Abstract
Systemic low-grade inflammation is consistently associated with functional status, cognitive functioning, multimorbidity, and survival in oldest olds. If inflammation is either a cause or a consequence of age-related pathology, genetic determinants of late-life survival can reside in cytokine genes polymorphisms, regulating inflammatory responses. The aim of this study was to test associations between commonly studied polymorphisms in interleukin (IL)6, IL10, IL15, and IL18, and tumor necrosis factor-α genes and late-life survival in a longitudinal cohort of nonagenarians: the Danish 1905 cohort. Additionally, associations were investigated between inflammatory markers and major predictors of mortality as cognitive and functional status. Modest sex-specific associations were found with survival, cognitive functioning, and handgrip strength. Evaluation of combined genotypes indicated that, in nonagenarian men, the balance of pro- and anti-inflammatory activity at IL18 and IL10 loci is protective against cognitive decline. In conclusion, in this large study with virtually complete follow-up, commonly studied polymorphisms in cytokine genes do not have a major impact on late-life survival or associated risk phenotypes.
Keywords: Inflammation, Cytokine, Late-life survival
OLD age survival has improved substantially since 1950 in developed countries. As a result, the number of octogenarians has increased 4-fold, the number of nonagenarians 8-fold, and the number of centenarians 20-fold (1). Longitudinal studies of nonagenarians have demonstrated that poor physical and cognitive performances predict mortality, whereas conventional risk factors in younger elderly people such as socio-demographic factors, smoking, and obesity have lost their importance (2). It has also become clear during the past decade that aging is accompanied by a chronic low-grade inflammatory state defined as twofold to fourfold increases in circulating levels of inflammatory mediators (3). Systemic low-grade inflammation is a strong independent predictor of all-cause and cardiovascular mortality (4,5) even in centenarians (6) as well as a risk factor/biomarker of age-related diseases, such as atherosclerosis (7), sarcopenia (8), diabetes (9), and neurodegenerative diseases (10).
From an evolutionary point of view, a high level of inflammatory responses helps the organism to avoid infections but the price may be an increased susceptibility to late-onset chronic diseases (11,12). Based on these considerations, the inflammatory hypothesis of aging proposes that structural damage accumulates from repeated episodes of acute inflammation until a threshold is crossed and function is affected (13). In addition, a link has been suggested between inflammatory events occurring in early periods of life and mortality at old age as data demonstrate that during the past century, the reduced burden of infections and inflammation in young ages may also have led to the decline in late-life mortality rate in Western countries (14). It has also been hypothesized that the age-related increased inflammatory activity depends on an imbalance between pro- and anti-inflammatory networks in old populations defined as “inflamm-aging” (15).
The initial triggers for cytokine production have not yet been identified in the old, and it is not known to which extent systemic low-grade inflammation induces, exaggerates, or reflects ongoing pathological processes (16). The balance between pro- and anti-inflammatory response can, however, be crucial in determining the onset and severity of age-related diseases, the rate of aging, and ultimately individual capability to attain longevity.
The production and release of inflammatory modulators are in part genetically influenced, and numerous genes coding for pro- or anti-inflammatory molecules are known. If systemic inflammation is a cause as well as a consequence of age-related pathology, it is possible that genetic polymorphisms, which determine the production rate of cytokines, will also be important risk factors in age-related diseases. In accordance with this, several studies have demonstrated associations between single nucleotide polymorphisms (SNPs) in cytokine genes and cardiovascular disease, type 2 diabetes, functional status, and cognitive functioning in younger elderly people, whereas others have not been able to confirm these reports (17–22). Additionally, several groups have reported associations between polymorphisms in cytokine genes and longevity (23–25). However, in this research area, inconsistent results and lack of reproducibility were often experienced and reported, claiming for the need of replication studies in larger cohorts (26). Moreover, only a few studies addressed inflammatory genes in longitudinal studies of the oldest old (27,28). Thus, inflammatory genes may have lost any clinical importance in the oldest old, or alternatively, genetic variations contribute more in nonagenarians/centenarians people, who often experience multimorbidity.
The aim of this study was to test the hypothesis that commonly studied polymorphisms in selected pro- and anti-inflammatory cytokine genes (tumor necrosis factor [TNF]-α and interleukin [IL]-6, IL-15, IL-18, and IL-10) predict late-life mortality in 1,651 participants from the Danish 1905 Cohort Study, initially visited in 1998 and followed for 10 years. Additionally, we also investigated if cytokine polymorphisms were associated with cognitive and physical functioning, representing the most important risk phenotypes in this population.
Polymorphisms analyzed in this study were mainly chosen for their functional relevance, like the occurrence in known or putative regulatory regions of the cytokine genes (29,30). Moreover, previous reports associated them with a wide range of age-related clinical disorders (31–34), and concentrations of the corresponding proteins are consistent risk factors of morbidity, disability, and functional status in cross-sectional and longitudinal cohort studies of old populations (35–38). A detailed description of the cytokine genes chosen and relative polymorphisms is reported in the Methods section.
METHODS
Samples
Participants in the study were drawn from the Danish 1905 cohort, which is a genetic–epidemiological nationwide survey carried out by the University of Southern Denmark (39). Participants of the survey are a nonselected group of the Danish 1905 cohort (2), representing all Danes born in 1905 and living in Denmark. In total, 2,262 out of 3,600 potential participants (recruitment rate 62.8%) initially agreed to participate in the survey, which included a home-based, 2-hour multidimensional interview with cognitive and physical tests as well as the collection of biologic material. One thousand six hundred and fifty-one participants (28% men) provided a DNA sample, which was collected by means of a finger prick or a cheek swab. Participants were visited in 1998, when they were 92–93 years old, and followed from the date of sampling until emigration or death up to the end of the study period (January 2008), at which time 1,474 (89.3%) of the 1,651 participants were dead.
Information on death or emigration for all the cohort members was retrieved from the Danish Central Population Register, which keeps a record of all those living in Denmark since 1968 and is continuously updated (40).
Measurements
All the measurements were assessed as described in previous reports (2,39,41).
Cognitive functions were assessed by Mini-Mental State Examination (MMSE) test and composite cognitive score. MMSE is a 30-item questionnaire that assesses orientation, episodic memory, attention, language, and construction functions and is widely used for the screening of the cognitive decline (42). Cognitive impairment is graded by a score that ranges between 0 and 30, and it is characterized as “severe impairment” if the score is below 17, “mild impairment” for scores between 18 and 23, and “no impairment” for scores above 24.
A composite cognitive score was computed by aggregating performance of five brief tests comprising fluency task (number of animals the person could name in 1 minute), forward and backward digit span, and immediate and delayed recall of a 12-item list.
Standardization of the cognitive score (indicated as ZCOG) was performed for facilitating the interpretation of the results, as suggested by McGue and Christensen (43).
Disability was tested by Katz Index of Activities of Daily Living (ADL). Five-item ADL indexes (bathing, dressing, toileting, transfer, and feeding) were used to obtain a three-level ADL scale; participants were defined as “not disabled” if independent in all items, “moderately disabled” if dependent in one or two items, “severely disabled” if dependent in three or more items, in accordance with the definition given in the article by Katz and colleagues (44).
Handgrip strength test was used as an estimate of physical functioning; handgrip (HG) represents one of the strongest predictors of disability in the elderly people and is highly correlated with mortality among old people (45). The measurement of grip strength was performed by a handheld dynamometer (SMEDLEY’S dynamometer TTM, Tokyo, Japan) for three performances with the strongest hand. The maximum value reported for each individual was considered for the analysis.
Cytokine Polymorphisms
The TNF-α gene is located on human chromosome 6p21.3, tandemly arranged with the TNF-β gene, and lies in the class III region of the major histocompatibility complex. Three markers were analyzed in this study: two SNPs in the promoter region, -308 (A/G) (rs1800629) and -238 (A/G) (rs361525), and a microsatellite, TNFa, located upstream of the whole TNF region, ∼3.5 kb from TNF-β and ∼10 kb from TNF-α transcription starting sites (46,47). The genetic variability at these positions is able to influence the levels of TNF-α production. In particular, the alleles A at -308 (A/G) locus and a2 at the TNFa microsatellite determine an increase in the gene expression (48,49). Although the effect of the TNF-238 (A/G) marker is less clear, a functional role has been suggested by the presence of a putative enhancer site close to this position (50).
