ICP4 promoter is active in Schwann cells, but not sensory neurons in TG [82]. As expected, the ICP4 promoter in transgenic mice is activated in TG neurons following infection with HSV-1. In contrast to the ICP4 promoter, transgenic mice containing the ICP0 or ICP27 promoters are active in certain neurons within the brain and TG [83]. The ICP0 promoter is also differentially regulated in TG neurons depending on the age of the mouse. The ICP0 promoter contains a cis-acting element that can bind a neuronal specific transcription factor, Olf-1, which is differentially and developmentally expressed in specific subsets of sensory neurons [84] suggesting that the Olf-1 site plays a role in activating ICP0 promoter activity in certain neurons.
All IE promoters contain a common cis-acting sequence (TAATGARAT) that is required for VP16-mediated trans-activation [34, 85]. VP16 must interact with two cellular proteins, Oct-1 and HCF, to efficiently induce IE promoter activity. A cellular transcription factor, Zhangfei, binds to HCF and prevents activation of the ICP0 promoter [86]. Another cellular transcription factor, Luman, also binds to HCF and sequesters HCF in the cytoplasm of sensory neurons, suggesting that Luman has a role in latency [86]. Zhangfei and Luman have basic domain-leucine zippers (bZIP) regions, acidic activation domains, and consensus HCF-binding motifs, yet have little amino acid similarity. In nonneuronal cells, HCF has a nuclear localization [87], but in sensory neurons it appears to be predominantly localized to the cytoplasm [88]. If the relative levels of Luman and Zhangfei are high, the availability of “free” HCF that could interact with VP16 would be reduced and consequently IE gene expression would be repressed. It has also been hypothesized that VP16 is not present in sufficient quantities in the nucleus of infected sensory neurons to stimulate efficient productive infection [88]. However, inducible expression of VP16 in the context of the viral genome or in transgenic mice did not lead to enhanced viral replication [89].
Other cellular transcription factors expressed in sensory neurons (Brn-3.0 and N-Oct3, e.g.) have the potential to regulate IE gene expression [90, 91]. Brn-3.0 binds to noncoding sequences in the HSV-1 genome, but the binding sites for Brn-3.0 are not identical to those for Oct-1 or other related transcription factors that also include Brn-3.1 and Brn-3.2 [92]. Brn-3.0 is important in the peripheral nervous system of mice because null mutations in the brn-3.0 locus result in neonatal death with defects in sensory ganglia and specific central nervous system nuclei [93, 94]. brn-3.2 is required for differentiation of certain retinal ganglion cells [95]. One study has concluded that Brn-3.1 and 3.2 have opposite effects on a target promoter [96]. Considering that the Brn3 family of transcription factors is expressed in the peripheral nervous system, these proteins may regulate HSV gene expression during the latency-reactivation cycle.
Following infection of primary neurons, ICP0 does not appear to accumulate in the nucleus of infected cells [97]. An independent study also concluded that the function of ICP0 is impaired in human neuronal-like cells because a nuclear structure (ND10) that ICP0 interacts with is different compared to nonneuronal cells [98]. The same neuronal-like cells do not support efficient viral replication, in part, because ICP0 expressing plasmids do not activate viral transcription efficiently. These studies argue that ICP0 does not function efficiently in neuronal cells and thus productive infection is inhibited.
2.2.4. The LAT Promoter Is Neuronal Specific
In sharp contrast to other HSV-1 promoters, the promoter that directs expression of the latency-associated transcript (LAT) is activated in sensory neurons (see Figure 2 for a schematic of the HSV-1 LAT promoter). Two separate promoter fragments that are upstream of the start site of LAT, latency-associated promoter 1 and 2 (LAP1 and LAP2), can cis-activate a reporter gene in transiently transfected cells [99, 100]. Several studies have demonstrated that sequences spanning the TATA box, LAP1, are critical for directing LAT expression in sensory neurons [99, 101–104]. LAP2 promoter has been proposed to promote expression of the stable 2 Kb LAT expression during productive infection of cultured cells. LAP2 may also play a role in promoting long-term expression of LAT in sensory neurons or may activate expression of novel transcripts during specific stages of infection in sensory neurons. Although the LAT promoter elements have neuronal specificity in transient transfection assays, they can also direct expression of a reporter gene in nonneural cells [105–109]. This may reflect the abundance of cellular transcription factor binding sites within the LAT promoter (Figure 2(c)). Many of these transcription factors are present in nonneural cells and can activate the LAT promoter in transiently transfected cells. For example, the two CRE binding sites in the LAT promoter are functional because cAMP activates the promoter [110, 111]. The CRE motif that is proximal to the TATA box is important for expression in neurons, and its presence has a positive effect on reactivation from latency [111–113]. Furthermore, Sp1, YY1, USF, and CAAT are frequently found in RNA polymerase II promoters that are not neural specific. Neuronal specific factors have been identified that bind to the LAT promoter [111–113]. The finding that the IE protein, ICP4, binds to DNA sequences downstream of the TATA box and represses the LAT promoter is one important reason why LAT is not an abundant transcript during productive infection [107].
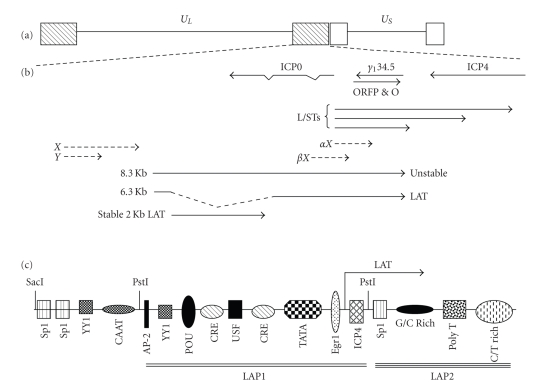
Figure 2.
Location of genes within the HSV-1 repeats. (a) UL and US denote the unique sequences of the long (L) and short (S) components of the genome. The boxes depict repeat sequences. (b) Transcription map of the repeat region. Location and orientation of LAT [114, 115], ICP0, γ134.5 [116, 117], ORFP [118], L/STs [119] are indicated by solid lines. Partially mapped transcripts (αX and βX) are denoted by dashed arrows [120, 121]. (c) The LAT promoter contains numerous cis-acting sites that can be bound by cellular transcription factors. Binding of ICP4 to the ICP4 binding site in the LAT promoter inhibits promoter activity [107]. In transient transfection assays, the LAT promoter can be divided into a strong promoter (LAP1) and a weaker promoter (LAP2) [99, 100]. For details of transcripts encoded by LAT, see Figure 3.
Long-term expression of LAT has also been examined in the context of the viral genome [122–125]. These studies have demonstrated that LAP2 sequences function as a long-term enhancer (Figure 2(c)) in latently infected mice. LAP2 also appears to maintain LAP1 promoter activity. Although DNA sequences within the LAT promoter activate RNA expression in sensory neurons, neuronal specificity does not appear to be contained into a single cis-acting motif. As expected, the LAT locus is transcriptionally active during latency and is associated with acetylated histones, whereas ICP0 expression is repressed and hypoacetylated [126].
2.3. Viral Gene Expression Is Restricted during Latency to the LAT Locus
2.3.1. LAT Is Abundantly Expressed in Sensory Neurons during Latency
LAT is abundantly transcribed in latently infected neurons of mice, rabbits, or humans [1, 104, 114, 115, 127–131]. Mice, rabbits, or humans latently infected with HSV-1 express LAT, and LAT is predominantly detected in the nucleus. LAT is complementary to ICP0 and overlaps the ICP0 transcript (Figure 2(b)), suggesting that LAT inhibits ICP0 expression by an antisense mechanism. Although the ability of LAT to repress ICP0 expression may be important, LAT sequences that promote spontaneous reactivation in a rabbit ocular model do not overlap ICP0 [132]. The simplest interpretations of these data are that LAT has more than one function or the ability of LAT to repress ICP0 expression is not that important in the small animal models used to study latency.
Detection of thymidine kinase and ICP4 transcripts, in addition to LAT, in TG of latently infected mice [133] appears to be the result of spontaneous reactivation or unsuccessful reactivation from latency [134, 135]. Viral genome positive neurons that are LAT negative can be detected in latently infected mice [24]. Since in situ PCR was used to detect viral DNA, but in situ hybridization was used to detect LAT, neurons expressing low levels of LAT were likely missed.
Splicing of the 8.5 Kb LAT transcript yields an abundant 2 Kb LAT and an unstable 6.5 Kb LAT [109, 114, 128] (Figure 3). Correct splicing of the 2 Kb LAT is necessary for establishment and maintenance of latency [139, 140]. In general, the stable 2 Kb LAT is not capped, is poly A-appears to be circular, and is a stable intron [141, 142]. Although LAT is predominantly detected in the nucleus, it is also present in the cytoplasm [143–145] and is associated with polyribosomes [143, 146] or splicing factors [143].
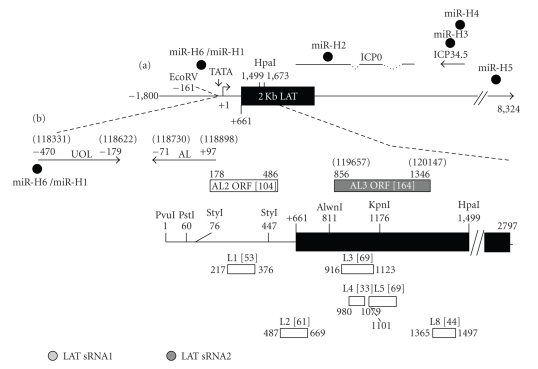
Figure 3.
Schematic of factors encoded within the LAT locus. (a) Schematic of genes within the long repeats that contain the LAT locus. The large arrow indicates the primary LAT transcript. The solid rectangle represents the very stable 2 kb LAT intron. The start of LAT transcription is indicated by the arrow at +1 (genomic nucleotide 118801). Several restriction enzyme sites and the relative locations of the ICP0 and ICP34.5 transcripts are shown for reference. The locations of the 6 micro-RNAs (miR-H1-6) that are located within the 8.3 kb LAT [136] are shown. (b) Partial restriction map of LAT and position of LAT open reading frames (L1-8) within the first 1.5 Kb of strain McKrae LAT coding sequences, which were based on previous studies [137]. The numbering system of the ORFs was consistent with a previous study [137]. Only the ORFs with at least 30 amino acids are shown (the number of amino acids in each ORF is denoted by the numbers in brackets). Open circles denote the position of two LAT small RNAs that are encoded within the first 1.5 kb LAT coding sequences [138]. Positions of UOL transcript, AL transcript, and ORFs located on the opposite strand of LAT (AL2 and AL3) are shown. The numbers of amino acids of AL2 and AL3 are in brackets. Nucleotide positions relative to the start of LAT transcription are not shown in parenthesis. Numbers in parentheses represent HSV-1 nucleotide positions.
2.3.2. Small Nonprotein Coding RNAs Are Encoded within the LAT Locus
Small noncoding RNAs can regulate gene expression [147, 148], promote neuronal differentiation [149], or inhibit apoptosis [150]. There are numerous types of small noncoding RNA: short interfering (si) RNA [151], small temporal RNA [152], heterochromatic siRNA [153], tiny noncoding RNAs [154], and micro-RNAs (miRNAs) [155]. miRNAs are nonprotein coding RNA molecules that are synthesized in the nucleus as 70–90 nucleotide precursors, and then processed into 21–23 nucleotide single-stranded RNA by the Dicer nuclease in the cytoplasm [155]. Dicer also processes siRNA. Following the discovery of 5 miRNAs encoded within the Epstein-Barr virus genome [156], miRNAs have been identified in Kaposis sarcoma-associated virus [157–159], mouse gammaherpesvirus 68 [157], human cytomegalovirus [157, 160, 161], HSV-1 [156, 162], Marek's disease virus [163], and simian virus (SV40) [164].
