Abstract
Purpose
Despite initial efficacy of imatinib mesylate (IM) in most gastrointestinal stromal tumor (GIST) patients, many experience primary/secondary drug resistance. Therefore, clinical management of GIST may benefit from further molecular characterization of tumors before and after IM treatment.
Experimental Design
As part of a recent Phase II Trial of neoadjuvant/adjuvant IM treatment for advanced primary and recurrent operable GISTs (RTOG-S0132), gene expression profiling using oligonucleotide microarrays was performed on tumor samples obtained before and after IM therapy. Patients were classified according to changes in tumor size after treatment based on CT scan measurements. Gene profiling data were evaluated with Statistical Analysis of Microarrays (SAM) to identify differentially expressed genes (in pre-treatment GIST samples).
Results
Based on SAM (FDR=10%), thirty-eight genes were expressed at significantly lower levels in the pre-treatment biopsy samples from tumors that significantly responded to 8 to 12 weeks of IM, i.e., ≥25% tumor reduction. Eighteen of these genes encoded KRAB domain containing zinc finger (KRAB-ZNF) transcriptional repressors. Importantly, ten KRAB-ZNF genes mapped to a single locus on chromosome 19p, and a subset predicted likely response to IM-based therapy in a naïve panel of GISTs. Furthermore, we found that modifying expression of genes within this predictive signature can enhance the sensitivity of GIST cells to IM.
Conclusions
Using clinical pre-treatment biopsy samples from a prospective neoadjuvant phase II trial we have identified a gene signature that includes KRAB-ZNF 91 subfamily members that may be both predictive of and functionally associated with likely response to short term IM treatment.
Keywords: GISTs, imatinib mesylate, microarray, KRAB-ZNF genes
Introduction
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors of the digestive tract, with between 3,300 to 6,000 new cases diagnosed each year in the US (1). The most common primary sites for these neoplasms are the stomach (60-70%) (2, 3), followed by the small intestine (25-35%) (4, 5), and to a much lesser degree the colon and rectum (10%) (6). GISTs have also been observed in the mesentery, omentum, esophagus, and the peritoneum (2, 7). GISTs occur most frequently in patients over 50, with a median age of presentation of 58 years; however, GISTs have also been observed in the pediatric population (8). These tumors contain smooth muscle and neural elements as described originally by Mazur and Clark in 1983, and are thought to arise from the interstitial cells of Cajal (9, 10). GISTs express and are clinically diagnosed by immunohistochemical staining of the 145 kDa transmembrane glycoprotein, KIT, by the CD117 antibody. The majority (~70%) of GISTs possess gain-of-function mutations in c-KIT in either exons 9, 11, 13 or 17, causing constitutive activation of the kinase receptor, whereas smaller subsets of GISTs possess either gain-of-function mutations in PDGFRA (exons 12, 14, or 18) (~10%) or no mutations in either KIT or PDGFRA and are therefore referred to as wild-type (WT) GISTs (~15-20%) (11-14). The primary treatment for GIST is surgical resection, which is often not curative in high risk GIST due to a high incidence of reoccurrence (15, 16). Since 2002, IM, an oral 2-phenylaminopyrimidine derivative that works as a selective inhibitor against mutant forms of type III tyrosine kinases such as KIT, PDGFRA, and BCR/ABL, has become a standard treatment for patients with metastatic and/or unresectable GIST, with objective responses or stable disease obtained in >80% of patients (17, 18). Response to IM has been correlated to the genotype of a given tumor (14). GIST patients with exon 11 KIT mutations have the best response and disease-free survival, while other KIT mutation types and WT GIST have worse prognoses. Despite the efficacy of IM, some patients experience primary and/or secondary resistance to the drug. [18F] fluorodeoxyglucose-positron emission tomography (PET) can be used to rapidly assess tumor response to IM (19); however, there are cases in which GISTs do not take up significant amounts of the glucose precursor and therefore this scanning method is of questionable value in evaluating response in this group of patients. Strategies for treatment of progressive disease can include: IM dose escalation (20), IM in combination with surgery, and alternative KIT/PDGFRA inhibitors including: sunitinib (21). There are also options to participate in clinical trials evaluating nilotinib (22), dasatinib (23) and HSP90 inhibitors (24). What may eventually prove to be the most effective paradigm in the clinical management of GIST is the development of individualized treatment approaches based on KIT and PDGFRA mutational status and/or predictive gene signatures of drug response. Ideally, in the future patients may be pre-selected for treatment with IM or additional first and second line therapies based on these tumor specific response markers.