The IL6 gene is located in humans on the short arm of chromosome 7 (7p21). The IL6-174 (G/C) (rs1800795) variation in the promoter region seems to be associated to variations in gene expression (51). With regard to the effect on plasma levels of IL6, reports have been very conflicting and two meta-analyses have reported no association between IL6-174 (G/C) and IL-6 levels but the C allele was associated with higher C-reactive protein levels (52,53).
The IL10 gene is located on chromosome 1 (1q31–32). Three major functional polymorphisms have been described in the proximal region of this gene, although the most important genetic factor in the regulation of constitutive IL10 messenger RNA (mRNA) levels is the promoter IL10-1082 (G/A) (rs1800896) SNP (32). The SNP shows allele-specific and dose effects on gene transcription, with the allele A associated with a lower protein production and a weak anti-inflammatory response (54).
The IL15 gene is located on chromosome 4 (4q31). Several SNPs are known for this gene, lying in both intronic and regulatory regions. Some of them, such as rs3806798 in the promoter region, rs1519551 and rs2322262 in the first intron, and rs2254514 in a regulatory region within the third exon of the gene, were identified and analyzed in population and family studies of inflammatory and allergic disorders (55,56) but never investigated in relation to aging.
The IL18 gene is located on chromosome 11 (11q22.2–q22.3) and is characterized by multiple transcription initiation sites. Promoter variants of the gene can influence circulating levels of the correspondent cytokine. The IL18-137(G/C) (rs187238), in particular, is a functional polymorphism located in a transcription factor–binding site in the promoter region, whose G allele leads to the increase of IL18 mRNA levels (57).
Genotyping
DNA was isolated from cheek swabs or blood spot cards using the QIAamp DNA Mini Kit (Qiagen, Germany).
Genotyping of all SNPs was performed by allelic discrimination using the Taqman technology. The IL15 SNPs rs3806798, rs1519511, rs2322262, and rs2254514 were genotyped by predesigned Taqman SNP genotyping assays (Applied Biosystems, Foster City, CA) using the conditions described by the manufacturer.
Primers and probes for the TNF-238 (A/G) and TNF-308 (A/G) SNPs were as described by Zambon and co-workers (58). For the remaining SNPs, primers and probes were designed using the Primer Express software (Applied Biosystems).
DNA was amplified in a total volume of 10 μL containing 5 μL Taqman Universal Master Mix (Applied Biosystems), 900 nM of each primer (DNA Technology, Denmark), 50–250 nM of each Vic- or FAM-labeled MGB probe (Applied Biosystems), and ∼10 ng template DNA.
Polymerase chain reaction (PCR) was performed in the ABI Prism 7700 (Applied Biosystems) and analyzed using the Sequence Detection System software (Applied Biosystems).
The TNFa microsatellite was genotyped by using the primers described in Udalova and colleagues (46). DNA amplification was performed in a total volume of 6 μL containing 1× PCR buffer, 200 μM of each dNTP (Roche, Switzerland), 167 nM of each primer (DNA Technology, Denmark), 0.18 U Taq DNA polymerase (Sigma, St. Louis, MO), and ∼10 ng template DNA. The PCR conditions were the following: denaturation at 94°C for 5 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 56°C for 1 minute 15 seconds, and extension at 72°C for 15 seconds. PCR was terminated by extension at 72°C for 6 minutes and cooled to 4°C. For detection, the sense primer was labeled with FAM. PCR products were resolved on the MegaBACE 1000 (GE Healthcare Biosciences, Pittsburgh, PA) according to the manufacturer’s instructions and analyzed by using the Fragment Profiler software (Amersham Biosciences, GE Healthcare Biosciences, Pittsburgh, PA). Alleles were named consecutively from 0 to 15, with the risk allele 2 corresponding to allele a2 in Culpan and colleagues (59).
All the primers used in this study are available on request to the authors.
Data Analysis
Allele frequencies of single markers were computed by gene counting from the observed genotypes, and Fisher’s exact test was applied to verify Hardy–Weinberg equilibrium (60). Linkage disequilibrium (LD) between pairs of markers was tested by a likelihood ratio test, as suggested by Weir (61). As for genotype data with unknown haplotype phase, an empirical distribution of haplotype frequencies obtained by a permutation procedure was used (62).
Kaplan–Meier analysis was performed to plot cumulative survival curves, and a log rank test was performed to test for difference in survival between the genotypes. Cox’s proportional hazards models were applied to assess the effect of differences, and the proportional hazard assumption was tested using the Schoenfeld residual test.
The effect of genotypes on disability was tested by a Fisher’s exact test. The effect of genotypes on cognitive functioning and handgrip strength was tested by analysis of variance (ANOVA), and Bartlett’s test (63) was used to verify the assumption of an equal distribution of variance between different groups. If the assumption of an equal distribution of variance was violated, an unequal version was conducted.
In case of aggregation into two classes, we controlled that no differences existed between subgroups before aggregating the genotypes. To control for gender-specific effects in the association between genetic polymorphism and age-related traits and mortality, all analyses were carried out separately in men and women.
Population genetic analyses were performed by Arlequin 3.1 software (64). Statistical analyses were carried out by using Stata 9.2 (STATA Corp., College Station, TX).
RESULTS
Genetic Analyses
The basic characteristics of the study population are illustrated in Table 1.
Table 1.
General Characteristics and Post-Survey Mortality in the Danish 1905 Cohort
| Men (n = 466) | Women (n = 1,185) | Total (n = 1,651) | |
| Survival | |||
| Deaths* [n (%)] | 431 (92.5) | 1,043 (88.0) | 1,474 (89.3) |
| Person-years | 1,439 | 4,642 | 6,081 |
| Mortality rate per 100 | 30.0 | 22.5 | 24.2 |
| Age (years) | |||
| Mean (SD) | 93.1 (0.3) | 93.1 (0.3) | 93.1 (0.3) |
| Range | 92.3–93.8 | 92.2–93.8 | 92.2–93.8 |
| Mini-Mental State Examination | |||
| Mean (SD) | 22.4 (5.8) | 21.5 (5.7) | 21.8 (5.7) |
| Range | 2–30 | 0–30 | 0–30 |
| Missing [n (%)] | 22 (4.7) | 46 (3.9) | 68 (4.1) |
| ZCOG | |||
| Mean (SD) | 0.09 (1.0) | 0.05 (1.0) | 0.06 (1.0) |
| Range | −2.1 to 3.4 | −2.3 to 3.3 | −2.3 to 3.4 |
| Missing [n (%)] | 24 (5.2) | 49 (4.1) | 73 (4.4) |
| Grip strength | |||
| Mean (SD) | 22.9 (6.5) | 13.5 (4.5) | 16.2 (6.7) |
| Range | 3–46 | 1–29 | 1–29 |
| Missing [n (%)] | 38 (8.2) | 109 (9.2) | 147 (8.9) |
| Activities of daily living [n (%)] | |||
| Not disabled | 265 (56.9) | 579 (48.9) | 844 (51.1) |
| Moderately disabled | 142 (30.5) | 467 (39.4) | 609 (36.9) |
| Disabled | 58 (12.4) | 138 (11.6) | 196 (11.9) |
| Missing | 0 (0) | 0 (0) | 0 (0) |
Note: *Survival data are calculated from a 10-year follow-up period.
Table 2 shows the genotypic distribution of cytokine gene polymorphisms at baseline in the Danish 1905 cohort. The distribution is in accordance with the predicted values assuming Hardy–Weinberg equilibrium. Regarding IL15 SNPs, we performed LD analysis between pairs of SNPs. LD was found between rs2254514 and rs2322262 (p = .002) as well as between rs3806798 and the other markers (rs1519551: p = .040; rs2254514: p = .009; rs2322262: p = .006) in our population.
Table 2.