A study by Umbach et al. [136] concluded LAT is a miRNA precursor that encodes four miRNAs, and two within LAT promoter sequences (Figure 3(a)). One of these miRNAs, LAT miR-H6, inhibits ICP4 protein levels but not ICP4 RNA levels. ICP0 protein levels, but not RNA levels, are inhibited by another LAT miRNA, miR-H2-3p. The authors conclude that suppression of ICP0 and ICP4 by these miRNAs “facilitates the establishment and maintenance of viral latency.” Since the six LAT-specific miRNAs are not located within the first 1.5 kb of LAT coding sequences, they may only play a supportive role during the latency-reactivation cycle in small animal models of infection. The fact that LAT-specific miRNAs inhibit ICP0 or ICP4 suggests that they enhance the establishment or maintenance of latency. In the context of the latency-reactivation cycle in small animal models, it is unlikely, they are crucial when compared to the first 1.5 kb of LAT coding sequences.
Two additional small RNAs (s-RNAs) are encoded within the first 1.5 kb of LAT coding sequences (LAT s-RNA1 and s-RNA2) [138] (Figure 3(b)). Expression of LAT s-RNA1 and s-RNA2 is readily detected in trigeminal ganglia of latently infected mice [165]. LAT s-RNA2 inhibits ICP4 protein expression, but not RNA expression. LAT s-RNA1 inhibits productive infection approximately 1,000-fold in transient transfections assays, whereas LAT s-RNA2 only inhibits productive infection 5-fold [165]. These LAT s-RNAs may not be miRNAs because they lack Dicer cleavage sites and a mature miRNA band that migrates between 21 and 23 nucleotides was not detected. LAT s-RNA1 and s-RNA2 would not have been identified using the methods described by Umbach et al. [136] because they size selected RNA species migrating between 17 and 30 nucleotides, and LAT s-RNA1 is 62 nt long and LAT s-RNA2 is 36 nt long.
2.3.3. Novel Transcripts Are Expressed within LAT Coding Sequences
Sequences that encompass LAT also encode several additional transcripts. For example, novel transcripts within the LAT promoter region have been reported [166]. More recently, a transcript and protein, UOL (Upstream of LAT), was identified that is encoded within the LAT promoter regulatory region [167]. Deletion of UOL does not dramatically reduce the spontaneous reactivation phenotype in rabbits [168]. Another transcript, antisense to LAT (AL), is expressed within the first 1.5 kb of LAT coding sequences and the start site of the LAT promoter and appears to encode a protein [169] (see Figure 3 for location of UOL and AL).
Two small ORFs that are also antisense to LAT (AL2 and AL3) have been identified within the first 1.5 kb of LAT coding sequences (Figure 3(b)). A transcript within the first 1.5 kb of LAT coding sequences (AL3) is expressed during productive infection and in trigeminal ganglia of latently infected mice [170]. Like AL, AL3 is antisense with respect to LAT. An AL3 protein was also detected in cells transfected with an AL3 expression vector, and in trigeminal ganglia of infected mice. Conversely, an AL3 protein was not detected during productive infection, in part, because the 5′ terminus of the AL3 transcript is downstream of the first in frame methionine of AL3. It is not currently known whether a transcript encompassing AL2 is expressed during productive infection or during latency. It remains to be seen whether AL2 or AL3 plays a role in the latency-reactivation cycle of HSV-1.
2.4. LAT Regulates the Latency-Reactivation Cycle
As discussed above, the latency-reactivation cycle of HSV-1 can be operationally defined in 3 steps: establishment of latency, maintenance of latency, and reactivation from latency (summarized in Figure 1). In a human being, latency is maintained for the life of the host, indicating that a well-conceived strategy exists that allows for periodic reactivation, while maintaining the viral genome in sensory neurons.
Numerous HSV-1 mutants that do not express detectable levels of LAT have been constructed and tested in animal models [17, 171]. Although a couple of studies have suggested that LAT plays no role in a latent infection [172, 173], most have concluded that LAT is important but not required. LAT enhances establishment of latency in mice [174, 175] because certain LAT-mutants contain lower levels of viral DNA in murine TG relative to wild type virus [21, 176]. Furthermore, LAT enhances establishment of latency in the rabbit eye model and consequently reduces reactivation from latency [177]. The finding that LAT represses productive viral gene expression in TG of mice during acute infection [178, 179] supports the studies concluding that LAT facilitates establishment of latency. When considering the role that LAT plays in reactivation from latency, its role in establishing latency must be taken into consideration.
LAT enhances establishment of latency in mice [174, 175] or rabbits [180] because certain LAT mutants contain lower levels of viral DNA in TG relative to wt virus [21, 176]. LAT represses productive viral gene expression in TG of mice during acute infection [178, 179] supporting the concept that LAT facilitates establishment of latency. The HSV-1 McKrae strain is frequently shed in tears of infected rabbits as a result of spontaneous reactivation, and LAT is crucial for spontaneous reactivation [177, 181–184]. Furthermore, HSV-1 17syn+ strains with deletions in LAT coding sequences do not reactivate efficiently using the rabbit eye model [185, 186]. Although LAT overlaps the ICP0 transcript, LAT sequences that promote the latency-reactivation cycle in rabbits do not overlap ICP0 [132].
LAT is also important for in vivo reactivation using two different rabbit eye infection models. The McKrae strain of HSV-1 is frequently shed in the tears of infected rabbits as a result of spontaneous reactivation [177, 181, 183, 184, 187]. In contrast, spontaneous reactivation is severely impaired if the LAT gene is deleted. However, these same LAT-mutants grow with the same efficiency as wild-type virus in cultured cells and in ocular tissue of infected rabbits. The first 1.5 Kb of the gene encoding LAT is sufficient for spontaneous reactivation from latency [177] (Figure 3). Since this region does not overlap ICP0, antisense repression of ICP0 expression by LAT does not appear to be required for spontaneous reactivation in the rabbit model. HSV-1 17syn+ strains that have deletions in the LAT promoter and 5′ region of the gene encoding LAT (approximately 1,200 base pair) also do not reactivate efficiently in a rabbit eye model [185, 186].
It is not clear whether LAT encodes a protein that regulates the latency-reactivation cycle. Although certain studies suggested that LAT does not encode a protein [137], several studies have concluded that a protein encoded within LAT sequences is expressed [118, 167, 188–192]. These proteins were suggested to substitute for ICP0 functions [191, 192], interfere with binding of ICP4 to DNA [190], or their functions were not described. These proposed LAT proteins map downstream of the critical first 1.5 kb of the primary LAT transcript, a region that appears both sufficient and necessary for LAT's antiapoptosis activity and its ability to support a wild type spontaneous reactivation phenotype [177, 193]. Within the first 1.5 kb of LAT coding sequences, 8 potential ORFs have been identified in the strain McKrae [137] (summarized in Figure 3(b)). A recent study has provided evidence that L2 (Figure 3(b)), which is located in the first 1.5 kb of LAT coding sequences, appears to be expressed in TG of latently infected mice [194]. In summary, the gene encoding LAT does not appear to be absolutely required for latency in small animal models. However, the importance of LAT may be underestimated using small animal models and measuring latency in terms of weeks or months, not decades. The involvement of a LAT encoded protein in the latency-reactivation cycle is unclear.
2.5. HSV-1 Encodes Several Genes That Regulate Apoptosis
2.5.1. Genes Expressed during Productive Infection Inhibit Apoptosis
Many viruses induce apoptosis in cultured cells [195–198]. Killing of infected cells by apoptosis in vivo can reduce inflammation, alter immune recognition, reduce burst size, and thus prevent virus spread. Members of the Alphaherpesvirinae subfamily induce apoptosis after infection of cultured cells [199–202]. HSV-1 can also induce or inhibit apoptosis in a cell type dependent manner after infection of cultured cells [200, 201, 203–205]. Several antiapoptotic genes encoded by HSV-1 (ICP27, Us3, Us5, gJ, gD, and LAT) have been identified [200, 201, 203, 204, 206–212]. Us3 is a protein kinase that, in the absence of other HSV-1 proteins, inhibits cleavage of BAD and formation of the proapoptotic form of BAD. Us3 is the only viral protein required for preventing caspase 3 activation, which is the “point of no return” following apoptosis induction. The presence of several HSV-1 antiapoptotic genes suggests that they have specific roles following infection of humans.
HSV infection can induce apoptosis by several distinct mechanisms. For example, HSV induces DNA damage, even in the absence of productive infection [213–217]. DNA damage is a potent stimulus for apoptosis [217]. When expressed from baculovirus expression vectors, Us1.5 and UL13 can activate caspase 3 [218]. As expected, Us3 can inhibit the proapoptotic activity of Us1.5 and UL13 because it can interfere with caspase 3 activation.
2.5.2. LAT Inhibits Apoptosis
LAT interferes with apoptosis in transiently transfected cells and TG of infected mice or rabbits [139, 219–221]. LAT expressing plasmids inhibit caspase 8- and caspase 9-induced apoptosis [222, 223], the two major apoptotic pathways in mammals [224–226]. LAT also inhibits caspase 3 activation [227]. The antiapoptosis functions of LAT correlate with promoting spontaneous reactivation [219, 222]. In fact, inhibiting apoptosis appears to be the most important function of LAT because three different antiapoptosis genes [228–231] restore wt levels of spontaneous reactivation to a LAT null mutant.
LAT s-RNA1 and s-RNA2 (Figure 3) cooperate to inhibit cold-shock-induced apoptosis in transiently transfected mouse neuroblastoma cells [165]. Introduction of ATG→TTG mutations in ORFs within the first 1.5 kb of LAT coding sequences impairs the antiapoptotic functions of LAT [232] suggesting that LAT encodes a functional protein or alters RNA structure. Two of these ATG→TTG mutations are within LAT sRNA1 and sRNA2, and introducing these mutations into both small RNAs inhibits their ability to inhibit apoptosis [165]. Although this suggests that the LAT sRNAs mediate the antiapoptotic functions of the first 1.5 kb of LAT coding sequences, there may be additional functions within this region that have antiapoptosis functions.
2.6. Model Describing How LAT Regulates the Latency-Reactivation Cycle
Based on published studies, a working model has been devised to explain how LAT regulates the latency-reactivation cycle. During acute infection of TG (1–4 dpi), extensive viral gene expression occurs [70–72]. The toxic effects of HSV-1 infection, in particular ICP0 [233, 234], US1.5, and UL13 [218], make neurons vulnerable to damage and death. The ability of HSV to induce DNA damage [213, 215, 216, 235] would also stimulate the mitochondrial pathway of apoptosis [217]. The antiapoptotic properties of US3, US5, gD, gJ, ICP27, and LAT would promote neuronal survival during acute infection [139, 165, 200, 201, 203, 204, 209, 219–223]. Deletion of LAT might not have a dramatic effect on apoptosis frequency during the early stages of acute infection because the other viral antiapoptotic genes are expressed.