The question of whether IM can be safe and effective as a rapid cytoreductive agent if administered prior to surgical resection has been evaluated in a recent novel Phase II trial (Radiation Therapy Oncology Group Study 0132) of 8 to 12 weeks of neoadjuvant followed by adjuvant IM for either locally advanced primary or metastatic operable GIST. In this study, biopsies were taken at time of enrollment, patients were treated with IM for 8 to 12 weeks prior to resection, followed by adjuvant IM treatment for 2 years. Contrast enhanced CT scans were performed before, 4-6 weeks into treatment, and after the neoadjuvant IM regimen in order to document classic tumor response by RECIST criteria. Based on CT response data, patients for this study were classified in to two groups, Group A (defined as ≥25% tumor shrinkage after 8-12 weeks of IM) and Group B (<25% tumor shrinkage, unchanged, or evidence of tumor enlargement after 8-12 weeks of IM). Microarray analysis of pre-treatment GIST biopsies identified a gene signature of 38 response genes. These included Krüppel-associated box (KRAB)-zinc finger (ZNF) genes that were significantly expressed in tumor biopsies from patients less responsive to short-term treatment of imatinib.
Methods
Patient Selection
63 patients (52 analyzable), with primary or recurrent operable GIST were enrolled onto the RTOG S0132 trial from 18 institutions. Patients' GIST samples were screened for CD117 (KIT) positivity by standard IHC prior to participation in the clinical trial. Patients were required to have adequate hematologic, renal, and hepatic function as well as measurable disease for response evaluation. All patients signed informed consent following IRB approval for this study and were consented to provide baseline biopsies and operative tissue.
Collection of samples
Tumor samples were obtained from pre-IM core needle biopsies (pre-treatment samples) and from the surgical specimen obtained at the time of resection following neoadjuvant/preoperative IM (post-treatment samples). A total of 48 pre- and 34 post-imatinib treated samples were collected and banked. All patients received IM at 600 mg daily by mouth which was continued daily until the day of surgery, with dose modifications for protocol defined toxicities. Fresh-frozen pre- and post-treatment GIST samples were collected from all participating institutions and shipped to the RTOG tissue bank prior to evaluation.
RNA isolation
Total RNA was isolated from all available pre- and post- frozen tissue samples using TRIzol reagent according to the protocols provided by the manufacturer (Invitrogen Corp., Carlsbad, CA). RNA quantification and quality assessment were performed on 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA). Due to the high variability in tissue collection and handling, storage and shipping procedures among the 18 institutions involved in the study and the tumor cellularity of the specimens, 35% (17 of 48) of pre- and 26% (9 of 34) of post-treatment samples were of limited quality and were therefore excluded from the gene profiling studies. Furthermore, one of the samples was excluded because the CT response data was lacking.
DNA Isolation
Genomic DNA was isolated as previously described (25). Quality DNA was isolated from 38 cases (2 pre-treatment biopsies and 36 post-treatment samples) and used for mutational analyses.
KIT and PDGFRA mutational status analysis
Mutational analysis was performed as previously described (26).
RNA amplification and microarray hybridization
Fifty nanograms (50 ng) of RNA from the various tissue samples, as well as 50 ng of Universal Human Reference RNA (Stratagene, La Jolla, CA) were amplified using Ovation Aminoallyl RNA amplification and labeling system (NuGEN Technologies, Inc., San Carlos, USA). Aminoallyl cDNA was purified with QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and yield was measured using Spectrophotometer ND-1000 (NanoDrop, Wilmington, DE). Sample aminoallyl cDNA was labeled with Alexa Fluor 647 dye (Invitrogen Corp) and reference aminoallyl cDNA was labeled with Alexa Fluor 555 dye (Invitrogen) as follows. Content of one vial from Alexa Fluor Reactive Dye Decapacks for Microarray Applications (Invitrogen) was resuspended in 2.5 μl of DMSO (Clontech, Mountain View, CA) and added to 2 mg of aminoallyl cDNA, which was previously dried down in vacuum centrifuge and resuspended in 7.5 μl of coupling buffer (66.5 mM NaHCO3, pH=9.0). After incubation for 1 hour in darkness at room temperature reaction was purified with QIAquick PCR Purification Kit (Qiagen). Labeling efficiency was assessed on Spectrophotometer ND-1000 (NanoDrop). Labeled sample and reference were combined and hybridized on 44K Whole Human Genome Oligo Microarray (Agilent) at 60°C for 17 hours. Washing was performed in 6 x SSPE buffer with 0.005% Sarcosine at room temperature for 1 min; 0.06 x SSPE buffer with 0.005% Sarcosine at room temperature for 1 min, and then treated with Agilent Stabilization and Drying Solution at room temperature for 30 seconds.
Data analysis
For the microarray studies we were able to obtain high quality RNA and array data from 28 pre-treatment samples and 25 post-treatment samples. For 17 we had matching pairs. Amplified and labeled RNAs were competitively hybridized against Stratagene Human Reference RNA using Agilent 4112a Whole Genome Human microarrays, scanned with an Agilent GMS 428 scanner, and preprocessed using the Functional Genomics Data Pipeline (27). These arrays were checked for quality by both Agilent quality control and by visual inspection of MA plots pre- and post-LOESS normalization (width = 0.7, no background correction). Arrays that were of poor quality (i.e., which showed signs of RNA degradation such as splitting of MA plots into two `wings') were repeated on a second RNA isolation from the same biopsy or tumor sample.