Genotypic Distribution of Cytokine Polymorphisms at Baseline in the Danish 1905 Cohort
| Women | Men | Hardy-Weinberg Equilibrium, p Value | |
| TNF-308 | |||
| AA | 31 | 11 | .86 |
| AG | 339 | 117 | |
| GG | 799 | 327 | |
| Total | 1,169 | 455 | |
| TNF-238 | |||
| AA | 2 | 0 | 1.00 |
| AG | 88 | 40 | |
| GG | 1,085 | 421 | |
| Total | 1,175 | 461 | |
| TNFa* | |||
| 0 | 619 | 236 | .90 |
| 1 | 458 | 184 | |
| 2 | 93 | 34 | |
| Total | 1,170 | 454 | |
| IL18-137 | |||
| CC | 609 | 36 | .72 |
| GC | 419 | 179 | |
| GG | 561 | 240 | |
| Total | 1,165 | 455 | |
| IL6-174 | |||
| CC | 267 | 99 | .34 |
| GC | 575 | 217 | |
| GG | 330 | 142 | |
| Total | 1,172 | 458 | |
| IL10-1082 | |||
| AA | 253 | 93 | 1.00 |
| AG | 586 | 228 | |
| GG | 331 | 137 | |
| Total | 1,170 | 458 | |
| IL15 rs1519551 | |||
| AA | 382 | 145 | .30 |
| AG | 577 | 215 | |
| GG | 210 | 98 | |
| Total | 1,169 | 458 | |
| IL15 rs2254514 | |||
| CC | 634 | 230 | .36 |
| CT | 452 | 195 | |
| TT | 87 | 33 | |
| Total | 1,173 | 458 | |
| IL15 rs2322262 | |||
| AA | 650 | 242 | .73 |
| AG | 441 | 183 | |
| GG | 84 | 35 | |
| Total | 1,175 | 460 | |
| IL15 rs3806798 | |||
| TT | 948 | 368 | .46 |
| AT | 215 | 86 | |
| AA | 16 | 7 | |
| Total | 1,179 | 461 | |
Notes: IL = interleukin; TNF = tumor necrosis factor.
TNFa locus: Group 2 identifies the homozygotes for risk allele 2, Group 1 the heterozygotes for risk allele 2, and Group 0 noncarriers of allele 2. p Values are referred to Hardy–Weinberg Equilibrium test in the whole population.
Cytokine Gene Polymorphisms and Mortality
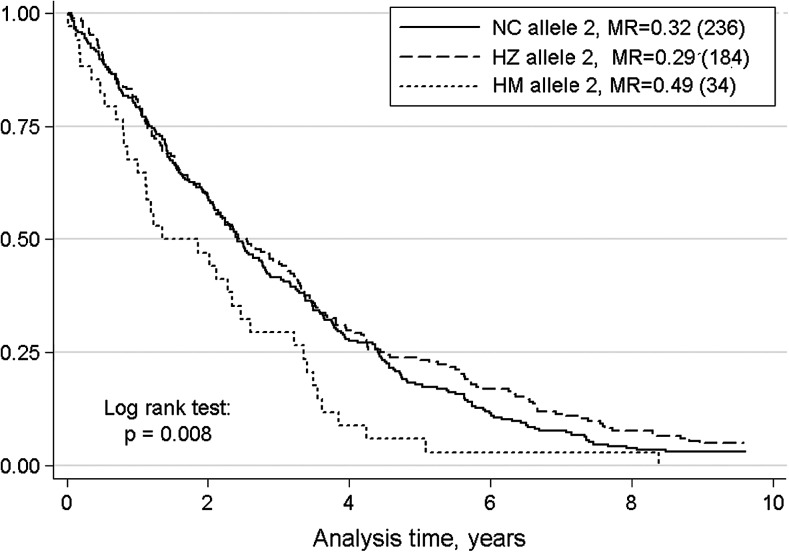
Association with mortality was found in men for the microsatellite TNFa at the TNF-α locus (Figure 1). Log rank test carried out on homozygote carriers (HM), heterozygote carriers (HZ), and noncarriers of the risk allele 2 (NC) showed significant differences between the three groups (p = .008). The aggregation of HZ and NC, which demonstrated more similar survival patterns, showed that homozygote male carriers of the risk allele 2 have a higher risk of death (hazard ratio [HR]: 1.6, 95% confidence interval [CI]: 1.10–2.28, p = .004) compared with heterozygote individuals or noncarriers of this allele.
Figure 1.
Kaplan–Meier survival estimates in men from the Danish 1905 cohort (N = 454) stratified by the occurrence of the risk allele 2 at TNFa locus. NC, noncarriers of the risk allele; HZ, heterozygote for the risk allele; HM, homozygote for the risk allele. Log rank test shows significant difference in the survival pattern between the three groups. Mortality rates and total numbers of individuals in each stratum are reported in legend. IL = interleukin; TNF = tumor necrosis factor.
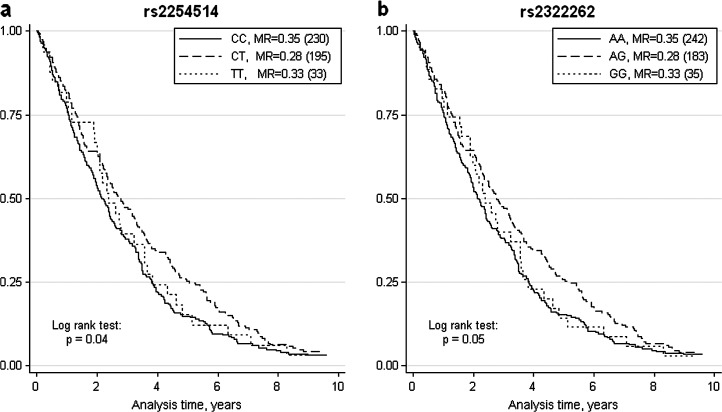
In men, a borderline association was found also between survival and the rs2254514 and rs2322262 markers of the IL15 gene (p = .04 and p = .05 to the log rank test on three genotypic groups; Figure 2a and b, respectively); as expected, results were very similar for these two SNPs due to the high level of LD between them. The aggregation in two genotypic groups, based on the similarity of survival patterns, showed that carriers of the variant allele (T for rs2254514 and G for rs2322262) have a lower risk of death compared with the homozygotes for the wild-type allele (HR: 0.8, 95% CI: 0.66–0.97, p = .020 in both cases). The proportional hazard assumption was not violated in the applied models (p > .05 in all cases).
Figure 2.
Kaplan–Meier survival estimates in men from the Danish 1905 cohort stratified by the genotypes at IL15 rs2254514 locus (a; N = 458) and rs2322262 locus (b; N = 460). Log rank test shows borderline differences in the survival pattern between the three groups. Mortality rates (MR) and total numbers of individuals in each stratum are reported in legend.
No association was found between rs2254514 and rs2322262 and mortality in women and between mortality and TNF-308, TNF-238, IL6-174, IL18-137, IL10-1082, and IL15 (rs1519551 and rs3806798) polymorphisms, neither in men nor in women from the Danish 1905 cohort.
Cytokine Gene Polymorphisms and Disability
None of the cytokine gene polymorphisms analyzed in this study was associated with disability at baseline in the Danish 1905 cohort (data not shown).
Cytokine Gene Polymorphisms and Handgrip Strength
Data for grip strength measurements were available in 1,504 of 1,651 participants, giving a total of 8.9% missing values.
The effect of genotypes on handgrip strength was tested by ANOVA test. An association was found between IL15 rs3806798 and handgrip strength in men (Table 3), with T carriers (TT + AT) having a significantly lower grip strength compared with the AA carriers (p = .014). For the same IL15 gene, the aggregation of rs1519551 genotypes in two groups showed that, among women, G carriers (AG + GG) had a borderline lower grip strength compared with AA carriers (p = .045). When missing values were taken into consideration (women who did not perform the test), we found a significant correlation between carrying the allele G at rs1519551 and missing measurement (p = .007), probably determining the borderline significance of the ANOVA test.
Table 3.