During transition from acute infection to latency (establishment of latency), viral gene expression is extinguished. The ability of the LAT micro-RNAs to inhibit ICP0 and ICP4 proteins expression [136] as well as the ability of LAT sRNA1 and LAT sRNA2 to inhibit productive infection [165] are likely to promote the establishment of latency. Furthermore, LAT would be the only viral antiapoptotic gene abundantly expressed during the establishment of latency. Neurons in which extensive viral gene expression had occurred during acute infection (permissive neurons) would be vulnerable to apoptosis in the absence of LAT expression. Nonpermissive neurons that harbor viral genomes would have suffered low levels of viral induced damage and thus would have a higher probability of survival in the absence of LAT. In mice, subsets of neurons have been identified in TG and the ability of HSV-1 to infect these neurons is different [236], supporting the concept that permissive and nonpermissive neurons exist.
The antiapoptosis functions of LAT would also appear to be crucial for protecting neurons from apoptotic stimuli during the maintenance of latency because it is the only viral gene that is abundantly expressed (Figure 1). In fact, during latency, LAT does have an effect on the number of surviving neurons following infection of mice [237]. Furthermore, the ability of LAT micro-RNAs [136] and LAT sRNA1 or LAT sRNA2 [165] to inhibit viral gene expression and/or productive infection would promote maintenance of latency. Since LAT sRNA1 and LAT sRNA2 [165] are located within the first 1.5 kb of LAT coding sequences, these small RNAs appear to be more important than the LAT micro-RNAs. However, the ability of the respective LAT noncoding RNAs to inhibit viral gene expression or productive infection is not as important as inhibiting apoptosis because three different antiapoptosis genes restore wt levels of spontaneous reactivation to a LAT null mutant [228–231].
The response of the central or peripheral nervous system to trauma, stress, or immunosuppression plays an important role during reactivation from latency. Stress leads to elevated corticosteriod levels, which has rapid effects on neural activity [238, 239]. Dexamethasone, a synthetic corticosteriod, induces viral gene expression [240], stimulates an HSV-1 origin of replication (Ori-L) in neuronal cells [50], and alters splicing patterns in the absence of protein synthesis [241]. Corticosteroids, or other forms of stress or trauma can induce neuronal neurodegeneration and/or apoptosis [242–248]. Since reactivation induces productive gene expression, all HSV-1 antiapoptotic genes would be expressed and should prolong neuronal survival, thus enhancing virus production.
2.7. Cell-Mediated Immune Responses Are Important for the Latency-Reactivation Cycle
2.7.1. Infiltration of Lymphocytes to TG during Acute Infection
Several independent studies have demonstrated that T cells, CD8+ T lymphocytes in particular, are crucial for controlling HSV infection in sensory ganglia [249, 250]. During acute infection, HSV antigen expression increases until 3 dpi in TG but is undetectable at 7 dpi [251]. Coincident with a decline of HSV antigen in TG there is an increase in Mac-1+ cells, macrophages, natural killer cells (NK), and certain CD8+ cells. No cells with characteristic lymphoid cell morphology can be detected in uninfected TG. After 5 dpi, the number of CD8+ T cells, F4/80+ cells (macrophages), and γδ T cells increases dramatically. At 3 dpi, TG neurons that are viral antigen positive can be detected that are surrounded by nonneural cells expressing TNF-α, IL-6, or IFN-γ [252]. Cells that express IL-2 or IL-4 are detected later after infection when viral antigens are difficult to detect. The number of cells producing IFN-γ and IL-4 increases between 3 and 7 dpi but the same cells do not appear to produce both factors [251]. At 7 days after infection, transcripts encoding IL-2, IL-10, IFN-γ, TNF-α, or RANTES (regulated upon activation, normal T cell expressed and secreted mRNA) are detected by RT-PCR [253]. By ELISA, IL-2, IL-6, IL-10, and IFN-γ are detected at the same time confirming the RT-PCR results. The same cellular antigens were not detected in TG from uninfected mice indicating that these changes were induced by infection.
2.7.2. Persistence of Lymphocytes in the Peripheral Nervous System during Latency
If true latency of HSV is established, cytokine expression in TG would not be detected. However, several studies have concluded that a persistent cell-mediated immune response occurs in TG during latency, and that T cells, CD8+ T lymphocytes in particular, inhibit reactivation from latency [249–251, 254–258].
The obvious explanation for persistence of immune effecter cells in TG is that low levels of viral proteins are expressed and an immune response occurs. A careful examination of TG neurons for viral gene expression in HSV-1 latently infected mice (37–47 days after infection) demonstrated that abundant viral transcripts, viral protein, and viral DNA replication occur in approximately 1 neuron per 10 TG [259]. Infectious virus is not detected in these mice confirming that they were latently infected. Neurons expressing high levels of HSV-1 transcripts are invariably surrounded by foci of infiltrating white blood cells. The term “spontaneous molecular reactivation” has been coined to describe these rare neurons [259].
2.7.3. Interferon Can Inhibit Reactivation from Latency
Persistence of the immune system in TG during latency is believed to play a role in the latency-reactivation cycle. CD8+ T cells that produce interferon-γ play an important role in preventing reactivation from latency in sensory neurons in mice latently infected with HSV-1 [256, 257]. Two independent studies have also concluded that interferon-α and interferon-γ control recurrent herpetic lesions [260, 261]. In addition to interferon, lymphocyte-mediated cytotoxicity could inhibit virus spread in TG. Lymphocyte-mediated cytotoxicity induces two potent apoptotic pathways: the granule exocytosis and the Fas-Fas ligand pathways [262, 263]. The granule exocytosis pathway is employed predominantly by CD8+, natural killer, and lymphokine-activated killer cells. A recent study has demonstrated that release of granzyme B from CD8+ T cells into latently infected neurons helps to inhibit reactivation from latency by cleaving ICP4 [264]. Since it is well established that granzyme B activates caspase 3 and the intrinsic pathway of apoptosis [265], the ability of LAT to inhibit apoptosis during maintenance of latency appears to be important.
3. Conclusions
HSV-1 latency is a complicated virus host interaction that is crucial for virus transmission, survival in nature, and recurrent disease. Numerous studies have indicated that sensory neurons are the primary site for latency. Since LAT is abundantly expressed in latently infected neurons, it is not surprising to find that LAT is important for the latency-reactivation cycle in small animal models. It is currently not clear whether expression of a LAT protein is important. Given the fact that several LAT small RNAs, including 6 known micro-RNAs, are expressed during latency implies that these small nonprotein coding RNAs are important for life-long ltency in humans. The finding that LAT sRNA1 and sRNA2 cooperate to inhibit apoptosis and also can inhibit productive infection supports a regulatory role for these small RNAs during the latency-reactivation cycle. It is also possible that additional transcripts encoded within LAT coding sequences (AL, AL3, or UOL) play a role in the latency-reactivation cycle. It will be necessary to design viral mutants that do not express these respective factors and then test the ability of these viruses to reactivate from latency in small animal models of infection.
Acknowledgments
The laboratory of C. Jones is supported by two USDA Grants (08-00891 and 09-01653), a PHS Grant (R21AI069176), and in part by a PHS Grant (1P20RR15635) to the Nebraska Center for Virology. Dr. Guey Chuen Perng's laboratory is supported by PHS Grant NINDS/NS049556.
References
- 1.Croen KD, Ostrove JM, Dragovic LJ, Smialek JE, Straus SE. Latent herpes simplex virus in human trigeminal ganglia. Detection of an immediate early gene “anti-sense” transcript by in situ hybridization. The New England Journal of Medicine. 1987;317(23):1427–1432. doi: 10.1056/NEJM198712033172302. [DOI] [PubMed] [Google Scholar]
- 2.Nahmias AJ, Roizman B. Infection with herpes-simplex viruses 1 and 2. 3. The New England Journal of Medicine. 1973;289(15):781–789. doi: 10.1056/NEJM197310112891505. [DOI] [PubMed] [Google Scholar]
- 3.Nesburn AB. Report of the Corneal Disease Panel: Vision Research- a National Plan, 1983–1987 Part III. Saint-Louis, Mo, USA: Mosby Co.; 1983. [Google Scholar]
- 4.Zhao Z-S, Granucci F, Yeh L, Schaffer PA, Cantor H. Molecular mimicry by herpes simplex virus-type 1: autoimmune disease after viral infection. Science. 1998;279(5355):1344–1347. doi: 10.1126/science.279.5355.1344. [DOI] [PubMed] [Google Scholar]
- 5.Gesser RM, Koo SC. Latent herpes simplex virus type 1 gene expression in ganglia innervating the human gastrointestinal tract. The Journal of Virology. 1997;71(5):4103–4106. doi: 10.1128/jvi.71.5.4103-4106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohr JM, Nelson JA, Oldstone MBA. Is herpes simplex virus associated with peptic ulcer disease? The Journal of Virology. 1990;64(5):2168–2174. doi: 10.1128/jvi.64.5.2168-2174.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corey L. Herpes simplex virus. In: Mandell GL, Bennett JE, Dolin R, editors. Principles and Practice of Infectious Diseases. Philadelphia, Pa, USA: Elsevier/Churchill Livingston; 2005. Month. pp. 1762–1780. [Google Scholar]
- 8.Stanberry LR, Oxman MN, Simmons A. Herpes simplex viruses. In: Gorbach SL, Bartlett JG, Blacklow NR, editors. Infectious Dieseases. Philadelphia, Pa, USA: Lippincott Williams and Wilkins; 2004. pp. 1905–1917. [Google Scholar]
- 9.Yamada S, Kameyama T, Nagaya S, Hashizume Y, Yoshida M. Relapsing herpes simplex encephalitis: pathological confirmation of viral reactivation. Journal of Neurology Neurosurgery and Psychiatry. 2003;74(2):262–264. doi: 10.1136/jnnp.74.2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lahat E, Barr J, Barkai G, Paret G, Brand N, Barzilai A. Long term neurological outcome of herpes encephalitis. Archives of Disease in Childhood. 1999;80(1):69–71. doi: 10.1136/adc.80.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGrath N, Anderson NE, Croxson MC, Powell KF. Herpes simplex encephalitis treated with acyclovir: diagnosis and long term outcome. Journal of Neurology Neurosurgery and Psychiatry. 1997;63(3):321–326. doi: 10.1136/jnnp.63.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoldenberg B. Herpes simplex encephalitis. Scandinavian Journal of Infectious Diseases, Supplement. 1991;23(80):40–46. [PubMed] [Google Scholar]
- 13.DeBiasi RL, Kleinschmidt-DeMasters BK, Richardson-Burns S, Tyler KL. Central nervous system apoptosis in human herpes simplex virus and cytomegalovirus encephalitis. Journal of Infectious Diseases. 2002;186(11):1547–1557. doi: 10.1086/345375. [DOI] [PubMed] [Google Scholar]
- 14.Kurt-Jones EA, Chan M, Zhou S, et al. Herpes simplex virus 1 interaction with toll-like receptor 2 contributes to lethal encephalitis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(5):1315–1320. doi: 10.1073/pnas.0308057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casrouge A, Zhang S-Y, Eidenschenk C, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314(5797):308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 16.Jones C. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Advances in Virus Research. 1998;51:81–133. doi: 10.1016/s0065-3527(08)60784-8. [DOI] [PubMed] [Google Scholar]
- 17.Wagner EK, Bloom DC. Experimental investigation of herpes simplex virus latency. Clinical Microbiology Reviews. 1997;10(3):419–443. doi: 10.1128/cmr.10.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baringer JR, Pisani P. Herpes simplex virus genomes in human nervous system tissue analyzed by polymerase chain reaction. Annals of Neurology. 1994;36(6):823–829. doi: 10.1002/ana.410360605. [DOI] [PubMed] [Google Scholar]