Clinical RECIST response is typically defined as a 30% decrease in the longest tumor diameter in the case of a primary target lesion or the sum of the longest diameters in the case of index tumors of metastatic disease. For the purpose of this analysis, as surgery occurred at a median of 65 days from the start of IM therapy, we arbitrarily divided these patients into Group A (≥25% tumor shrinkage after 8-12 weeks of IM) or Group B (<25% tumor shrinkage, unchanged, or evidence of tumor enlargement after 8-12 weeks of IM). In the seminal phase II metastatic GIST study the median time to partial response (PR) (≥30% reduction) was 16 weeks, therefore, we concluded that the duration of pre-op IM was probably too short to expect a significant number of patients having a classic PR per RECIST. We therefore chose an arbitrary grouping of CT measured response for patients in Group A of ≥25% close to the 30% RECIST criteria for PR. Had we selected ≥30% decreased in tumor dimension there would have been too few patients in Group A for any meaningful analysis. All other patient's gene array samples that correlated clinically to ≤25% decrease in tumor measurements as determined by the study clinical parameters were then placed in Group B. It should be noted that gene profiling for predictive biomarkers of response was a post-hoc analysis and not a primary or secondary endpoint of this original study. The 28 pre-treatment samples were analyzed with Significance Analysis of Microarrays (SAM) (28) implemented in the Multi-Experiment Viewer (MEV) (29) to identify genes that showed significant pre-treatment differential expression between the two groups. A false discovery rate of 10% was used. Microarrays were annotated using the most recent (20 Aug 2007) Agilent annotation file. The most current accession number corresponding to Agilent IDs were retrieved from the file. Ensembl accession numbers were annotated with gene symbols and descriptions on June 6, 2008. Genebank accession numbers or gene names were annotated with NCBI Entrez information on June 9, 2008.
Since 10 of the differentially expressed genes mapped to the same locus (HSA19p12-19p13.1), we also analyzed all of the genes in this locus for response upon treatment (25 post-treatment samples, with 13 samples from Group B and 12 from Group A) with IM. We performed this test by looking at each gene individually and looking for its average response in four categories: Group A pre-treatment, Group B pre-treatment, Group A post-treatment, Group B post-treatment. Microarray data including original Agilent scanner output files for all samples used in this study are available through the Gene Expession Omnibus (GEO) (accession number GSE15966).
Quantitative RT-PCR
To confirm the microarray data, RNA was freshly isolated from 9 of the trial's pre-IM samples (RTOG19, 22, 31, 39, 47, 56) including 3 samples (RTOG25, 35, and 53) not included in the original microarray analyses and reverse transcribed to cDNA by SuperScript II reverse transcriptase (Invitrogen). Expression of RNA for three KRAB-ZNF genes (ZNF 91, ZNF 43 and ZNF 208) and two endogenous control genes (HPRT and 18S) was measured in each pre-sample by real-time PCR (with TaqMan Gene Expression Assay products on an ABI PRISM 7900 HT Sequence Detection System, Applied Biosystems, Foster, CA) following protocols recommended by the manufacturer and as previously described (30). The relative mRNA expressions of ZNF 91, ZNF 43 and ZNF 208 were adjusted with either HPRT or actin. The primer/probe (FAM) sets for ZNF 91, ZNF 43, ZNF 208, HPRT and 18S were obtained from Applied Biosystems.
siRNA transfection and IM sensitivity
Two siRNAs against each ZNF of interest (Qiagen) were pooled together and GIST cells were reverse transfected in four 96-well plates as described according to the protocols provided by the manufacturer (Qiagen). In addition, siRNA smart pools against KIT and GL-2 (Dharmacon) were used as positive and negative controls, respectively, and used for Z-score calculations. Forty-eight hours later vehicle only or vehicle + IM (45 nM) were added to two plates. After twenty-four hours cell viability was assessed using the cell titer blue assay. This assay is based on the ability of living cells to convert the redox dye, resazurin, into the fluorescent end product, resorufin. Cell titer blue was added to all wells and incubated for four hours followed by data recording using an EnVision microplate reader (PerkinElmer).