Distribution of Handgrip Strength Values in the Danish 1905 Cohort at Baseline Divided According to Cytokine Genotypes
| Women |
Men |
|||||
| Grip Strength | N | p Value | Grip Strength | N | p Value | |
| TNF-308 | ||||||
| AA | 14.7 (4.2) | 25 | .34 | 24.1 (5.1) | 11 | .33 |
| AG | 13.6 (4.5) | 303 | 22.2 (6.2) | 109 | ||
| GG | 13.4 (4.5) | 736 | 23.1 (6.6) | 298 | ||
| TNF-238 | ||||||
| AA | 16.0 (0) | 2 | .36 | — | 0 | .61 |
| AG | 14.1 (4.3) | 82 | 23.4 (6.2) | 38 | ||
| GG | 13.4 (4.5) | 984 | 22.8 (6.5) | 385 | ||
| TNFa* | ||||||
| 0 | 13.3 (4.5) | 561 | .17 | 23.0 (6.6) | 212 | .43 |
| 1 | 13.8 (4.4) | 419 | 22.9 (6.3) | 173 | ||
| 2 | 13.0 (4.6) | 84 | 21.4 (6.5) | 31 | ||
| IL18-137 | ||||||
| CC | 13.7 (4.6) | 561 | .59 | 24.1 (7.2) | 36 | .50 |
| GC | 13.7 (4.6) | 419 | 22.7 (6.6) | 160 | ||
| GG | 13.4 (4.4) | 80 | 22.9 (6.3) | 222 | ||
| IL6-174 | ||||||
| CC | 13.6 (4.4) | 241 | .95 | 22.2 (6.1) | 93 | .40 |
| GC | 13.5 (4.4) | 521 | 22.9 (6.7) | 199 | ||
| GG | 13.5 (4.7) | 303 | 23.4 (6.3) | 128 | ||
| IL10-1082 | ||||||
| AA | 13.5 (4.7) | 232 | .30 | 23.5 (6.3) | 85 | .45 |
| AG | 13.7 (4.3) | 532 | 22.5 (6.7) | 211 | ||
| GG | 13.2 (4.6) | 301 | 23.1 (6.2) | 124 | ||
| IL15 rs1519551 | ||||||
| AA | 13.9 (4.5) | 325 | .13 | 23.4 (6.9) | 120 | .26 |
| AG | 13.3 (4.5) | 444 | 22.2 (6.3) | 185 | ||
| GG | 13.3 (4.0) | 173 | 23.2 (6.6) | 79 | ||
| IL15 rs2254514 | ||||||
| CC | 13.5 (4.6) | 506 | .86 | 22.6 (7.0) | 191 | .66 |
| CT | 13.4 (4.3) | 368 | 23.1 (5.9) | 165 | ||
| TT | 13.3 (3.7) | 73 | 22.1 (7.0) | 28 | ||
| IL15 rs2322262 | ||||||
| AA | 13.5 (4.6) | 519 | .80 | 22.7 (7.0) | 203 | .77 |
| AG | 13.4 (4.2) | 358 | 23.0 (5.8) | 155 | ||
| GG | 13.3 (3.7) | 71 | 22.1 (7.0) | 28 | ||
| IL15 rs3806798 | ||||||
| TT | 13.4 (4.4) | 762 | .57 | 22.5 (6.6) | 313 | .01 |
| AT | 13.5 (4.4) | 177 | 23.7 (5.7) | 66 | ||
| AA | 14.8 (3.9) | 12 | 28.8 (8.5) | 7 | ||
Notes: IL = interleukin; TNF = tumor necrosis factor.
TNFa locus: Group 2 identifies homozygotes for risk allele 2, Group 1 heterozygotes for risk allele 2, and Group 0 noncarriers of allele 2. Means and standard deviations are shown. p Values refer to analysis of variance test among the three groups. Underlined values are considered significant.
Cytokine Gene Polymorphisms and Cognitive Functioning
Data for MMSE and ZCOG assessments were available in 1,583 and 1,578 participants, giving a total of 4.1% and 4.4% missing values, respectively.
Also in this case, the effect of genotypes on cognitive functioning was tested by ANOVA test.
Sex-specific associations were found for cytokine gene polymorphisms in relation to cognitive functioning at baseline. In particular, IL10 and IL15 SNPs were associated with cognitive functioning in women, whereas borderline associations for TNF-α and IL18 were found in men (Table 4).
Table 4.
Distribution of MMSE Values and Composite Cognitive Score in the Danish 1905 Cohort at Baseline Divided According to Cytokine Genotypes
| Women |
Men |
|||||||||||
| MMSE | N | p Value | ZCOG | N | p Value | MMSE | N | p Value | ZCOG | N | p Value | |
| TNF-308 | ||||||||||||
| AA | 21.7 (6.3) | 28 | .86 | 0.09 (0.9) | 28 | .95 | 24.0 (4.9) | 11 | .25 | 0.08 (0.5) | 11 | .95 |
| AG | 21.4 (5.7) | 324 | 0.04 (0.9) | 324 | 23.0 (5.6) | 112 | 0.12 (1.0) | 111 | ||||
| GG | 21.6 (5.6) | 772 | 0.05 (0.9) | 769 | 22.2 (5.8) | 311 | 0.09 (1.0) | 310 | ||||
| TNF-238 | ||||||||||||
| AA | 24.5 (3.5) | 2 | .55 | 0.72 (0.7) | 2 | .12 | — | 0 | .44 | — | 0 | .11 |
| AG | 22.0 (5.5) | 86 | 0.24 (1.0) | 86 | 21.7 (6.0) | 39 | −0.15 (1.0) | 39 | ||||
| GG | 21.5 (5.7) | 1,042 | 0.03 (0.9) | 1,039 | 22.5 (5.8) | 400 | 0.12 (1.0) | 398 | ||||
| TNFa* | ||||||||||||
| 0 | 21.3 (5.8) | 593 | .25 | 0.03 (1.0) | 590 | .44 | 22.0 (5.8) | 221 | .12 | 0.08 (0.9) | 221 | .13 |
| 1 | 21.9 (5.5) | 445 | 0.09 (0.9) | 445 | 23.0 (5.7) | 179 | 0.15 (1.0) | 177 | ||||
| 2 | 21.3 (5.6) | 88 | −0.03 (0.9) | 88 | 21.2 (6.6) | 32 | −0.24 (0.8) | 32 | ||||
| IL18-137 | ||||||||||||
| CC | 21.4 (6.1) | 87 | .94 | 0.14 (0.9) | 87 | .55 | 24.2 (4.0) | 36 | .10 | 0.48 (0.9) | 36 | .05 |
| GC | 21.6 (5.5) | 448 | 0.07 (0.9) | 447 | 22.0 (6.0) | 167 | 0.03 (1.0) | 166 | ||||
| GG | 21.5 (5.8) | 586 | 0.03 (1.0) | 584 | 22.4 (5.9) | 231 | 0.09 (1.0) | 230 | ||||
| IL6-174 | ||||||||||||
| CC | 21.2 (5.9) | 254 | .25 | −0.03 (0.9) | 254 | .32 | 22.1 (6.0) | 95 | .48 | 0.04 (0.9) | 95 | .58 |
| GC | 21.8 (5.7) | 550 | 0.07 (0.9) | 548 | 22.2 (5.8) | 204 | 0.06 (0.9) | 203 | ||||
| GG | 21.3 (5.5) | 323 | 0.09 (1.0) | 322 | 22.9 (5.7) | 137 | 0.15 (1.0) | 136 | ||||
| IL10-1082 | ||||||||||||
| AA | 22.1 (5.3) | 244 | .01 | 0.10 (0.9) | 244 | .01 | 22.2 (5.7) | 91 | .96 | −0.06 (1.0) | 90 | .22 |
| AG | 21.7 (5.6) | 564 | 0.10 (0.9) | 561 | 22.4 (6.0) | 218 | 0.12 (1.0) | 217 | ||||
| GG | 20.8 (6.0) | 319 | −0.08 (1.0) | 319 | 22.4 (5.5) | 127 | 0.16 (0.9) | 127 | ||||
| IL15rs1519551 | ||||||||||||
| AA | 21.8 (5.6) | 373 | .35 | 0.05 (0.9) | 371 | .69 | 22.8 (5.6) | 136 | .16 | 0.15 (0.9) | 136 | .67 |
| AG | 21.3 (5.8) | 550 | 0.06 (0.9) | 549 | 21.8 (6.2) | 207 | 0.05 (1.0) | 206 | ||||
| GG | 21.7 (5.5) | 202 | −0.001 (0.9) | 202 | 22.8 (5.2) | 93 | 0.06 (0.8) | 92 | ||||
| IL15rs2254514 | ||||||||||||
| CC | 21.5 (5.8) | 610 | .06 | 0.06 (1.0) | 609 | .02 | 22.0 (6.3) | 217 | .26 | 0.09 (1.0) | 216 | .73 |
| CT | 21.2 (5.6) | 435 | −0.02 (0.9) | 434 | 22.8 (5.1) | 188 | 0.10 (0.9) | 187 | ||||
| TT | 22.8 (5.7) | 84 | 0.29 (0.9) | 83 | 21.6 (5.5) | 31 | −0.05 (0.8) | 31 | ||||
| IL15rs2322262 | ||||||||||||
| AA | 21.6 (5.7) | 624 | .15 | 0.07 (0.9) | 622 | .03 | 22.1 (6.3) | 229 | .47 | 0.08 (1.0) | 228 | .64 |
| AG | 21.3 (5.7) | 425 | −0.02 (0.9) | 425 | 22.8 (5.3) | 176 | 0.13 (1.0) | 175 | ||||
| GG | 22.6 (5.7) | 81 | 0.26 (0.8) | 80 | 22.0 (5.4) | 33 | −0.05 (0.9) | 33 | ||||
| IL15rs3806798 | ||||||||||||
| TT | 21.6 (5.6) | 909 | .83 | 0.05 (0.9) | 907 | .75 | 22.2 (6.0) | 354 | .28 | 0.07 (1.0) | 352 | .78 |
| AT | 21.4 (6.2) | 209 | 0.05 (0.9) | 208 | 22.5 (5.5) | 78 | 0.15 (1.0) | 78 | ||||
| AA | 21.0 (5.2) | 16 | −0.13 (0.7) | 16 | 25.7 (2.4) | 7 | 0.18 (0.4) | 7 | ||||
Notes: IL = interleukin; TNF = tumor necrosis factor.