- 19.Baringer JR, Swoveland P. Recovery of herpes-simplex virus from human trigeminal ganglions. The New England Journal of Medicine. 1973;288(13):648–650. doi: 10.1056/NEJM197303292881303. [DOI] [PubMed] [Google Scholar]
- 20.Bastian FO, Rabson AS, Yee CL, Tralka TS. Herpesvirus hominis: isolation from human trigeminal ganglion. Science. 1972;178(4058):306–307. doi: 10.1126/science.178.4058.306. [DOI] [PubMed] [Google Scholar]
- 21.Maggioncalda J, Mehta A, Su YH, Fraser NW, Block TM. Correlation between herpes simplex virus type 1 rate of reactivation from latent infection and the number of infected neurons in trigeminal ganglia. Virology. 1996;225(1):72–81. doi: 10.1006/viro.1996.0576. [DOI] [PubMed] [Google Scholar]
- 22.Mehta A, Maggioncalda J, Bagasra O, et al. In situ DNA PCR and RNA hybridization detection of herpes simplex virus sequences in trigeminal ganglia of latently infected mice. Virology. 1995;206(1):633–640. doi: 10.1016/s0042-6822(95)80080-8. [DOI] [PubMed] [Google Scholar]
- 23.Ramakrishnan R, Fink DJ, Jiang G, Desai P, Glorioso JC, Levine M. Competitive quantitative PCR analysis of herpes simplex virus type 1 DNA and latency-associated transcript RNA in latently infected cells of the rat brain. The Journal of Virology. 1994;68(3):1864–1873. doi: 10.1128/jvi.68.3.1864-1873.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramakrishnan R, Levine M, Fink DJ. PCR-based analysis of herpes simplex virus type 1 latency in the rat trigeminal ganglion established with a ribonucleotide reductase-deficient mutant. The Journal of Virology. 1994;68(11):7083–7091. doi: 10.1128/jvi.68.11.7083-7091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawtell NM. Comprehensive quantification of herpes simplex virus latency at the single-cell level. The Journal of Virology. 1997;71(7):5423–5431. doi: 10.1128/jvi.71.7.5423-5431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itzhaki RF, Lin WR, Shang D, Wilcock GK, Faragher B, Jamieson GA. Herpes simplex virus type 1 in brain and risk of Alzheimer’s disease. The Lancet. 1997;349(9047):241–244. doi: 10.1016/S0140-6736(96)10149-5. [DOI] [PubMed] [Google Scholar]
- 27.Liedtke W, Opalka B, Zimmermann CW, Lignitz E. Age distribution of latent herpes simplex virus 1 and varicella-zoster virus genome in human nervous tissue. Journal of the Neurological Sciences. 1993;116(1):6–11. doi: 10.1016/0022-510x(93)90082-a. [DOI] [PubMed] [Google Scholar]
- 28.Spear PG. Herpes simplex virus: receptors and ligands for cell entry. Cellular Microbiology. 2004;6(5):401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 29.Spear PG, Manoj S, Yoon M, Jogger CR, Zago A, Myscofski D. Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry. Virology. 2006;344(1):17–24. doi: 10.1016/j.virol.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 30.Spear PG, Longnecker R. Herpesvirus entry: an update. The Journal of Virology. 2003;77(19):10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87(3):427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 32.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280(5369):1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 33.Honess RW, Roizman B. Regulation of herpesvirus macromolecular synthesis. I: cascade regulation of the synthesis of three groups of viral proteins. The Journal of Virology. 1974;14(1):8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Hare P. The virion transactivator of herpes simplex virus. Seminars in Virology. 1993;4(3):145–155. [Google Scholar]
- 35.Carrozza MJ, DeLuca NA. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Molecular and Cellular Biology. 1996;16(6):3085–3093. doi: 10.1128/mcb.16.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLuca NA, McCarthy AM, Schaffer PA. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. The Journal of Virology. 1985;56(2):558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLuca NA, Schaffer PA. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Molecular and Cellular Biology. 1985;5(8):1997–2008. doi: 10.1128/mcb.5.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixon RAF, Schaffer PA. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. The Journal of Virology. 1980;36(1):189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy AM, McMahan L, Schaffer PA. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. The Journal of Virology. 1989;63(1):18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McMahan L, Schaffer PA. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. The Journal of Virology. 1990;64(7):3471–3485. doi: 10.1128/jvi.64.7.3471-3485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sacks WR, Greene CC, Aschman DP, Schaffer PA. Herpes simpex virus type 1 ICP27 is an essential regulatory protein. The Journal of Virology. 1985;55(3):796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts MS, Boundy A, O’Hare P, Pizzorno MC, Ciufo DM, Hayward GS. Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 (α4) promoter and a specific binding site for the IE175 (ICP4) protein. The Journal of Virology. 1988;62(11):4307–4320. doi: 10.1128/jvi.62.11.4307-4320.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Hare P, Hayward GS. Three trans-acting regulatory proteins of herpes simplex virus modulate immediate-early gene expression in a pathway involving positive and negative feedback regulation. The Journal of Virology. 1985;56(3):723–733. doi: 10.1128/jvi.56.3.723-733.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michael N, Roizman B. Repression of the herpes simplex virus 1 α4 gene by its gene product occurs within the context of the viral genome and is associated with all three identified cognate sites. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(6):2286–2290. doi: 10.1073/pnas.90.6.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu B, Rivera-Gonzalez R, Smith CA, DeLuca NA. Herpes simplex virus infected cell polypeptide 4 preferentially represses Sp1-activated over basal transcription from its own promoter. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(20):9528–9532. doi: 10.1073/pnas.90.20.9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu B, DeLuca N. Requirements for activation of the herpes simplex virus glycoprotein C promoter in vitro by the viral regulatory protein ICP4. The Journal of Virology. 1994;68(12):7953–7965. doi: 10.1128/jvi.68.12.7953-7965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith CA, Bates P, Rivera-Gonzalez R, Gu B, DeLuca NA. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. The Journal of Virology. 1993;67(8):4676–4687. doi: 10.1128/jvi.67.8.4676-4687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandri-Goldin RM, Mendoza GE. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes and Development. 1992;6(5):848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 49.Sandri-Goldin RM, Hibbard MK, Hardwicke MA. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. The Journal of Virology. 1995;69(10):6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hardwicke MA, Schaffer PA. Differential effects of nerve growth factor and dexamethasone on herpes simplex virus type 1 oriL- and oriS-dependent DNA replication in PC12 cells. The Journal of Virology. 1997;71(5):3580–3587. doi: 10.1128/jvi.71.5.3580-3587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hardy WR, Sandri-Goldin RM. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. The Journal of Virology. 1994;68(12):7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hill A, Juovic P, York I, et al. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375(6530):411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 53.Goldsmith K, Chen W, Johnson DC, Hendricks RL. Infected cell protein (ICP)47 enhances herpes simplex virus neurovirulence by blocking the CD8+ T cell response. Journal of Experimental Medicine. 1998;187(3):341–348. doi: 10.1084/jem.187.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jordan R, Schaffer PA. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. The Journal of Virology. 1997;71(9):6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α1; regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. The Journal of Virology. 1997;71(2):1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. The Journal of Virology. 1997;71(10):7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meredith M, Orr A, Everett R. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology. 1994;200(2):457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 58.Meredith M, Orr A, Elliott M, Everett R. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology. 1995;209(1):174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 59.Everett R, O’Hare P, O’Rourke D, Barlow P, Orr A. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. The Journal of Virology. 1995;69(11):7339–7344. doi: 10.1128/jvi.69.11.7339-7344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Everett RD, Lomonte P, Sternsdorf T, van Driel R, Orr A. Cell cycle regulation of PML modification and ND10 composition. Journal of Cell Science. 1999;112(24):4581–4588. doi: 10.1242/jcs.112.24.4581. [DOI] [PubMed] [Google Scholar]
- 61.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. The EMBO Journal. 1997;16(3):566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. The EMBO Journal. 1997;16(7):1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poon APW, Liang Y, Roizman B. Herpes simplex virus 1 gene expression is accelerated by inhibitors of histone deacetylases in rabbit skin cellls infected with a mutant carrying a cDNA copy of the infected cell protein no. 0. The Journal of Virology. 2003;77(23):12671–12678. doi: 10.1128/JVI.77.23.12671-12678.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hobbs WE, II, DeLuca NA. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. The Journal of Virology. 1999;73(10):8245–8255. doi: 10.1128/jvi.73.10.8245-8255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu H, Liang Y, Mandel G, Roizman B. Components of the REST/CoREST/histone deacytlase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lomonte P, Seigneurin-Berny D, Everett RD, Khochbin S, Epstein AL. Interaction of HSV-1 ICP0 with class II histone deacytlases. In: Proceedings of the 26th International Herpesvirus Workshop; 2001; Regensburg, Germany. [Google Scholar]
- 67.Poon APW, Gu H, Roizman B. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(26):9993–9998. doi: 10.1073/pnas.0604142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cliffe AR, Knipe DM. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. The Journal of Virology. 2008;82(24):12030–12038. doi: 10.1128/JVI.01575-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cai W, Schaffer PA. A cellular function can enhance gene expression and plating efficiency of a mutant defective in the gene for ICP0, a transactivating protein of herpes simplex virus type 1. The Journal of Virology. 1991;65(8):4078–4090. doi: 10.1128/jvi.65.8.4078-4090.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knotts FB, Cook ML, Stevens JG. Pathogenesis of herpetic encephalitis in mice after ophthalmic inoculation. Journal of Infectious Diseases. 1974;130(1):16–27. doi: 10.1093/infdis/130.1.16. [DOI] [PubMed] [Google Scholar]
- 71.Kramer MF, Chen SH, Knipe DM, Coen DM. Accumulation of viral transcripts and DNA during establishment of latency by herpes simplex virus. The Journal of Virology. 1998;72(2):1177–1185. doi: 10.1128/jvi.72.2.1177-1185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Speck PG, Simmons A. Divergent molecular pathways of productive and latent infection with a virulent strain of herpes simplex virus type 1. The Journal of Virology. 1991;65(8):4001–4005. doi: 10.1128/jvi.65.8.4001-4005.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coen DM, Kosz-Vnenchak M, Jacobson JG, et al. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(12):4736–4740. doi: 10.1073/pnas.86.12.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leib DA, Coen DM, Bogard CL, et al. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. The Journal of Virology. 1989;63(2):759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Efstathiou S, Kemp S, Darby G, Minson AC. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. Journal of General Virology. 1989;70(4):869–879. doi: 10.1099/0022-1317-70-4-869. [DOI] [PubMed] [Google Scholar]