Results
RTOG-S0132 trial design and patient response to IM
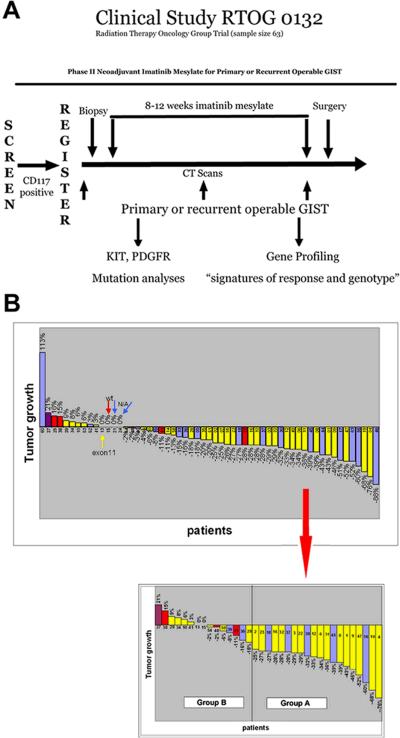
Sixty-three (63) patients with primary or recurrent potentially resectable malignant GIST, from 18 institutions, were originally enrolled onto the trial beginning in February 2002 and ending in June 2006 (15). A tumor positive for KIT (CD117) staining by IHC was the necessary prerequisite for patient enrollment. Fifty-three percent (53%) of primary tumors were located in the stomach, 27% in small bowel, and 20% in GI other sites. Metastatic tumors were primarily located in the abdomen/peritoneum. Additional clinical information is shown in Table 1. Prior to the start of the 8-12 week IM regimen, a CT scan was performed and a tumor biopsy (pre-treatment sample) was obtained. CT scans were repeated ~4-6 weeks into IM therapy and again immediately prior to surgical resection (after 8-12 weeks IM therapy) (Figure 1A). CT measurements, taken from the longest cross sectional diameter of the primary GIST or the index metastatic lesion(s), were used to assess tumor response (i.e. tumor shrinkage, no measurable change, or tumor enlargement) to IM therapy (Figure 1B). Of the 52 analyzable patients, 58% (30 of 52) had surgical resection of primary locally advanced GIST, whereas 42% (22 of 52) had recurrent/metastatic GIST resected. Genomic DNA was isolated from available large biopsies (pre-treatment samples) or resected tumor (post-treatment samples) and KIT and PDGFRA mutational analysis was performed (Figure 1B). Mutational analysis was performed on 39 of the 52 patients and the most frequent mutations occurred in exon 11 (82%, 32 of 39), followed by exon 9 (3%, 1/39). No mutations were found in exons 13 and 17 of KIT or in exons 12, 14 and 18 of PDGFRA. Fifteen percent (15%, 6 of 39) of the patients tested lacked mutations in both KIT and PDGFRA. Similar frequencies have been observed previously (12).
Table 1.
Patients' and tumors' characteristics.
| n (%) | |
|---|---|
| Median age (range), years | 58.5 (24 to 84) |
| Sex | |
| Female | 24 (46) |
| Male | 28 (54) |
| Primary tumor | 30 (58) |
| Metastatic/recurrent tumor | 22 (42) |
| Site of primary tumor | |
| Stomach | 16 (53) |
| Small bowel | 8 (27) |
| Other | 6 (20) |
| Site of metastatic tumor | |
| Abdomen/peritoneum | 15 (68) |
| Liver only | 6 (27) |
| Liver/peritoneum | 1 (5) |
| Size of tumor (cm) | |
| ≤ 10 | 37 (71) |
| >10 | 15 (29) |
| Mutation | |
| Exon 11 KIT | 32 (62) |
| Exon 9 KIT | 1 (2) |
| Exon 17 KIT | 0 (0) |
| PDGFRα(exons 18 and 12) | 0 (0) |
| Wild-type | 6 (12) |
| N/A* | 13 (25) |
Not enough tissue for mutational analysis.
Figure 1. RTOG-S0132 trial design and patient response to IM.
A) Patients with primary or recurrent operable GISTs were screened for KIT (CD117) expression by IHC for eligibility. Prior to IM treatment, a CT was performed and biopsies were collected by core needle aspiration. Patients were then treated with an 8-12 week regimen of IM, followed by cytoreductive surgery. A CT was also performed once during treatment (~4-6 weeks into IM treatment) and immediately prior to surgery. B) (Top) Percentage of tumor growth based on CT measurements taken from the longest cross sectional diameter of the primary GIST or the index metastatic lesion(s) for each RTOG-S0132 patient. (Bottom) Specific samples (pre-, post- or both) used for microarray analysis classified as Group A or B based on the percent of tumor shrinkage/growth visualized by CT. Mutational analysis of most patients was performed and is denoted by color of bar (yellow = KIT exon 11 mutants, red = wild-type GISTs, purple = KIT exon 9 mutants, blue = not enough DNA available for mutational analysis). Group A is defined as ≥25% tumor shrinkage after 8-12 weeks of IM and Group B contains tumors demonstrating <25% tumor reduction, no change, or evidence of tumor enlargement after 8-12 weeks of IM.