TNFa locus: Group 2 identifies homozygotes for risk allele 2, Group 1 heterozygotes for risk allele 2, and Group 0 noncarriers of allele 2. Means and standard deviations are shown. p Values refer to analysis of variance test among the three groups. Underlined values are considered significant.
Thus, IL10-1082GG female carriers had a significantly lower cognitive functioning compared with noncarriers (AA + AG; MMSE: p = .005, ZCOG: p = .003). Additionally, female C carriers (CC + CT) of the IL15 rs2254514 marker have a significantly lower cognitive functioning compared with TT carriers (MMSE: p = .027, ZCOG: p = .018). As a result of the LD, rs2322262 showed a very similar pattern, with A carriers (AA + AG) having a lower ZCOG compared with GG carriers (p = .032) and a similarly, although not significant, negative association with MMSE scores (p = .087).
In women, a borderline association was also found for TNF-238 A carriers (AA + AG) and cognitive functioning (ZCOG: p = .052). The low frequency of the rare A allele in the Danish population probably affected the result.
In men, IL18-137 G carriers (GG + GC) had a significantly lower cognitive performance compared with CC carriers (MMSE: p = .009, ZCOG: p = .017). In men, HC of the risk allele 2 at the microsatellite TNFa had also a lower cognitive functioning compared with non-homozygote carriers (MMSE: p = .001, ZCOG: p = .050).
Cumulative effect of risk combinations.—
With the aim of testing whether the simultaneous presence of single markers associated with cognitive functioning increased the associations found, we tested the cumulative effect of protective/risk combinations at these loci on cognitive scores by ANOVA test.
A total of 266 women carried the genotypic combination suggested to be risky for cognitive functioning TNF-238GG/IL10-1082GG/IL15rs2254514(CC + CT)/IL15rs2322262(AA + AG) and had an MMSE and ZCOG mean values of 20.7 and −0.1, respectively. In comparison, women carrying all the other possible combinations of alleles at these loci had an MMSE and ZCOG mean value of 21.8 and 0.1, respectively. The difference in cognitive functioning was highly significant between the two groups (MMSE: p = .005, ZCOG: p = .002).
In men, 34 carriers of the protective combination TNFa1/IL18-137CC had a significant higher performance in cognitive functioning compared with noncarriers (MMSE: p = .008, ZCOG: 0.009). Men with the protective combination had an MMSE and ZCOG mean value of 24.3 and 0.5, respectively, while the noncarriers reported an MMSE and ZCOG mean value of 22.2 and 0.05, respectively.
Consistent with these results, men carrying the risk combination TNFa2/IL18-137(GG + GC) have a significantly increased risk of death (HR: 1.8, 95% CI: 1.2–2.6, p = .001) compared with those carrying protective/neutral genotypes.
Interaction Between Pro- and Anti-Inflammatory Genotypes in Cognitive Functioning and Grip Strength
With the aim of testing if a balance between pro- and anti-inflammatory cytokine can be associated with a better preservation of cognitive and functional status in the elderly people from the 1905 cohort, we tested the additive contribution of inherited variants at TNFa and IL18-137 (G/C) pro-inflammatory loci, together with the -1082 (A/G) marker at IL10 anti-inflammatory gene, to cognitive and grip strength scores. The other markers considered in this article were excluded for different reasons. In particular, TNF-308 (A/G) and TNF-238 (A/G) SNPs were excluded because of the low frequency of the allele A in both markers, which influenced the sample sizes in risk combinations. As for IL16-174 (G/C), considering both the ambivalent role of IL6 as pro- and anti-inflammatory cytokine and the controversial associations between the allelic variants at this SNP with gene expression and protein levels previously discussed, we excluded the marker from the risk combinations. Finally, because data about the influence of IL15 markers on protein expression were not available, this cytokine was excluded too.
Risk genotypes were considered those supposed to be associated with high inflammatory status and specifically IL18-137(GG + GC), IL10-1082AA, and TNFa2. Comparing the performances of people carrying the pairwise combination of one marker at pro-inflammatory TNF-α or IL18 locus with that at the anti-inflammatory IL10 locus by ANOVA test, in men, we found a significant correlation between the low-producing inflammatory genotype IL18-137CC/IL10-1082(AG + GG) and better cognitive functioning, both measured by MMSE and composite cognitive tests (MMSE: p = .024, ZCOG: p = .03). In particular, 32 men carriers of the protective combination reported an MMSE and ZCOG mean values of 24 and 0.46, respectively, while the 399 noncarriers had an MMSE and ZCOG mean values of 22.2 and 0.06, respectively. When we considered the whole three-loci risk combination, both for cognitive functioning and for handgrip, we did not find association in our population.
No association with inflammatory genetic profiles at these polymorphisms was found in women, both for handgrip and for cognitive functioning.
DISCUSSION
Previous studies in the Danish 1905 cohort have revealed that mortality risk factors important in younger elderly people (ie, smoking and obesity) are not important in the 90+ subjects, whereas factors such as disability level and cognitive and physical performance are highly relevant (2). Inflammatory markers have been suggested to play a causative role in the susceptibility to age-related diseases and mortality even at very old age (6).
Accordingly, we tested the hypothesis that commonly studied polymorphisms in cytokine genes predict late-life mortality in the Danish 1905 cohort. However, we found that the chosen variants had only a minor impact on survival and risk phenotypes, including cognitive and functional impairment that are associated with survival in the 1905 cohort. In addition, effects often showed a sex difference. The existence of sex-specific predisposition to age-related diseases and longevity, as well as differences in mortality trajectories in the two sexes, is documented in literature (65,66). However, the minor associations found in this study may indicate that these polymorphisms do not have a major clinical impact in late life, and associations between systemic low-grade inflammation, morbidity, and mortality are, to a larger extent, related to external factors, ongoing pathological processes, or other candidate variations/genes. In accordance with this evidence is the poor correlation often found between cytokine polymorphisms and circulating levels of the corresponding protein in the blood (67).
Regarding late-life survival, we found only weak associations for a few polymorphisms and effects were restricted to men only.
Thus, the risk allele 2 at TNFa microsatellite was associated with a higher risk of death in homozygote men. This allele has previously been associated with high TNF-α secretion (48) and higher risk of inflammatory diseases (68). In our study, the risk allele 2 at TNFa locus was also associated with a lower performance in cognitive tests (MMSE and composite cognitive score), further supporting the hypothesis that TNFa is a risk factor in the old. Moreover, the observation of a better cognitive functioning in people carrying one copy of the risk allele suggests a moderate dose effect of the TNFa polymorphism on cognitive functioning and fits with the hypothesis that heterozygosity for a high-producing allele at TNF-α locus can exert a protective effect on age-related degeneration due to an optimal balance of the inflammatory response (16,33). We found no association between the TNF-308 (A/G) polymorphism and survival or associated risk phenotypes in the 1905 cohort, confirming the population specificity previously observed for this polymorphism in relation to survival (25,69,70). Moreover, we found only a borderline significant effect of the TNF-238 (A/G) polymorphism on cognitive performances in women. In contrast to our findings in the 1905 cohort, the TNF-308 polymorphism has been previously related to cognitive functioning in smaller studies of elderly populations (21), dementia in centenarians (33), and patients with Alzheimer’s disease (48,71,72), although others have reported no association (67,73). It is possible that discrepancies among studies are due to minor effects of the gene or differences in the severity of cognitive decline in the populations studied (manifest dementia vs age-related cognitive decline). Furthermore, the low frequency of the rare allele A at TNF-308 (A/G) and TNF-238 (A/G) markers in the Danish population probably affected the results. On the whole, our findings indicate a modest impact of the TNF-α gene on late-life mortality and associated risk phenotypes in our cohort.