- 76.Katz JP, Bodin ET, Coen DM. Quantitative polymerase chain reaction analysis of herpes simplex virus DNA in ganglia of mice infected with replication-incompetent mutants. The Journal of Virology. 1990;64(9):4288–4295. doi: 10.1128/jvi.64.9.4288-4295.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Margolis TP, Sedarati F, Dobson AT, Feldman LT, Stevens JG. Pathways of viral gene expression during acute neuronal infection with HSV-1. Virology. 1992;189(1):150–160. doi: 10.1016/0042-6822(92)90690-q. [DOI] [PubMed] [Google Scholar]
- 78.Sedarati F, Izumi KM, Wagner EK, Stevens JG. Herpes simplex virus type 1 latency-associated transcription plays no role in establishment or maintenance of a latent infection in murine sensory neurons. The Journal of Virology. 1989;63(10):4455–4458. doi: 10.1128/jvi.63.10.4455-4458.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sedarati F, Margolis TP, Stevens JG. Latent infection can be established with drastically restricted transcription and replication of the HSV-1 genome. Virology. 1993;192(2):687–691. doi: 10.1006/viro.1993.1089. [DOI] [PubMed] [Google Scholar]
- 80.Steiner I, Spivack JG, Deshmane SL, Ace CI, Preston CM, Fraser NW. A herpes simplex virus type 1 mutant containing a nontransinducing Vmw65 protein establishes latent infection in vivo in the absence of viral replication and reactivates efficiently from explanted trigeminal ganglia. The Journal of Virology. 1990;64(4):1630–1638. doi: 10.1128/jvi.64.4.1630-1638.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valyi-Nagy T, Deshmane SL, Raengsakulrach B, et al. Herpes simplex virus type 1 mutant strain in1814 establishes a unique, slowly progressing infection in SCID mice. The Journal of Virology. 1992;66(12):7336–7345. doi: 10.1128/jvi.66.12.7336-7345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taus NS, Mitchell WJ. The transgenic ICP4 promoter is activated in Schwann cells in trigeminal ganglia of mice latently infected with herpes simplex virus type 1. The Journal of Virology. 2001;75(21):10401–10408. doi: 10.1128/JVI.75.21.10401-10408.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Loiacono CM, Myers R, Mitchell WJ. Neurons differentially activate the herpes simplex virus type 1 immediate-early gene ICP0 and ICP27 promoters in transgenic mice. The Journal of Virology. 2002;76(5):2449–2459. doi: 10.1128/jvi.76.5.2449-2459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Devireddy LR, Jones CJ. Olf-1, a neuron-specific transcription factor, can activate the herpes simplex virus type 1-infected cell protein 0 promoter. The Journal of Biological Chemistry. 2000;275(1):77–81. doi: 10.1074/jbc.275.1.77. [DOI] [PubMed] [Google Scholar]
- 85.O’Hare P, Goding CR. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell. 1988;52(3):435–445. doi: 10.1016/s0092-8674(88)80036-9. [DOI] [PubMed] [Google Scholar]
- 86.Lu R, Misra V. Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Research. 2000;28(12):2446–2454. doi: 10.1093/nar/28.12.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kristie TM, Pomerantz JL, Twomey TC, Parent SA, Sharp PA. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. The Journal of Biological Chemistry. 1995;270(9):4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 88.Kristie TM, Vogel JL, Sears AE. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(4):1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sears AE, Hukkanen V, Labow MA, Levine AJ, Roizman B. Expression of the herpes simplex virus 1 α transinducing factor (VP16) does not induce reactivation of latent virus or prevent the establishment of latency in mice. The Journal of Virology. 1991;65(6):2929–2935. doi: 10.1128/jvi.65.6.2929-2935.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hagmann M, Georgiev O, Schaffner W, Douville P. Transcription factors interacting with herpes simplex virus α gene promoters in sensory neurons. Nucleic Acids Research. 1995;23(24):4978–4985. doi: 10.1093/nar/23.24.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Turner EE, Rhee JM, Feldman LT. The POU-domain factor Brn-3.0 recognizes characteristic sites in the herpes simplex virus genome. Nucleic Acids Research. 1997;25(13):2589–2594. doi: 10.1093/nar/25.13.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gruber CA, Rhee JM, Gleiberman A, Turner EE. POU domain factors of the Brn-3 class recognize functional DNA elements which are distinctive, symmetrical, and highly conserved in evolution. Molecular and Cellular Biology. 1997;17(5):2391–2400. doi: 10.1128/mcb.17.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McEvilly RJ, Erkman L, Luo L, Sawchenko PE, Ryan AF, Rosenfeld MG. Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature. 1996;384(6609):574–577. doi: 10.1038/384574a0. [DOI] [PubMed] [Google Scholar]
- 94.Schonemann MD, Ryan AK, Erkman L, McEvilly RJ, Bermingham J, Rosenfeld MG. POU domain factors in neural development. Advances in Experimental Medicine and Biology. 1998;449:39–53. doi: 10.1007/978-1-4615-4871-3_4. [DOI] [PubMed] [Google Scholar]
- 95.Erkman L, McEvilly RJ, Luo L, et al. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381(6583):603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- 96.Morris PJ, Theil T, Ring CJA, Lillycrop KA, Moroy T, Latchman DS. The opposite and antagonistic effects of the closely related POU family transcription factors Brn-3a and Brn-3b on the activity of a target promoter are dependent on differences in the POU domain. Molecular and Cellular Biology. 1994;14(10):6907–6914. doi: 10.1128/mcb.14.10.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen X-P, Li J, Mata M, et al. Herpes simplex virus type 1 ICP0 protein does not accumulate in the nucleus of primary neurons in culture. The Journal of Virology. 2000;74(21):10132–10141. doi: 10.1128/jvi.74.21.10132-10141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hsu WL, Everett RD. Human neuron-committed teratocarcinoma NT2 cell line has abnormal ND10 structures and is poorly infected by herpes simplex virus type 1. The Journal of Virology. 2001;75(8):3819–3831. doi: 10.1128/JVI.75.8.3819-3831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen X, Schmidt MC, Goins WF, Glorioso JC. Two herpes simplex virus type 1 latency-active promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections. The Journal of Virology. 1995;69(12):7899–7908. doi: 10.1128/jvi.69.12.7899-7908.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goins WF, Sternberg LR, Croen KD, et al. A novel latency-active promoter is contained within the herpes simplex virus type 1 U(L) flanking repeats. The Journal of Virology. 1994;68(4):2239–2252. doi: 10.1128/jvi.68.4.2239-2252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Deshmane SL, Nicosia M, Valyi-Nagy T, Feldman LT, Dillner A, Fraser NW. An HSV-1 mutant lacking the LAT TATA element reactivates normally in explant cocultivation. Virology. 1993;196(2):868–872. doi: 10.1006/viro.1993.1548. [DOI] [PubMed] [Google Scholar]
- 102.Dobson AT, Margolis TP, Gomes WA, Feldman LT. In vivo deletion analysis of the herpes simplex virus type 1 latency- associated transcript promoter. The Journal of Virology. 1995;69(4):2264–2270. doi: 10.1128/jvi.69.4.2264-2270.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dobson AT, Sederati F, Devi-Rao G, et al. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. The Journal of Virology. 1989;63(9):3844–3851. doi: 10.1128/jvi.63.9.3844-3851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mitchell WJ, Lirette RP, Fraser NW. Mapping of low abundance latency-associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. Journal of General Virology. 1990;71(1):125–132. doi: 10.1099/0022-1317-71-1-125. [DOI] [PubMed] [Google Scholar]
- 105.Batchelor AH, O’Hare P. Regulation and cell-type-specific activity of a promoter located upstream of the latency-associated transcript of herpes simplex virus type 1. The Journal of Virology. 1990;64(7):3269–3279. doi: 10.1128/jvi.64.7.3269-3279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Batchelor AH, O’Hare P. Localization of cis-acting sequence requirements in the promoter of the latency-associated transcript of herpes simplex virus type 1 required for cell-type-specific activity. The Journal of Virology. 1992;66(6):3573–3582. doi: 10.1128/jvi.66.6.3573-3582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Batchelor AH, Wilcox KW, O’Hare P. Binding and repression of the latency-associated promoter of herpes simplex virus by the immediate early 175K protein. Journal of General Virology. 1994;75(4):753–767. doi: 10.1099/0022-1317-75-4-753. [DOI] [PubMed] [Google Scholar]
- 108.Zwaagstra JC, Ghiasi H, Nesburn AB, Wechsler SL. Identification of a major regulatory sequence in the latency associated transcript (LAT) promoter of herpes simplex virus type 1 (HSV-1) Virology. 1991;182(1):287–297. doi: 10.1016/0042-6822(91)90672-x. [DOI] [PubMed] [Google Scholar]
- 109.Zwaagstra JC, Ghiasi H, Slanina SM, et al. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. The Journal of Virology. 1990;64(10):5019–5028. doi: 10.1128/jvi.64.10.5019-5028.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kenny JJ, Krebs FC, Hartle HT, et al. Identification of a second ATF/CREB-like element in the herpes simplex virus type 1 (HSV-1) latency-associated transcript (LAT) promoter. Virology. 1994;200(1):220–235. doi: 10.1006/viro.1994.1180. [DOI] [PubMed] [Google Scholar]
- 111.Leib DA, Nadeau KC, Rundle SA, Schaffer PA. The promoter of the latency-associated transcripts of herpes simplex virus type 1 contains a functional cAMP-response element: role of the latency-associated transcripts and cAMP in reactivation of viral latency. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(1):48–52. doi: 10.1073/pnas.88.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bloom DC, Stevens JG, Hill JM, Tran RK. Mutagenesis of a cAMP response element within the latency-associated transcript promoter of HSV-1 reduces adrenergic reactivation. Virology. 1997;236(1):202–207. doi: 10.1006/viro.1997.8723. [DOI] [PubMed] [Google Scholar]
- 113.Rader KA, Ackland-Berglund CE, Miller JK, Pepose JS, Leib DA. In vivo characterization of site-directed mutations in the promoter of the herpes simplex virus type 1 latency-associated transcripts. Journal of General Virology. 1993;74(9):1859–1869. doi: 10.1099/0022-1317-74-9-1859. [DOI] [PubMed] [Google Scholar]