Gene expression profiles associated with response to IM
RNA was isolated from both pre- and post-treatment samples and those deemed of adequate amount and quality were evaluated by using Agilent oligonucleotide microarrays (see Methods). GIST specimens (pre-, post- or both) used for microarray analysis are shown in Figure 1B (bottom). CT measurements were used to classify patients as either “immediate responders” (Group A) if the patient's tumor demonstrated a 25% or greater reduction in size during the 8 to 12 weeks of IM treatment. The other GIST samples were combined and will subsequently be referred to as Group B. The index used for these latter tumors ranged from an 18% diameter reduction to a 21% tumor enlargement after 8-12 weeks of IM. Mutation status alone was not a sole predictor of response to short-term IM treatment. Seventy-five percent (15/20) of biopsy samples from Group A possessed an exon 11 KIT mutation. The remaining 25% (5/20) had no mutational analysis available. In comparison, 53% (8/15) of samples from Group B had exon 11 KIT mutations, whereas the remaining samples either possessed an exon 9 KIT mutant (7%, 1/15), were KIT/PDGFRA mutation negative (27%, 4/15), or were undetermined (13%, 2/15). Using the array data generated from the pre-treatment biopsy specimen RNAs, SAM identified 38 genes as differentially expressed at a false discovery rate of 10% between the two groups, with all gene transcripts present at higher levels in patients within Group B (Table 2). Thirty-two (32) of these corresponded to known genes, 18 of these are Krüppel-associated box (KRAB)-zinc finger (ZNF) genes, 10 of which mapped to the same locus (HSA19p12-19p13.1). Two additional genes, LOC646825 and LOC388523, showed similarity to ZNF 91 and ZNF 208 (Figure 2). Other genes within this signature encode for the zinc finger-containing proteins, ZMYND11 and ZMAT1 and transcription factors, GTF2I and GABPAP. The remaining genes encode the following proteins: RASSF8, WDR90, SF3B1, UGT2B7 and four hypothetical proteins (Table 2).
Table 2.
SAM (significance analysis of microarrays) analysis of genes differentially expressed between rapid responders and stable disease.
| Accession | Gene symbol | Description | Cytoband | SAM score |
|---|---|---|---|---|
| NM_178549 | ZNF678 | zinc finger protein 678 | 1q42.13 | 4.10 |
| NM_212479 | ZMYND11 | zinc finger, MYND domain containing 11 | 10p15.3 | 4.11 |
| NM_007211 | RASSF8 | Ras association (RalGDS/AF-6) domain family 8 | 12p12.1 | 3.55 |
| A_24_P75888 | n/a | n/a | 14q11.1 | 4.35 |
| AK126622 | WDR90 | WD repeat domain 90 | 16p13.3 | 3.61 |
| A_24_P717262 | n/a | n/a | 19p12 | 4.23 |
| ENST00000341262 | ZNF56 | zinc finger protein 56 (Fragment) | 19p12 | 3.97 |
| AK131420 | ZNF66 | zinc finger protein 66 | 19p12 | 4.06 |
| NM_003429 | ZNF85 | zinc finger protein 85 | 19p12 | 4.36 |
| NM_133473 | ZNF431 | zinc finger protein 431 | 19p12 | 3.90 |
| NM_001001415 | ZNF429 | zinc finger protein 429 | 19p12 | 4.29 |
| NM_003423 | ZNF43 | zinc finger protein 43 | 19p12 | 4.10 |
| NM_007153 | ZNF208 | zinc finger protein 208 | 19p12 | 3.69 |
| NM_001001411 | ZNF676 | zinc finger protein 676 | 19p12 | 4.08 |
| ENST00000357491 | LOC646825 | DISCONTINUED: similar to zinc finger protein 91 | 19p12 | 4.14 |
| NM_001080409 | ZNF99 | zinc finger protein 99 | 19p12 | 3.84 |
| XR_017338 | LOC388523 | similar to zinc finger protein 208 | 19p12 | 4.10 |
| NM_003430 | ZNF91 | zinc finger protein 91 | 19p12 | 3.95 |
| ENST00000334564 | ZNF528 | zinc finger protein 528 | 19q13.33 | 3.92 |
| NM_024733 | ZNF665 | zinc finger protein 665 | 19q13.41 | 3.74 |
| NM_001004301 | ZNF813 | zinc finger protein 813 | 19q13.41 | 3.86 |
| AK001808 | n/a | CDNA FLJ10946 fis, clone PLACE1000005 | 2q24.3 | 4.21 |