The IL15 gene showed the highest number of associations in our study including survival in men, cognitive performance in women, and handgrip strength in both sexes. Thus, two polymorphisms in linkage disequilibrium (rs2254514 and rs2322262) showed associations with survival in men and cognitive functioning at baseline in women. A borderline sex-specific association was found also between handgrip strength and the IL15 marker rs1519551 in women and rs3806798 in men.
Thus, IL15 seems to have some effect on old age survival and related risk phenotypes. However, the four SNPs analyzed in this study were chosen as candidates for the association with allergic disorders (55,56) but never investigated in relation to aging. Considering the involvement of IL15 in the remodeling of T-cell compartment in the elderly people (74), an influence of the IL15 gene in survival and age-related phenotypes can be hypothesized. Despite this, we cannot rule out that these polymorphisms are not the causal variants and that the associations found are due to other functional variations within the same gene, in linkage disequilibrium with them. In any case, because this is the first article reporting an association between IL15 gene variability and survival and related traits, results here reported need confirmation in replication studies. The modest association between cognitive decline and IL18-137 G allele in men in this study is in accordance with reports of increased transcription of the pro-inflammatory cytokine and associations between this allele and the risk of cognitive decline in octogenarians and late-onset Alzheimer’s disease (67,75). On the contrary, the association between IL10-1082 GG genotype with a decreased cognitive functioning in women contrasted our hypothesis of a protective role of this marker in neurodegeneration (76). Despite the association with cognitive functioning, neither IL18 nor IL10 polymorphisms were associated with mortality in this study.
The IL6 polymorphism had no effect on cognitive and physical functioning parameters. Consistent with this, a large meta-analysis recently performed does not support a major role for this marker in achieving longevity across European populations (26).
Moreover, there were no associations between all the chosen cytokine SNPs and ADL in our study.
The sex-specific effects observed in this study may, on the one hand, be random and reflect minor clinical effects of the chosen markers at advanced age. On the other hand, sex-specific associations have been reported for genes involved in human longevity (77–79), and it has been suggested that Genotype × Sex interactions contribute to differences in the prevalence, course, and severity of diseases, probably through the regulatory network controlled by hormones that interacts with functional gene variation in a sex-specific way (65).
On the whole, taking into account the differences in the physiological setup between the two sexes, sex-specific Gene × Environment interactions can exist. Furthermore, the result of association with survival found in men adds a further piece of evidence to the idea that the genetic component that modulates the probability of reaching extreme old age is gender specific and likely more important in men than in women (66). Studies that replicate these findings would be required to confirm or refute the gender-specific associations here presented.
Taking into account the complexity of the inflammatory cascade, where cytokines act in networks rather than on a single protein basis, we hypothesized the existence of an interaction between different cytokines also at gene level. Accordingly, we investigated the effect of high- and low-grade inflammatory combinations of cytokine genotypes on cognitive and physical functioning. We found an association between a low-grade inflammatory combination (IL18-137 CC/IL10-1082 AG + GG) with a preserved cognitive function in men from the Danish 1905 cohort. Under the hypothesis that this genotype is associated with low levels of the pro-inflammatory cytokine IL18 and high level of the anti-inflammatory IL10, this result indicates that a low inflammatory status obtained by a balance between pro- and anti-inflammatory cytokines may protect against cognitive decline and be advantageous, at least in nonagenarian men. The hypothesis of an association between high inflammatory status and reduced cognitive functioning is supported also by the finding that in the Danish 1905 cohort, men carriers of the high inflammatory combination IL18-137 GG + GC/TNFa2 show lower cognitive scores. Although the role of inflammation on cognitive functioning in the elderly people is still debated (80–82), our results suggests that the economical combination of pro- and anti-inflammatory response protects against decreased cognitive function, at least in nonagenarian men.
It could be argued that Bonferroni adjustment should be performed to correct for multiple testing when considering the aggregation of genotypes. However, we believe that this correction is suitable when searching for associations without a priori hypotheses but not when assessing specific questions, as in our case (83).
In conclusion, commonly studied polymorphisms in cytokine genes had only minor effects on late-life survival and related risk phenotypes including cognitive decline and disability. Moreover, effects were often restricted to one sex only. Accordingly, other factors may explain the strong associations between systemic low-grade inflammation, mortality, and morbidity in very old populations. This may reflect that the chosen cytokine markers have lost their clinical importance at very advanced age or the multifunctionality of inflammatory cytokines.
FUNDING
The study was supported by U.S. National Institute on Aging Research grant NIA-PO1-AG08761. The Danish Aging Research Center is supported by a grant from the VELUX Foundation. The Centre of Inflammation and Metabolism is supported by a grant from the Danish National Research Foundation (DG 02-512-555).
Acknowledgments
We thank S. Li, U. Munk, and S. Gregersen for technical assistance.
References
- 1.Vaupel JW, Carey JR, Christensen K, et al. Biodemographic trajectories of longevity. Science. 1998;280:855–860. doi: 10.1126/science.280.5365.855. [DOI] [PubMed] [Google Scholar]
- 2.Nybo H, Petersen HC, Gaist D, et al. Predictors of mortality in 2,249 nonagenarians—the Danish 1905-Cohort Survey. J Am Geriatr Soc. 2003;51:1365–1373. doi: 10.1046/j.1532-5415.2003.51453.x. [DOI] [PubMed] [Google Scholar]
- 3.Krabbe KS, Pedersen M, Bruunsgaard H. Inflammatory mediators in the elderly. Exp Gerontol. 2004;39:687–699. doi: 10.1016/j.exger.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Störk S, Feelders RA, Van den Beld AW, et al. Prediction of mortality risk in the elderly. Am J Med. 2006;119:519–525. doi: 10.1016/j.amjmed.2005.10.062. [DOI] [PubMed] [Google Scholar]
- 5.Harris TB, Ferrucci L, Tracy RP, et al. Associations of elevated interleukin-6 and C-reactive protein levels with mortality in the elderly. Am J Med. 1999;106:506–512. doi: 10.1016/s0002-9343(99)00066-2. [DOI] [PubMed] [Google Scholar]
- 6.Bruunsgaard H, Andersen-Ranberg K, Hjelmborg JB, Pedersen BK, Jeune B. Elevated levels of tumor necrosis factor alpha and mortality in centenarians. Am J Med. 2003;115:278–283. doi: 10.1016/s0002-9343(03)00329-2. [DOI] [PubMed] [Google Scholar]
- 7.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 8.Visser M, Pahor M, Taaffe DR, et al. Relationship of interleukin-6 and tumor necrosis factor-alpha with muscle mass and muscle strength in elderly men and women: the Health ABC Study. J Gerontol A Biol Sci Med Sci. 2002;57:M326–M332. doi: 10.1093/gerona/57.5.m326. [DOI] [PubMed] [Google Scholar]
- 9.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 10.Ray S, Britschgi M, Herbert C, et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 11.Tracey KJ. The inflammatory reflex. Nature. 2002;420:853–859. doi: 10.1038/nature01321. [DOI] [PubMed] [Google Scholar]