- 114.Rock DL, Nesburn AB, Ghiasi H, et al. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. The Journal of Virology. 1987;61(12):3820–3826. doi: 10.1128/jvi.61.12.3820-3826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to a herpesvirus α gene mRNA is prominent in latently infected neurons. Science. 1987;235(4792):1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 116.Chou J, Kern ER, Whitley RJ, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in culture. Science. 1990;250(4985):1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 117.Chou J, Roizman B. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+ Journal of Virology. 1990;64(3):1014–1020. doi: 10.1128/jvi.64.3.1014-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lagunoff M, Roizman B. Expression of a herpes simplex virus 1 open reading frame antisense to the γ(1)34.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. Journal of Virology. 1994;68(9):6021–6028. doi: 10.1128/jvi.68.9.6021-6028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yeh L, Schaffer PA. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. Journal of Virology. 1993;67(12):7373–7382. doi: 10.1128/jvi.67.12.7373-7382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bohenzky RA, Lagunoff M, Roizman B, Wagner EK, Silverstein S. Two overlapping transcription units which extend across the L-S junction of herpes simplex virus type 1. Journal of Virology. 1995;69(5):2889–2897. doi: 10.1128/jvi.69.5.2889-2897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bohenzky RA, Papavassiliou AG, Gelman IH, Silverstein S. Identification of a promoter mapping within the reiterated sequences that flank the herpes simplex virus type 1 UL region. Journal of Virology. 1993;67(2):632–642. doi: 10.1128/jvi.67.2.632-642.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Berthomme H, Lokensgard J, Yang L, Margolis T, Feldman LT. Evidence for a bidirectional element located downstream from the herpes simplex virus type 1 latency-associated promoter that increases its activity during latency. The Journal of Virology. 2000;74(8):3613–3622. doi: 10.1128/jvi.74.8.3613-3622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lokensgard JR, Berthomme H, Feldman LT. The latency-associated promoter of herpes simplex virus type 1 requires a region downstream of the transcription start site for long-term expression during latency. The Journal of Virology. 1997;71(9):6714–6719. doi: 10.1128/jvi.71.9.6714-6719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lokensgard JR, Berthomme H, Feldman LT. The latency-associated promoter of herpes simplex virus type 1 requires a region downstream of the transcription start site for long-term expression during latency. The Journal of Virology. 1997;71(9):6714–6719. doi: 10.1128/jvi.71.9.6714-6719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lokensgard JR, Bloom DC, Dobson AT, Feldman LT. Long-term promoter activity during herpes simplex virus latency. The Journal of Virology. 1994;68(11):7148–7158. doi: 10.1128/jvi.68.11.7148-7158.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kubat NJ, Tran RK, McAnany P, Bloom DC. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. The Journal of Virology. 2004;78(3):1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Deatly AM, Spivack JG, Lavi E, Fraser NW. RNA from an immediate early region of the type 1 herpes simplex virus genome is present in the trigeminal ganglia of latently infected mice. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(10):3204–3208. doi: 10.1073/pnas.84.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Deatly AM, Spivack JG, Lavi E, O’Boyle DR, II, Fraser NW. Latent herpes simplex virus type 1 transcripts in peripheral and central nervous system tissues of mice map to similar regions of the viral genome. The Journal of Virology. 1988;62(3):749–756. doi: 10.1128/jvi.62.3.749-756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Krause PR, Croen KD, Straus SE, Ostrove JM. Detection and preliminary characterization of herpes simplex virus type 1 transcripts in latently infected human trigeminal ganglia. The Journal of Virology. 1988;62(12):4819–4823. doi: 10.1128/jvi.62.12.4819-4823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wagner EK, Flanagan WM, Devi-Rao G, et al. The herpes simplex virus latency-associated transcript is spliced during the latent phase of infection. The Journal of Virology. 1988;62(12):4577–4585. doi: 10.1128/jvi.62.12.4577-4585.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wagner EK, Devi-Rao G, Feldman LT, et al. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. The Journal of Virology. 1988;62(4):1194–1202. doi: 10.1128/jvi.62.4.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Perng GC, Ghiasi H, Slanina SM, Nesburn AB, Wechsler SL. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. The Journal of Virology. 1996;70(2):976–984. doi: 10.1128/jvi.70.2.976-984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kramer MF, Coen DM. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. The Journal of Virology. 1995;69(3):1389–1399. doi: 10.1128/jvi.69.3.1389-1399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shimeld C, Hill TJ, Blyth WA, Easty DL. Reactivation of latent infection and induction of recurrent herpetic eye disease in mice. Journal of General Virology. 1990;71(2):397–404. doi: 10.1099/0022-1317-71-2-397. [DOI] [PubMed] [Google Scholar]
- 135.Green MT, Courtney RJ, Dunkel EC. Detection of an immediate early herpes simplex virus type 1 polypeptide in trigeminal ganglia from latently infected animals. Infection and Immunity. 1981;34(3):987–992. doi: 10.1128/iai.34.3.987-992.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Drolet BS, Perng G-C, Cohen J, et al. The region of the herpes simplex virus type 1 LAT gene involved in spontaneous reactivation does not encode a functional protein. Virology. 1998;242(1):221–232. doi: 10.1006/viro.1997.9020. [DOI] [PubMed] [Google Scholar]
- 138.Peng W, Vitvitskaia O, Carpenter D, Wechsler SL, Jones C. Identification of two small RNAs within the first 1.5-kb of the herpes simplex virus type 1-encoded latency-associated transcript. Journal of NeuroVirology. 2008;14(1):41–52. doi: 10.1080/13550280701793957. [DOI] [PubMed] [Google Scholar]
- 139.Kang W, Mukerjee R, Fraser NF. Establishment and maintenance of HSV latent infection is mediated through correct splicing of the LAT primary transcript. Virology. 2003;312(1):233–244. doi: 10.1016/s0042-6822(03)00201-0. [DOI] [PubMed] [Google Scholar]
- 140.Mador N, Panet A, Latchman D, Steiner I. Expression and splicing of the latency-associated transcripts of herpes simplex virus type 1 in neuronal and non-neuronal cell lines. Journal of Biochemistry. 1995;117(6):1288–1297. doi: 10.1093/oxfordjournals.jbchem.a124857. [DOI] [PubMed] [Google Scholar]
- 141.Krummenacher C, Zabolotny JM, Fraser NW. Selection of a nonconsensus branch point is influenced by an RNA stem-loop structure and is important to confer stability to the herpes simplex virus 2-kilobase latency-associated transcript. The Journal of Virology. 1997;71(8):5849–5860. doi: 10.1128/jvi.71.8.5849-5860.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Farrell MJ, Dobson AT, Feldman LT. Herpes simplex virus latency-associated transcript is a stable intron. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(3):790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ahmed M, Fraser NW. Herpes simplex virus type 1 2-kilobase latency-associated transcript intron associates with ribosomal proteins and splicing factors. The Journal of Virology. 2001;75(24):12070–12080. doi: 10.1128/JVI.75.24.12070-12080.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nicosia M, Zabolotny JM, Lirette RP, Fraser NW. The HSV-1 2-kb latency-associated transcript is found in the cytoplasm comigrating with ribosomal subunits during productive infection. Virology. 1994;204(2):717–728. doi: 10.1006/viro.1994.1587. [DOI] [PubMed] [Google Scholar]
- 145.Thomas DL, Lock M, Zabolotny JM, Mohan BR, Fraser NW. The 2-kilobase intron of the herpes simplex virus type 1 latency-associated transcript has a half-life of approximately 24 hours in SY5Y and COS-1 cells. Journal of Virology. 2002;76(2):532–540. doi: 10.1128/JVI.76.2.532-540.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Goldenberg D, Mador N, Ball MJ, Panet A, Steiner I. The abundant latency-associated transcripts of herpes simplex virus type 1 are bound to polyribosomes in cultured neuronal cells and during latent infection in mouse trigeminal ganglia. Journal of Virology. 1997;71(4):2897–2904. doi: 10.1128/jvi.71.4.2897-2904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hannon GJ. RNA interference. Nature. 2002;418(6894):244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 148.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nature Reviews Molecular Cell Biology. 2003;4(6):457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 149.Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116(6):779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- 150.Xu P, Guo M, Hay BA. microRNAs and the regulation of cell death. Trends in Genetics. 2004;20(12):617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 151.Finnegan EJ, Matzke MA. The small RNA world. Journal of Cell Science. 2003;116(23):4689–4693. doi: 10.1242/jcs.00838. [DOI] [PubMed] [Google Scholar]
- 152.Banerjee D, Slack F. Control of developmental timing by small temporal rnas: a paradigm for rna-mediated regulation of gene expression. BioEssays. 2002;24(2):119–129. doi: 10.1002/bies.10046. [DOI] [PubMed] [Google Scholar]
- 153.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297(5588):1831–1835. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 154.Fraser A. Worms in LA. Nature Genetics. 2003;35:3–5. doi: 10.1038/ng0903-3. [DOI] [PubMed] [Google Scholar]
- 155.Bartel DP. microRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 156.Pfeffer S, Zavolan M, Grasser FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 157.Pfeffer S, Sewer A, Lagos-Quintana M, et al. Identification of microRNAs of the herpesvirus family. Nature Methods. 2005;2(4):269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 158.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(15):5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kapos’s sarcoma-associated herpesvirus. Journal of Virology. 2005;79(14):9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Grey F, Antoniewicz A, Allen E, et al. Identification and characterization of human cytomegalovirus-encoded microRNAs. Journal of Virology. 2005;79(18):12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dunn W, Trang P, Zhong Q, Yang E, van Belle C, Liu F. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cellular Microbiology. 2005;7(11):1684–1695. doi: 10.1111/j.1462-5822.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- 162.Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2006;442(7098):82–85. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- 163.Burnside J, Bernberg E, Anderson A, et al. Marek’s disease virus encodes microRNAs that map to meq and the latency-associated transcript. Journal of Virology. 2006;80(17):8778–8786. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435(7042):682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 165.Shen W, Silva MS, Jaber T, et al. Two small RNAs encoded within the first 1.5 kilobases of the herpes simplex virus type 1 latency-associated transcript can inhibit productive infection and cooperate to inhibit apoptosis. Journal of Virology. 2009;83(18):9131–9139. doi: 10.1128/JVI.00871-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Singh J, Wagner E. Transcriptional analysis of the herpes simplex virus type 1 region containing the TRL/UL junction. Virology. 1993;196(1):220–231. doi: 10.1006/viro.1993.1470. [DOI] [PubMed] [Google Scholar]
- 167.Naito J, Mukerjee R, Mott KR, et al. Identification of a protein encoded in the herpes simplex virus type 1 latency associated transcript promoter region. Virus Research. 2005;108(1-2):101–110. doi: 10.1016/j.virusres.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 168.Chan D, Cohen J, Naito J, et al. A mutant deleted for most of the herpes simplex virus type 1 (HSV-1) UOL gene does not affect the spontaneous reactivation phenotype in rabbits. Journal of NeuroVirology. 2006;12(1):5–16. doi: 10.1080/13550280500516401. [DOI] [PubMed] [Google Scholar]
- 169.Perng G-C, Maguen B, Jin L, et al. A novel herpes simplex virus type 1 transcript (AL-RNA) antisense to the 5′ end of the latency-associated transcript produces a protein in infected rabbits. Journal of Virology. 2002;76(16):8003–8010. doi: 10.1128/JVI.76.16.8003-8010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jaber T, Henderson G, Li S, et al. Identification of a novel herpes simplex virus type 1 transcript and protein (AL3) expressed during latency. Journal of General Virology. 2009;90(10):2342–2352. doi: 10.1099/vir.0.013318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jones C. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clinical Microbiology Reviews. 2003;16(1):79–95. doi: 10.1128/CMR.16.1.79-95.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Block TM, Spivack JG, Steiner I, et al. A herpes simplex virus type 1 latency-associated transcript mutant reactivates with normal kinetics from latent infection. Journal of Virology. 1990;64(7):3417–3426. doi: 10.1128/jvi.64.7.3417-3426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ho DY, Mocarski ES. β-galactosidase as a marker in the peripheral and neural tissues of the herpes simplex virus-infected mouse. Virology. 1988;167(1):279–283. doi: 10.1016/0042-6822(88)90079-7. [DOI] [PubMed] [Google Scholar]