| BE168511 | SF3B1 | Splicing factor 3b, subunit 1, 155kDa | 2q33.1 | 3.86 |
| NM_138402 | LOC93349 | hypothetical protein BC004921 | 2q37.1 | 4.42 |
| ENST00000305570 | LOC727867 | similar to PRED65 | 21q11.2 | 3.57 |
| ENST00000341087 | n/a | n/a | 4p16.3 | 4.53 |
| NM_001074 | UGT2B7 | UDP glucuronosyltransferase 2 family, polypeptide B7 | 4q13.2 | 3.66 |
| NM_182524 | ZNF595 | zinc finger protein 595 | 4p16.3 | 3.71 |
| THC2708803 | n/a | n/a | 4q22.3 | 3.85 |
| A_24_P492885 | n/a | n/a | 7q11.21 | 4.39 |
| XM_001127354 | LOC728376 | similar to hCG1996858 | 7p11.2 | 4.48 |
| AF277624 | ZNF479 | zinc finger protein 479 | 7p11.2 | 4.19 |
| NR_002723 | GABPAP | GA binding protein TF, alpha subunit pseudogene | 7q11.21 | 4.14 |
| XM_001128828 | LOC728927 | similar to hCG40110 | 7q11.21 | 4.05 |
| NM_178558 | ZNF680 | zinc finger protein 680 | 7q11.21 | 3.59 |
| NM_001518 | GTF2I | general transcription factor II, i | 7q 11.23 | 4.09 |
| NM_197977 | ZNF189 | zinc finger protein 189 | 9q31.1 | 3.64 |
| NM_032441 | ZMAT1 | zinc finger, matrin type 1 | Xq22.1 | 3.74 |
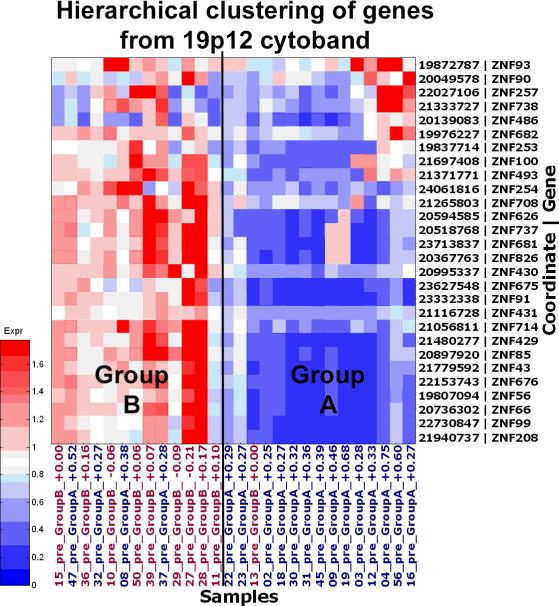
Figure 2. Gene expression profiles associated with response to IM.
A heat map showing the HSA19p12-13.1 KRAB-ZNF hierarchical cluster. In the image blue represents down-regulation, whereas red represents up-regulation. Patients who initially responded rapidly to IM clearly show decreased KRAB-ZNF expression compared to the others.
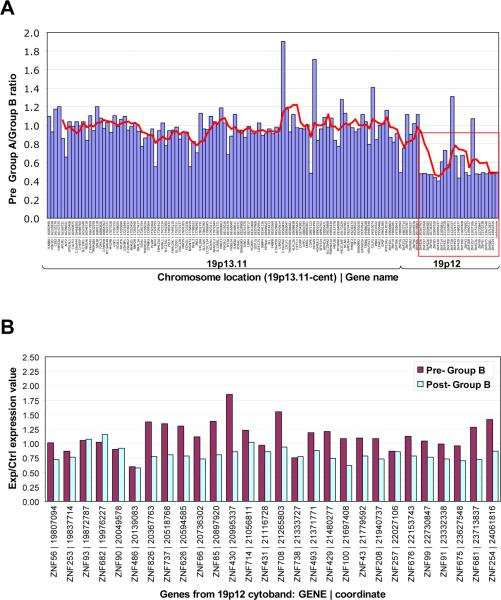
The observation that the majority of the genes within this predictive signature were KRAB-ZNF genes (18 of 32), ten of which are located within the same chromosomal region (HSA19p12-19p13.1) was intriguing and warranted further study. Analysis of pre-treatment sample expression differences for all genes within the 19p12-13.1 locus showed a consistent difference (Figure 3A, red box). All the ZNF genes showed higher overall expression in samples from patients within Group B across the locus, even though adjoining genes showed equal expression between the two groups. Of additional interest, these KRAB-ZNFs appear to be coordinately regulated in response to IM therapy in that KRAB-ZNF mRNA levels decrease in tumors from patients in Group B after IM. In order to rule out the possibility that an enrichment of other non-tumor cells, such as endothelial and inflammatory cells may be contributing to the observed expression patterns we examined the cellular content of the post-IM samples and used only those that displayed >70% tumor cellularity (Figure 3B). We also observed a very similar pattern of decreased ZNF expression in the Group B post-IM samples with lower tumor (<70%) cellularity (data not shown), suggesting that the observed trend is likely associated with tumor cell response to IM. Analysis of the pre- and post-treatment samples from Group A showed an opposing trend in that the level of ZNF genes increased following the 8-12 week IM regimen; however, since the cellularity was <70% for all but one of these samples we cannot rule out the effect of non-tumor cells on these expression patterns (data not shown).
Figure 3. KRAB-ZNF gene expression on chromosome 19p12-13.1 before and after IM therapy.
A: Analysis of pre-treatment ratios of tumors showing >25% (Group A) or <25 reduction (Group B) using data from 28 patients for all genes in the 19p12-13.1 locus. All genes, in this locus (red box) showed higher mean ZNF expression levels in Group B samples (i.e. lower Group A/Group B ratio) while adjoining genes showed roughly equal expression between the two groups. B: Analysis of changes in expression of genes in this locus upon IM treatment in Group B samples with >70% tumor cellularity. Red bars represent means of pre-treatment samples from Group B and blue bars represent means of post-treatment samples from Group B.