- 12.Van Den Biggelaar AH, De Craen AJ, Gussekloo J, et al. Inflammation underlying cardiovascular mortality is a late consequence of evolutionary programming. FASEB J. 2004;18:1022–1024. doi: 10.1096/fj.03-1162fje. [DOI] [PubMed] [Google Scholar]
- 13.Tracy R. Contribution to NIA Workshop on Inflammation, Inflammatory Mediators and Aging. Bethesda, MD: 2004. Inflammation and aging: a biologist’s perspective. [Google Scholar]
- 14.Finch CE, Crimmins EM. Inflammatory exposure and historical changes in human life-spans. Science. 2004;305:1736–1739. doi: 10.1126/science.1092556. [DOI] [PubMed] [Google Scholar]
- 15.Franceschi C, Bonafe M, Valensin S, et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 16.Wiss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med. 2006;12:1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- 17.Bernard V, Pillois X, Dubus I, et al. The -308 G/A tumor necrosis factor-alpha gene dimorphism: a risk factor for unstable angina. Clin Chem Lab Med. 2003;41:511–516. doi: 10.1515/CCLM.2003.077. [DOI] [PubMed] [Google Scholar]
- 18.Tso AR, Merino JG, Warach S. Interleukin-6 174G/C polymorphism and ischemic stroke: a systematic review. Stroke. 2007;38:3070–3075. doi: 10.1161/STROKEAHA.107.492231. [DOI] [PubMed] [Google Scholar]
- 19.Heijmans BT, Westendorp RG, Droog S, Kluft C, Knook DL, Slagboom PE. Association of the tumour necrosis factor alpha -308G/A polymorphism with the risk of diabetes in an elderly population-based cohort. Genes Immun. 2007;3:225–228. doi: 10.1038/sj.gene.6363859. [DOI] [PubMed] [Google Scholar]
- 20.Liu D, Metter EJ, Ferrucci L, Roth SM. TNF promoter polymorphisms associated with muscle phenotypes in humans. J Appl Physiol. 2008;105:859–867. doi: 10.1152/japplphysiol.90655.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baune BT, Ponath G, Rothermundt M, Riess O, Funke H, Berger K. Association between genetic variants of IL-1beta, IL-6 and TNF-alpha cytokines and cognitive performance in the elderly general population of the MEMO-study. Psychoneuroendocrinology. 2008;33:68–76. doi: 10.1016/j.psyneuen.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Tedde A, Putignano AL, Nacmias B, Bagnoli S, Cellini E, Sorbi S. Lack of association between TNF-alpha polymorphisms and Alzheimer’s disease in an Italian cohort. Neurosci Lett. 2008;446:139–142. doi: 10.1016/j.neulet.2008.09.044. [DOI] [PubMed] [Google Scholar]
- 23.Bonafè M, Olivieri F, Cavallone L, et al. A gender-dependent genetic predisposition to produce high levels of IL-6 is detrimental for longevity. Eur J Immunol. 2001;31:2357–2361. doi: 10.1002/1521-4141(200108)31:8<2357::aid-immu2357>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 24.Lio D, Scola L, Crivello A, et al. Inflammation, genetics, and longevity: further studies on the protective effects in men of IL-10 -1082 promoter SNP and its interaction with TNF-alpha-308 promoter SNP. J Med Genet. 2003;40:296–299. doi: 10.1136/jmg.40.4.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cardelli M, Cavallone L, Marchegiani F, et al. A genetic-demographic approach reveals male-specific association between survival and tumor necrosis factor (A/G)-308 polymorphism. J Gerontol A Biol Sci Med Sci. 2008;63:454–460. doi: 10.1093/gerona/63.5.454. [DOI] [PubMed] [Google Scholar]
- 26.Di Bona D, Vasto S, Capurso C, et al. Effect of interleukin-6 polymorphisms on human longevity: a systematic review and meta-analysis. Ageing Res Rev. 2009;8:36–42. doi: 10.1016/j.arr.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Van den Biggelaar AH, Huizinga TW, de Craen AJ, et al. Impaired innate immunity predicts frailty in old age. The Leiden 85-plus study. Exp Gerontol. 2004;39:1407–1414. doi: 10.1016/j.exger.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Bruunsgaard H, Christiansen L, Pedersen AN, Schroll M, Jørgensen T, Pedersen BK. The IL-6 -174G>C polymorphism is associated with cardiovascular diseases and mortality in 80-year-old humans. Exp Gerontol. 2004;39:255–261. doi: 10.1016/j.exger.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Bidwell JL, Wood NA, Morse HR, Olomolaiye OO, Keen LJ, Laundy GJ. Human cytokine gene nucleotide sequence alignments: supplement 1. Eur J Immunogenet. 1999;26:135–223. doi: 10.1046/j.1365-2370.1999.00143.x. [DOI] [PubMed] [Google Scholar]
- 30.Bidwell J, Keen L, Gallagher G, et al. Cytokine gene polymorphism in human disease: on-line databases, supplement 1. Genes Immun. 2001;2:61–70. doi: 10.1038/sj.gene.6363733. [DOI] [PubMed] [Google Scholar]
- 31.Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crawley E, Kay R, Sillibourne J, Patel P, Hutchinson I, Woo P. Polymorphic haplotypes of the interleukin-10 5′ flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile rheumatoid arthritis. Arthritis Rheum. 1999;42:1101–1108. doi: 10.1002/1529-0131(199906)42:6<1101::AID-ANR6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 33.Bruunsgaard H, Benfield TL, Andersen-Ranberg K, et al. The tumor necrosis factor alpha -308G>A polymorphism is associated with dementia in the oldest old. J Am Geriatr Soc. 2004;52:1361–1366. doi: 10.1111/j.1532-5415.2004.52369.x. [DOI] [PubMed] [Google Scholar]
- 34.Frayling TM, Rafiq S, Murray A, et al. An interleukin-18 polymorphism is associated with reduced serum concentrations and better physical functioning in older people. J Gerontol A Biol Sci Med Sci. 2007;62:73–78. doi: 10.1093/gerona/62.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrucci L, Harris TB, Guralnik JM, et al. Serum IL-6 level and the development of disability in older persons. J Am Geriatr Soc. 1999;47:639–646. doi: 10.1111/j.1532-5415.1999.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 36.Ershler WB, Keller ET. Age-associated increased interleukin-6 gene expression, late-life diseases, and frailty. Annu Rev Med. 2000;51:245–270. doi: 10.1146/annurev.med.51.1.245. [DOI] [PubMed] [Google Scholar]
- 37.Cesari M, Penninx BW, Pahor M, et al. Inflammatory markers and physical performance in older persons: the InCHIANTI study. J Gerontol A Biol Sci Med Sci. 2004;59:242–248. doi: 10.1093/gerona/59.3.m242. [DOI] [PubMed] [Google Scholar]
- 38.Danesh J, Kaptoge S, Mann AG, et al. Long-term interleukin-6 levels and subsequent risk of coronary heart disease: two new prospective studies and a systematic review. PLoS Med. 2008;5:e78. doi: 10.1371/journal.pmed.0050078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nybo H, Gaist D, Jeune B, et al. The Danish 1905 cohort: a genetic-epidemiological nationwide survey. J Aging Health. 2001;13:32–46. doi: 10.1177/089826430101300102. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen CB, Gotzsche H, Moller JO, Mortensen PB. The Danish Civil Registration System. A cohort of eight million persons. Dan Med Bull. 2006;53:441–449. [PubMed] [Google Scholar]
- 41.Christensen K, McGue M, Petersen I, Jeune B, Vaupel JW. Exceptional longevity does not result in excessive levels of disability. Proc Natl Acad Sci U S A. 2008;105:13274–13279. doi: 10.1073/pnas.0804931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state.” A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 43.McGue M, Christensen K. The heritability of cognitive functioning in very old adults: evidence from Danish twins aged 75 years and older. Psychol Aging. 2001;16:272–280. doi: 10.1037//0882-7974.16.2.272. [DOI] [PubMed] [Google Scholar]
- 44.Katz S, Downs TD, Cash HR, Grotz RC. Progress in development of the index of ADL. Gerontologist. 1970;10:20–30. doi: 10.1093/geront/10.1_part_1.20. [DOI] [PubMed] [Google Scholar]
- 45.Jeune B, Skytthe A, Cournil A, et al. Handgrip strength among nonagenarians and centenarians in three European regions. J Gerontol A Biol Sci Med Sci. 2006;61:707–712. doi: 10.1093/gerona/61.7.707. [DOI] [PubMed] [Google Scholar]