- 174.Thompson RL, Sawtell NM. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. Journal of Virology. 1997;71(7):5432–5440. doi: 10.1128/jvi.71.7.5432-5440.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sawtell NM, Thompson RL. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. Journal of Virology. 1992;66(4):2157–2169. doi: 10.1128/jvi.66.4.2157-2169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Devi-Rao GB, Bloom DC, Stevens JG, Wagner EK. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. Journal of Virology. 1994;68(3):1271–1282. doi: 10.1128/jvi.68.3.1271-1282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Perng G-C, Ghiasi H, Slanina SM, Nesburn AB, Wechsler SL. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. Journal of Virology. 1996;70(2):976–984. doi: 10.1128/jvi.70.2.976-984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chen S-H, Kramer MF, Schaffer PA, Coen DM. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. Journal of Virology. 1997;71(8):5878–5884. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Garber DA, Schaffer PA, Knipe DM. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. Journal of Virology. 1997;71(8):5885–5893. doi: 10.1128/jvi.71.8.5885-5893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Perng G-C, Slanina SM, Yukht A, Ghiasi H, Nesburn AB, Wechsler SL. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. Journal of Virology. 2000;74(4):1885–1891. doi: 10.1128/jvi.74.4.1885-1891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Perng G-C, Dunkel EC, Geary PA, et al. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. Journal of Virology. 1994;68(12):8045–8055. doi: 10.1128/jvi.68.12.8045-8055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Perng G-C, Chokephaibulkit K, Thompson RL, et al. The region of the herpes simplex virus type 1 LAT gene that is colinear with the ICP34.5 gene is not involved in spontaneous reactivation. Journal of Virology. 1996;70(1):282–291. doi: 10.1128/jvi.70.1.282-291.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Perng G-C, Slanina SM, Ghiasi H, Nesburn AB, Wechsler SL. A 371-nucleotide region between the herpes simplex virus type 1 (HSV-1) LAT promoter and the 2-kilobase LAT is not essential for efficient spontaneous reactivation of latent HSV-1. Journal of Virology. 1996;70(3):2014–2018. doi: 10.1128/jvi.70.3.2014-2018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Perng G-C, Slanina SM, Yukht A, et al. A herpes simplex virus type 1 latency-associated transcript mutant with increased virulence and reduced spontaneous reactivation. Journal of Virology. 1999;73(2):920–929. doi: 10.1128/jvi.73.2.920-929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Trousdale MD, Steiner I, Spivack JG, et al. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. Journal of Virology. 1991;65(12):6989–6993. doi: 10.1128/jvi.65.12.6989-6993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hill JM, Sedarati F, Javier RT, Wagner EK, Stevens JG. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology. 1990;174(1):117–125. doi: 10.1016/0042-6822(90)90060-5. [DOI] [PubMed] [Google Scholar]
- 187.Perng G-C, Slanina SM, Ghiasi H, Nesburn AB, Wechsler SL. The effect of latency-associated transcript on the herpes simplex virus type 1 latency-reactivation phenotype is mouse strain-dependent. Journal of General Virology. 2001;82(5):1117–1122. doi: 10.1099/0022-1317-82-5-1117. [DOI] [PubMed] [Google Scholar]
- 188.Doerig C, Pizer LI, Wilcox CL. An antigen encoded by the latency-associated transcript in neuronal cell cultures latently infected with herpes simplex virus type 1. Journal of Virology. 1991;65(5):2724–2727. doi: 10.1128/jvi.65.5.2724-2727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lock M, Miller C, Fraser NW. Analysis of protein expression from within the region encoding the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1. Journal of Virology. 2001;75(7):3413–3426. doi: 10.1128/JVI.75.7.3413-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Randall G, Lagunoff M, Roizman B. The product of ORF O located within the domain of herpes simplex virus 1 genome transcribed during latent infection binds to and inhibits in vitro binding of infected cell protein 4 to its cognate DNA site. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(19):10379–10384. doi: 10.1073/pnas.94.19.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Thomas SK, Gough G, Latchman DS, Coffin RS. Herpes simplex virus latency-associated transcript encodes a protein which greatly enhances virus growth, can compensate for deficiencies in immediate-early gene expression, and is likely to function during reactivation from virus latency. Journal of Virology. 1999;73(8):6618–6625. doi: 10.1128/jvi.73.8.6618-6625.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Thomas SK, Lilley CE, Latchman DS, Coffin RS. A protein encoded by the herpes simplex virus (HSV) type 1 2-kilobase latency-associated transcript is phosphorylated, localized to the nucleus, and overcomes the repression of expression from exogenous promoters when inserted into the quiescent HSV genome. Journal of Virology. 2002;76(8):4056–4067. doi: 10.1128/JVI.76.8.4056-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Perng G-C, Esmaili D, Slanina SM, et al. Three herpes simplex virus type 1 latency-associated transcript mutants with distinct and asymmetric effects on virulence in mice compared with rabbits. Journal of Virology. 2001;75(19):9018–9028. doi: 10.1128/JVI.75.19.9018-9028.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Henderson G, Jaber T, Carpenter D, Wechsler SL, Jones C. Identification of herpes simplex virus type 1 (HSV-1) proteins encoded within the first 1.5 kb of the latency-associated transcript (LAT) doi: 10.3109/13550280903296353. Journal of Neurovirology. In press. [DOI] [PubMed] [Google Scholar]
- 195.Hardwick JM. Viral interference with apoptosis. Seminars in Cell and Developmental Biology. 1998;9(3):339–349. doi: 10.1006/scdb.1998.0243. [DOI] [PubMed] [Google Scholar]
- 196.Razvi ES, Welsh RM. Apoptosis in viral infections. Advances in Virus Research. 1995;45:1–60. doi: 10.1016/s0065-3527(08)60057-3. [DOI] [PubMed] [Google Scholar]
- 197.Shen Y, Shenk TE. Viruses and apoptosis. Current Opinion in Genetics and Development. 1995;5(1):105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 198.Teodoro JG, Branton PE. Regulation of apoptosis by viral gene products. Journal of Virology. 1997;71(3):1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Devireddy LR, Jones CJ. Activation of caspases and p53 by bovine herpesvirus 1 infection results in programmed cell death and efficient virus release. Journal of Virology. 1999;73(5):3778–3788. doi: 10.1128/jvi.73.5.3778-3788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Galvan V, Brandimarti R, Roizman B. Herpes simplex virus 1 blocks caspase-3-independent and caspase-dependent pathways to cell death. Journal of Virology. 1999;73(4):3219–3226. doi: 10.1128/jvi.73.4.3219-3226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Galvan V, Roizman B. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(7):3931–3936. doi: 10.1073/pnas.95.7.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sadzot-Delvaux C, Thonard P, Schoonbroodt S, Piette J, Rentier B. Varicella-zoster virus induces apoptosis in cell culture. Journal of General Virology. 1995;76(11):2875–2879. doi: 10.1099/0022-1317-76-11-2875. [DOI] [PubMed] [Google Scholar]
- 203.Asano S, Honda T, Goshima F, et al. US3 protein kinase of herpes simplex virus type 2 plays a role in protecting corneal epithelial cells from apoptosis in infected mice. Journal of General Virology. 1999;80(1):51–56. doi: 10.1099/0022-1317-80-1-51. [DOI] [PubMed] [Google Scholar]
- 204.Aubert M, Blaho JA. The herpes simplex virus type 1 regulatory protein ICP27 is required for the prevention of apoptosis in infected human cells. Journal of Virology. 1999;73(4):2803–2813. doi: 10.1128/jvi.73.4.2803-2813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Leopardi R, Roizman B. The herpes simplex virus major regulatory protein ICP4 blocks apoptosis induced by the virus or by hyperthermia. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9583–9587. doi: 10.1073/pnas.93.18.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ahmed M, Fraser NW. Herpes simplex virus type 1 2-kilobase latency-associated transcript intron associates with ribosomal proteins and splicing factors. Journal of Virology. 2001;75(24):12070–12080. doi: 10.1128/JVI.75.24.12070-12080.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Blaho JA, Aubert M. Modulation of apoptosis during herpes simplex virus infection in human cells. Microbes and Infection. 2001;3:1–8. doi: 10.1016/s1286-4579(01)01444-7. [DOI] [PubMed] [Google Scholar]
- 208.Jerome KR, Chen Z, Lang R, et al. HSV and glycoprotein J inhibit caspase activation and apoptosis induced by granzyme B or Fas. Journal of Immunology. 2001;167(7):3928–3935. doi: 10.4049/jimmunol.167.7.3928. [DOI] [PubMed] [Google Scholar]
- 209.Jerome KR, Fox R, Chen Z, Sears AE, Lee H-Y, Corey L. Herpes simplex virus inhibits apoptosis through the action of two genes, Us5 and Us3. Journal of Virology. 1999;73(11):8950–8957. doi: 10.1128/jvi.73.11.8950-8957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Jiang Y, Inman M, Zhang Y, Posadas NA, Jones C. A mutation in the latency-related gene of bovine herpesvirus 1 inhibits protein expression from open reading frame 2 and an adjacent reading frame during productive infection. Journal of Virology. 2004;78(6):3184–3189. doi: 10.1128/JVI.78.6.3184-3189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Munger J, Chee AV, Roizman B. The US3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. Journal of Virology. 2001;75(12):5491–5497. doi: 10.1128/JVI.75.12.5491-5497.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Munger J, Roizman B. The US3 protein kinase of herpes simplex virus 1 mediates the posttranslational modification of BAD and prevents BAD-induced programmed cell death in the absence of other viral proteins. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10410–10415. doi: 10.1073/pnas.181344498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chenet-Monte C, Mohammad F, Celluzzi CM, Schaffer PA, Farber FE. Herpes simplex virus gene products involved in the induction of chromosomal aberrations. Virus Research. 1986;6(3):245–260. doi: 10.1016/0168-1702(86)90073-0. [DOI] [PubMed] [Google Scholar]
- 214.Ellison BH, Ellison SA. Chromosomal aberrations induced by an animal virus. Nature. 1961;192(4798):145–147. doi: 10.1038/192145a0. [DOI] [PubMed] [Google Scholar]
- 215.Heilbronn R, zur Hausen H. A subset of herpes simplex virus replication genes induces DNA amplification within the host cell genome. Journal of Virology. 1989;63(9):3683–3692. doi: 10.1128/jvi.63.9.3683-3692.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Pilon L, Langelier Y, Royal A. Herpes simplex virus type 2 mutagenesis: characterization of mutants induced at the hprt locus of nonpermissive XC cells. Molecular and Cellular Biology. 1986;6(8):2977–2983. doi: 10.1128/mcb.6.8.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Soengas MS, Alarcon RM, Yoshida H, et al. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284(5411):156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- 218.Hagglund R, Munger J, Poon APW, Roizman B. US3 protein kinase of herpes simplex virus 1 blocks caspase 3 activation induced by the products of US1.5 and UL13 genes and modulates expression of transduced US1.5 open reading frame in a cell type-specific manner. Journal of Virology. 2002;76(2):743–754. doi: 10.1128/JVI.76.2.743-754.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Inman M, Perng G-C, Henderson G, et al. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. Journal of Virology. 2001;75(8):3636–3646. doi: 10.1128/JVI.75.8.3636-3646.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Perng G-C, Jones C, Ciacci-Zanella J, et al. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287(5457):1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 221.Ahmed M, Lock M, Miller CG, Fraser NW. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. Journal of Virology. 2002;76(2):717–729. doi: 10.1128/JVI.76.2.717-729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Jin L, Peng W, Perng G-C, et al. Identification of herpes simplex virus type 1 latency-associated transcript sequences that both inhibit apoptosis and enhance the spontaneous reactivation phenotype. Journal of Virology. 2003;77(11):6556–6561. doi: 10.1128/JVI.77.11.6556-6561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Henderson G, Peng W, Jin L, et al. Regulation of caspase 8- and caspase 9-induced apoptosis by the herpes simplex virus type 1 lantency-associated transcript. Journal of NeuroVirology. 2002;8(supplement 2):103–111. doi: 10.1080/13550280290101085. [DOI] [PubMed] [Google Scholar]