Validation with qRT-PCR
We used qRT-PCR to validate the differential expression pattern of the predictor genes. For this analysis, four genes were selected from the list of 18 KRAB-ZNF genes identified in the microarray analysis based on availability of commercial qRT-PCR assays. We found the assays for ZNF 43, ZNF 208 and ZNF 91 to work reliably. All three were expressed significantly higher in Group B prior to IM treatment compared to the immediate response group. The expression of each gene was evaluated in a small validation panel consisting of nine pre-treatment samples from patients on the trial for which high quality RNAs could be isolated (see Methods). ZNF 43, ZNF 208, and ZNF 91 mRNA levels were significantly lower in patients whose tumors rapidly shrunk in response to IM than in those who did not (Figure 4). Expression levels of the three genes were highly correlated with each other (all pairwise correlations were greater than 0.93 with p values < 0.0003).
Figure 4. Validation of ZNF Gene Expression by qRT-PCR.
Fold expression changes of three of the ZNFs within the predictive signature gene panel, i.e., ZNF 43, ZNF 208 and ZNF 91, were measured using qRT-PCR. The ratios of each gene to control (HPRT or actin) were measured using total RNAs from nine pre-treatment samples (5 in Group A and 4 in Group B) and universal human reference RNA. The relative median mRNA levels for ZNF 43, ZNF 208, and ZNF 91 in Group A were 412-, 257- and 77-fold higher as compared to controls, whereas the median levels in Group B were 21-, 18-, and 11-fold normalized to controls, respectively. Two-sided Wilcoxon rank sum tests were used to compare the distribution of ZNF 43, ZNF 208, and ZNF 91 mRNA expression between the two groups and Pearson's coefficients were used to measure the pairwise correlation of the ZNF gene expression. Tests were conducted using a 5% type I error. The predictive value of ZNF 43 and ZNF 208 were found to be statistically significant (*p=0.02). Results are representative of three independent experiments.
We next sought to determine if modifying the expression of a subset of the genes within this predictive signature could alter the sensitivity of GIST cells to IM. We selected ZNF 208, ZNF 91, ZNF 85 and ZNF 43 for siRNA targeted knockdown. From these screens, we demonstrated that depletion of each of the four ZNFs were able to sensitize GIST cells to varying degrees of IM (Sensitization Index = viability with drug/viability with vehicle only was 0.58 to 0.85). These findings suggest that some members of this gene signature may not only have predictive value but functional relevance to IM activity in vivo. We also developed genomic-based qPCR analysis to assess gene copy number of these KRAB-ZNF genes. We found that upregulation of these ZNFs in patients within Group B was not associated with gene amplification (data not shown), indicating that the changes in mRNA were independent of gene copy number.
Discussion
In this study, we set out to obtain a gene expression profile that could be predictive of likely IM induced cytoreduction in GIST patients prior to therapy. Because several alternative options for progressive disease treatment are currently being evaluated, such as new kinase inhibitors or combination therapy with IM, such a profile may be useful in determining appropriate personalized clinical treatment of GIST patients.
The clinical trial from which tissue samples were obtained for this study has yielded some interesting findings. The majority of patients on this trial had apparent clinical benefit from IM therapy prior to surgery. Forty-nine percent (49%) of all patients enrolled onto the trial manifested ≥25% tumor size reduction following the initiation of 8-12 weeks of IM therapy, with 75.4% having at least some degree of tumor response (Figure 1B). In addition, pre-operative IM therapy was associated with minimal drug related toxicity and surgical morbidity (31). We observed benefit from the neoadjuvant use of IM for downsizing tumors prior to surgical resection. Using pre-IM samples from this study we were able to perform microarray analysis to obtain a gene expression profile that may be indicative of the likely response to short-term IM therapy. Although expression of several interesting genes, such as RASSF8, SF3B1, and UGT2B7 were found to be associated with differential response to IM, we were drawn to the observation that nearly a third of the genes clustered in one locus on chromosome 19p12 near the centromere (Figure 2). These differentially expressed ZNFs are KRAB-ZNF genes that are members of the ZNF 91 subfamily (32, 33). In addition, we demonstrated that expression of these ZNFs appeared to be coordinately regulated by IM treatment (Figure 3B and data not shown).
The ZNF 91 subfamily includes 64 genes, 37 of which are found on chromosome 19 (32). These KRAB-ZNF proteins are characterized by the presence of a DNA-binding domain composed of between 4 and 30 zinc-finger motifs and a KRAB domain near the amino terminus. They form one of the largest families of transcriptional regulators. Many members of this family are still uncharacterized and the specific functions of many members are unknown; however, some of these ZNFs have been associated with undifferentiated cells and also implicated in cancers. Lovering and Trowsdale showed that expression of ZNF 43 was increased in lymphoid cell lines and that inducing terminal differentiation in vitro in one of these cell lines led to reduced ZNF 43 expression (34). Another study using microarrays comparing normal controls to mononuclear cells of AML patients, showed ZNF 91 expression was increased in 93% of AML cases and that inhibiting expression of ZNF 91 induced apoptosis of these cells (35). Eight other ZNFs, not found to reach significance in our tests for differential expression in our studies, have been denoted as “candidate cancer genes” or CAN-genes by large-scale mutagenesis screens in breast and colorectal cancers (36).