- 46.Udalova IA, Nedospasov SA, Webb GC, Chaplin DD, Turetskaya RL. Highly informative typing of the human TNF locus using six adjacent polymorphic markers. Genomics. 1993;16:180–186. doi: 10.1006/geno.1993.1156. [DOI] [PubMed] [Google Scholar]
- 47.Hajeer AH, Hutchinson IV. TNF-alpha gene polymorphism: clinical and biological implications. Microsc Res Tech. 2000;50:216–228. doi: 10.1002/1097-0029(20000801)50:3<216::AID-JEMT5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 48.Pociot F, Briant L, Jongeneel CV, et al. Association of tumor necrosis factor (TNF) and class II major histocompatibility complex alleles with the secretion of TNF-alpha and TNF-beta by human mononuclear cells: a possible link to insulin-dependent diabetes mellitus. Eur J Immunol. 1993;23:224–231. doi: 10.1002/eji.1830230135. [DOI] [PubMed] [Google Scholar]
- 49.Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. PNAS. 1997;94:3195–3199. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D’Alfonso S, Richiardi PM. A polymorphic variation in a putative regulation box of the TNFA promoter region. Immunogenetics. 1994;39:150–154. doi: 10.1007/BF00188619. [DOI] [PubMed] [Google Scholar]
- 51.Terry CF, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275:18138–18144. doi: 10.1074/jbc.M000379200. [DOI] [PubMed] [Google Scholar]
- 52.Huth C, Illig T, Herder C, et al. Joint analysis of individual participants’ data from 17 studies on the association of the IL6 variant -174G>C with circulating glucose levels, interleukin-6 levels, and body mass index. Ann Med. 2009;41:128–138. doi: 10.1080/07853890802337037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sie MP, Sayed-Tabatabaei FA, Oei HH, et al. Interleukin 6 -174 g/c promoter polymorphism and risk of coronary heart disease: results from the Rotterdam study and a meta-analysis. Arterioscler Thromb Vasc Biol. 2006;26:212–217. doi: 10.1161/01.ATV.0000194099.65024.17. [DOI] [PubMed] [Google Scholar]
- 54.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 55.Kurz T, Strauch K, Dietrich H, et al. Multilocus haplotype analyses reveal association between 5 novel IL-15 polymorphisms and asthma. J Allergy Clin Immunol. 2004;113:896–901. doi: 10.1016/j.jaci.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Christensen U, Haagerup A, Binderup HG, Vestbo J, Kruse TA, Børglum AD. Family based association analysis of the IL2 and IL15 genes in allergic disorders. Eur J Hum Genet. 2006;14:227–235. doi: 10.1038/sj.ejhg.5201541. [DOI] [PubMed] [Google Scholar]
- 57.Giedraitis V, He B, Huang WX, Hillert J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol. 2001;112:146–152. doi: 10.1016/s0165-5728(00)00407-0. [DOI] [PubMed] [Google Scholar]
- 58.Zambon CF, Basso D, Navaglia F, et al. Pro- and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine. 2005;29:141–152. doi: 10.1016/j.cyto.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 59.Culpan D, MacGowan SH, Ford JM, et al. Tumour necrosis factor-alpha gene polymorphisms and Alzheimer’s disease. Neurosci Lett. 2003;350:61–65. doi: 10.1016/s0304-3940(03)00854-1. [DOI] [PubMed] [Google Scholar]
- 60.Levene H. On a matching problem arising in genetics. Ann Math Stat. 1949;20:91–94. [Google Scholar]
- 61.Weir BS. Genetic Data Analysis II. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- 62.Slatkin M, Excoffier L. Testing for linkage disequilibrium in genotypic data using the expectation–maximization algorithm. Heredity. 1996;76:377–383. doi: 10.1038/hdy.1996.55. [DOI] [PubMed] [Google Scholar]
- 63.Snedecor GW, Cochran WG. Statistical Methods. 8th ed. Ames, IA: Iowa State University Press; 1989. [Google Scholar]
- 64.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 65.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008;9:911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Franceschi C, Motta L, Valensin S, et al. Do men and women follow different trajectories to reach extreme longevity? Italian Multicenter Study on Centenarians (IMUSCE) Aging. 2000;12:77–84. doi: 10.1007/BF03339894. [DOI] [PubMed] [Google Scholar]
- 67.Krabbe KS, Mortensen EL, Avlund K, et al. Genetic priming of a proinflammatory profile predicts low IQ in octogenarians. Neurobiol Aging. 2009;30:769–781. doi: 10.1016/j.neurobiolaging.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 68.Mulcahy B, Waldron-Lynch F, McDermott MF, et al. Genetic variability in the tumor necrosis factor-lymphotoxin region influences susceptibility to rheumatoid arthritis. Am J Hum Genet. 1996;59:676–683. [PMC free article] [PubMed] [Google Scholar]
- 69.Wang XY, Hurme M, Jylhä M, Hervonen A. Lack of association between human longevity and polymorphisms of IL-1 cluster, IL-6, IL-10 and TNF-alpha genes in Finnish nonagenarians. Mech Ageing Dev. 2001;123:29–38. doi: 10.1016/s0047-6374(01)00338-4. [DOI] [PubMed] [Google Scholar]
- 70.Ross OA, Curran MD, Rea IM, et al. HLA haplotypes and TNF polymorphism do not associate with longevity in the Irish. Mech Ageing Dev. 2003;124:563–567. doi: 10.1016/s0047-6374(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 71.Collins JS, Perry RT, Watson B, Jr., et al. Association of a haplotype for tumor necrosis factor in siblings with late-onset Alzheimer disease: the NIMH Alzheimer Disease Genetics Initiative. Am J Med Genet. 2000;96:823–830. doi: 10.1002/1096-8628(20001204)96:6<823::aid-ajmg26>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 72.Alvarez V, Mata IF, Gonzalez P, et al. Association between the TNFalpha-308 A/G polymorphism and the onset-age of Alzheimer disease. Am J Med Genet. 2002;114:574–577. doi: 10.1002/ajmg.10515. [DOI] [PubMed] [Google Scholar]
- 73.Laws SM, Perneczky R, Wagenpfeil S, et al. TNF polymorphisms in Alzheimer disease and functional implications on CSF beta-amyloid levels. Hum Mutat. 2005;26:29–35. doi: 10.1002/humu.20180. [DOI] [PubMed] [Google Scholar]
- 74.Holland AM, van den Brink MR. Rejuvenation of the aging T cell compartment. Curr Opin Immunol. 2009;21:454–459. doi: 10.1016/j.coi.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu JT, Tan L, Song JH, et al. Interleukin-18 promoter polymorphisms and risk of late onset Alzheimer’s disease. Brain Res. 2009;1253:169–175. doi: 10.1016/j.brainres.2008.11.083. [DOI] [PubMed] [Google Scholar]
- 76.Suárez A, Castro P, Alonso R, Mozo L, Gutiérrez C. Interindividual variations in constitutive interleukin-10 messenger RNA and protein levels and their association with genetic polymorphisms. Transplantation. 2003;75:711–717. doi: 10.1097/01.TP.0000055216.19866.9A. [DOI] [PubMed] [Google Scholar]
- 77.Passarino G, Montesanto A, Dato S, et al. Sex and age specificity of susceptibility genes modulating survival at old age. Hum Hered. 2006;62:213–220. doi: 10.1159/000097305. [DOI] [PubMed] [Google Scholar]
- 78.Ordovas JM. Gender, a significant factor in the cross talk between genes, environment, and health. Gend Med. 2007;4(suppl B):S111–S122. doi: 10.1016/s1550-8579(07)80052-0. [DOI] [PubMed] [Google Scholar]
- 79.Cederholm T, Persson M, Andersson P, et al. Polymorphisms in cytokine genes influence long-term survival differently in elderly male and female patients. J Intern Med. 2007;262:215–223. doi: 10.1111/j.1365-2796.2007.01803.x. [DOI] [PubMed] [Google Scholar]
- 80.Allan SM, Rothwell NJ. Cytokines and acute neurodegeneration. Nat Rev Neurosci. 2001;2:734–744. doi: 10.1038/35094583. [DOI] [PubMed] [Google Scholar]
- 81.Lucas SM, Rothwell NJ, Gibson RM. The role of inflammation in CNS injury and disease. Br J Pharmacol. 2006;147:S232–S240. doi: 10.1038/sj.bjp.0706400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alley DE, Crimmins EM, Karlamangla A, Hu P, Seeman TE. Inflammation and rate of cognitive change in high-functioning older adults. J Gerontol A Biol Sci Med Sci. 2008;63:50–55. doi: 10.1093/gerona/63.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pernerger TV. What’s wrong with Bonferroni adjustments? BMJ. 1998;316:1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]