- 224.Schmitz I, Kirchhoff S, Krammer PH. Regulation of death receptor-mediated apoptosis pathways. International Journal of Biochemistry and Cell Biology. 2000;32(11-12):1123–1136. doi: 10.1016/s1357-2725(00)00048-0. [DOI] [PubMed] [Google Scholar]
- 225.Wang X. The expanding role of mitochondria in apoptosis. Genes and Development. 2001;15(22):2922–2933. [PubMed] [Google Scholar]
- 226.Krueger A, Baumann S, Krammer PH, Kirchhoff S. FLICE-inhibitory proteins: regulators of death receptor-mediated apoptosis. Molecular and Cellular Biology. 2001;21(24):8247–8254. doi: 10.1128/MCB.21.24.8247-8254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Carpenter D, Hsiang C, Jin L, et al. Stable cell lines expressing high levels of the herpes simplex virus type 1 LAT are refractory to caspase 3 activation and DNA laddering following cold shock induced apoptosis. Virology. 2007;369(1):12–18. doi: 10.1016/j.virol.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mott KR, Osorio N, Jin L, et al. The bovine herpesvirus-1 LR ORF2 is critical for this gene’s ability to restore the high wild-type reactivation phenotype to a herpes simplex virus-1 LAT null mutant. Journal of General Virology. 2003;84(11):2975–2985. doi: 10.1099/vir.0.19421-0. [DOI] [PubMed] [Google Scholar]
- 229.Perng G-C, Maguen B, Jin L, et al. A gene capable of blocking apoptosis can substitute for the herpes simplex virus type 1 latency-associated transcript gene and restore wild-type reactivation levels. Journal of Virology. 2002;76(3):1224–1235. doi: 10.1128/JVI.76.3.1224-1235.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jin L, Perng G-C, Mott KR, et al. A herpes simplex virus type 1 mutant expressing a baculovirus inhibitor of apoptosis gene in place of latency-associated transcript has a wild-type reactivation phenotype in the mouse. Journal of Virology. 2005;79(19):12286–12295. doi: 10.1128/JVI.79.19.12286-12295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Jin L, Carpenter D, Moerdyk-Schauwecker M, et al. Cellular FLIP can substitute for the herpes simplex virus type 1 latency-associated transcript gene to support a wild-type virus reactivation phenotype in mice. Journal of NeuroVirology. 2008;14(5):389–400. doi: 10.1080/13550280802216510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Carpenter D, Henderson G, Hsiang C, et al. Introducing point mutations into the ATGs of the putative open reading frames of the HSV-1 gene encoding the latency associated transcript (LAT) reduces its anti-apoptosis activity. Microbial Pathogenesis. 2008;44(2):98–102. doi: 10.1016/j.micpath.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Samaniego LA, Neiderhiser L, DeLuca NA. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. Journal of Virology. 1998;72(4):3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Samaniego LA, Wu N, DeLuca NA. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. Journal of Virology. 1997;71(6):4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Schlehofer JR, zur Hausen H. Induction of mutations within the host cell genome by partially inactivated herpes simplex virus type 1. Virology. 1982;122(2):471–475. doi: 10.1016/0042-6822(82)90247-1. [DOI] [PubMed] [Google Scholar]
- 236.Yang L, Voytek CC, Margolis TP. Immunohistochemical analysis of primary sensory neurons latently infected with herpes simplex virus type 1. Journal of Virology. 2000;74(1):209–217. doi: 10.1128/jvi.74.1.209-217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Branco FJ, Fraser NW. Herpes simplex virus type 1 latency-associated transcript expression protects trigeminal ganglion neurons from apoptosis. Journal of Virology. 2005;79(14):9019–9025. doi: 10.1128/JVI.79.14.9019-9025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Joels M, de Kloet ER. Control of neuronal excitability by corticosteroid hormones. Trends in Neurosciences. 1992;15(1):25–30. doi: 10.1016/0166-2236(92)90345-9. [DOI] [PubMed] [Google Scholar]
- 239.McEwen BS. Non-genomic and genomic effects of steroids on neural activity. Trends in Pharmacological Sciences. 1991;12(4):141–147. doi: 10.1016/0165-6147(91)90531-v. [DOI] [PubMed] [Google Scholar]
- 240.Halford WP, Gebhardt BM, Carr DJJ. Mechanisms of herpes simplex virus type 1 reactivation. Journal of Virology. 1996;70(8):5051–5060. doi: 10.1128/jvi.70.8.5051-5060.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Collett JW, Steele RE. Alternative splicing of a neural-specific Src mRNA (Src+) is a rapid and protein synthesis-independent response to neural induction in Xenopus laevis. Developmental Biology. 1993;158(2):487–495. doi: 10.1006/dbio.1993.1206. [DOI] [PubMed] [Google Scholar]
- 242.Busser J, Geldmacher DS, Herrup K. Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer’s disease brain. Journal of Neuroscience. 1998;18(8):2801–2807. doi: 10.1523/JNEUROSCI.18-08-02801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gill JS, Windebank AJ. Cisplatin-induced apoptosis in rat dorsal root ganglion neurons is associated with attempted entry into the cell cycle. Journal of Clinical Investigation. 1998;101(12):2842–2850. doi: 10.1172/JCI1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hu S, Peterson PK, Chao CC. Cytokine-mediated neuronal apoptosis. Neurochemistry International. 1997;30(4-5):427–431. doi: 10.1016/s0197-0186(96)00078-2. [DOI] [PubMed] [Google Scholar]
- 245.Park DS, Farinell SE, Greene LA. Inhibitors of cyclin-dependent kinases promote survival of post-mitotic neuronally differentiated PC12 cells and sympathetic neurons. The Journal of Biological Chemistry. 1996;271(14):8161–8169. doi: 10.1074/jbc.271.14.8161. [DOI] [PubMed] [Google Scholar]
- 246.Park DS, Levine B, Ferrari G, Greene LA. Cyclin dependent kinase inhibitors and dominant negative cyclin dependent kinase 4 and 6 promote survival of NGF-deprived sympathetic neurons. Journal of Neuroscience. 1997;17(23):8975–8983. doi: 10.1523/JNEUROSCI.17-23-08975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Park DS, Morris EJ, Greene LA, Geller HM. G1/S cell cycle blockers and inhibitors of cyclin-dependent kinases suppress camptothecin-induced neuronal apoptosis. Journal of Neuroscience. 1997;17(4):1256–1270. doi: 10.1523/JNEUROSCI.17-04-01256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Park DS, Morris EJ, Stefanis L, et al. Multiple pathways of neuronal death induced by DNA-damaging agents, NGF deprivation, and oxidative stress. Journal of Neuroscience. 1998;18(3):830–840. doi: 10.1523/JNEUROSCI.18-03-00830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Nash AA, Jayasuriya A, Phelan J, Cobbold SP, Waldmann H, Prospero T. Different roles for L3T4+ and Lyt 2+ T cell subsets in the control of an acute herpes simplex virus infection of the skin and nervous system. Journal of General Virology. 1987;68(3):825–833. doi: 10.1099/0022-1317-68-3-825. [DOI] [PubMed] [Google Scholar]
- 250.Simmons A, Tscharke D, Speck P. The role of immune mechanisms in control of herpes simplex virus infection of the peripheral nervous system. Current Topics in Microbiology and Immunology. 1992;179:31–56. doi: 10.1007/978-3-642-77247-4_3. [DOI] [PubMed] [Google Scholar]
- 251.Liu T, Tang Q, Hendricks RL. Inflammatory infiltration of the trigeminal ganglion after herpes simplex virus type 1 corneal infection. Journal of Virology. 1996;70(1):264–271. doi: 10.1128/jvi.70.1.264-271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shimeld C, Whiteland JL, Williams NA, Easty DL, Hill TJ. Cytokine production in the nervous system of mice during acute and latent infection with herpes simplex virus type 1. Journal of General Virology. 1997;78(12):3317–3325. doi: 10.1099/0022-1317-78-12-3317. [DOI] [PubMed] [Google Scholar]
- 253.Halford WP, Gebhardt BM, Carr DJJ. Persistent cytokine expression in trigeminal ganglion latently infected with herpes simplex virus type 1. Journal of Immunology. 1996;157(8):3542–3549. [PubMed] [Google Scholar]
- 254.Simmons A, Tscharke DC. Anti-CD8 impairs clearance of herpes simplex virus from the nervous systemml: implications for the fate of virally infected neurons. Journal of Experimental Medicine. 1992;175(5):1337–1344. doi: 10.1084/jem.175.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Khanna KM, Bonneau RH, Kinchington PR, Hendricks RL. Herpes simplex virus-specific memory CD8+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity. 2003;18(5):593–603. doi: 10.1016/s1074-7613(03)00112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Liu T, Khanna KM, Carriere BN, Hendricks RL. Gamma interferon can prevent herpes simplex virus type 1 reactivation from latency in sensory neurons. Journal of Virology. 2001;75(22):11178–11184. doi: 10.1128/JVI.75.22.11178-11184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Liu T, Khanna KM, Chen X, Fink DJ, Hendricks RL. CD8+ T cells can block herpes simplex virus type 1 (HSV-1) reactivation from latency in sensory neurons. Journal of Experimental Medicine. 2000;191(9):1459–1466. doi: 10.1084/jem.191.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Prabhakaran K, Sheridan BS, Kinchington PR, et al. Sensory neurons regulate the effector functions of CD8+ T cells in controlling HSV-1 latency ex vivo. Immunity. 2005;23(5):515–525. doi: 10.1016/j.immuni.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 259.Feldman LT, Ellison AR, Voytek CC, Yang L, Krause P, Margolis TP. Spontaneous molecular reactivation of herpes simplex virus type 1 latency in mice. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(2):978–983. doi: 10.1073/pnas.022301899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Cunningham AL, Mikloska Z. The Holy Grail: immune control of human herpes simplex virus infection and disease. Herpes. 2001;8(supplement 1):6A–10A. [PubMed] [Google Scholar]
- 261.Mikloska Z, Cunningham AL. Alpha and gamma interferons inhibit herpes simplex virus type 1 infection and spread in epidermal cells after axonal transmission. Journal of Virology. 2001;75(23):11821–11826. doi: 10.1128/JVI.75.23.11821-11826.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kagi D, Hengartner H. Different roles for cytotoxic T cells in the control of infections with cytopathic versus noncytopathic viruses. Current Opinion in Immunology. 1996;8(4):472–477. doi: 10.1016/s0952-7915(96)80033-1. [DOI] [PubMed] [Google Scholar]
- 263.Shresta S, Pham CT, Thomas DA, Graubert TA, Ley TJ. How do cytotoxic lymphocytes kill their targets? Current Opinion in Immunology. 1998;10(5):581–587. doi: 10.1016/s0952-7915(98)80227-6. [DOI] [PubMed] [Google Scholar]
- 264.Knickelbein JE, Khanna KM, Yee MB, Baty CJ, Kinchington PR, Hendricks RL. Noncytotoxic lytic granule-mediated CD8+ T cell inhibition of HSV-1 reactivation from neuronal latency. Science. 2008;322(5899):268–271. doi: 10.1126/science.1164164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Yang X, Stennicke HR, Wang B, et al. Granzyme B mimics apical caspases: description of a unified pathway for trans-activation of executioner caspase-3 and -7. The Journal of Biological Chemistry. 1998;273(51):34278–34283. doi: 10.1074/jbc.273.51.34278. [DOI] [PubMed] [Google Scholar]