In addition, KRAB-ZNF expression has been associated with resistance to IM. Using DNA microarrays, Chung et al. (2006) showed that 22 genes, 2 of which are ZNFs, were positively correlated with increasing IM dosage in chronic myelogenous leukemia cell lines (37). Therefore, our study is not the first to link response to IM with KRABZNF expression, but is the first to establish this connection in GIST patients and to the genes within the HSA19p12-19p13.1 locus. The ultimate goal of this work was to identify a profile that is indicative of immediate response to IM so that in the future, expression of these ZNFs can be examined in patient biopsies prior to treatment, allowing for the most effective therapeutic regimen to be employed, particularly in relation to planned surgical resection. As there is significant overexpression of these KRAB-ZNF's and other genes within the predictive signature in patients who are not as responsive to IM, our study suggests that IHC-based or qRT-PCR expression analyses of these genes could potentially serve as a rapid means for prescreening GIST patients prior to treatment. However, it should be re-iterated that this predictive gene signature was established using a 25% decrease in tumor size cutoff for response rather than RECIST or Choi criteria since this was a neoadjuvant trial and the design of the trial (8 to 12 weeks of imatinib) was not to assess short-term imatinib response by standard criteria. Nevertheless, it will be important to further evaluate these predictive biomarkers using independent cohorts of GIST samples with clinical outcome information.
We have shown that qRT-PCR assays are informative when adequate RNA samples can be obtained either from small needle biopsies or resected tumor samples. Our studies also highlight the need for additional studies to assess the role of these KRAB-ZNFs in potentially mediating IM-response. In preliminary studies we have found that siRNA mediated targeted knockdown of ZNF 208, ZNF 91, ZNF 85 and ZNF 43 can enhance the sensitivity of GIST cells to IM, albeit to varying degrees. Further functional studies are currently underway to determine how these genes may be influencing IM activity in GISTs and their potential clinical therapeutic value.
We also searched for links as to why many of these ZNF genes within a single locus are coordinately regulated at the expression level. Using transcription factor binding site analysis, from advanced biomedical computing center and viewed using CIMminer software, we sought to identify common transcription factors (TFs) that could explain why, in some samples, all the genes are either upregulated or downregulated. The analysis showed that there are a number of TFs that regulate these ZNFs (data not shown). One TF, HinfA, appeared to be associated with 12 of the ZNFs of interest. HinfA is a TF known to bind to A/T rich repeats in the promoters of human histone (H3 and H4) genes (38). However, HinfA was not measured on our array. Vogel and colleagues have found that the heterochromatin binding proteins, CBX1 and SUV39H1 have been associated with co-expression of ZNF genes (39). However, our analysis of the three probes for CBX1 and one probe for SUV39H1 did not detect significant differences in expression between these two groups.
In summary, we were able to elucidate a gene expression profile that is unique to patients whose tumors are less responsive to IM in comparison to those that rapidly respond. This profile consists of 32 genes, 18 of which are KRAB-ZNFs. We feel that these results have potential clinical relevance and could help stratify patients most responsive to IM, and potentially design more effective treatment regimens particularly in neoadjuvant use for GIST patients in the future.
Acknowledgement
We would like to acknowledge the valuable input of Drs. Chi Tarn, Chong Xu, Harsh Pathak, Yan Zhou, and Eric Ross on this manuscript and Ms. Lisa Vanderveer for technical support. The Biosample Repository Core Facility, the Clinical Molecular Genetics Laboratory, and the Genome Core Facility at FCCC and the RTOG Tissue Bank were essential for the tissue studies. We would like to acknowledge the support of Ms. Tania Stutman and the GIST Cancer Research Fund for providing a fellowship to L.R.
Grant supports: This work was supported in part by NIH grants (CA106588 and a supplement from the Radiation Therapy Oncology Group to U10 CA21661) to A.K.G., LM009382 to M.F.O., an award by the FCCC Translational Research Committee to M.v.M. and A.K.G as part of the FCCC core grant (P30 CA006927), and by the NIH Training Grant (Institutional NRSA) Appointment (CA009035-31) to L.R.
References
- 1.Corless CL, Heinrich MC. Molecular pathobiology of gastrointestinal stromal sarcomas. Annual review of pathology. 2008;3:557–86. doi: 10.1146/annurev.pathmechdis.3.121806.151538. [DOI] [PubMed] [Google Scholar]
- 2.El-Rifai W, Sarlomo-Rikala M, Andersson LC, Knuutila S, Miettinen M. DNA sequence copy number changes in gastrointestinal stromal tumors: tumor progression and prognostic significance. Cancer research. 2000;60:3899–903. [PubMed] [Google Scholar]
- 3.Miettinen M, Lasota J. Gastrointestinal stromal tumors: review on morphology, molecular pathology, prognosis, and differential diagnosis. Archives of pathology & laboratory medicine. 2006;130:1466–78. doi: 10.5858/2006-130-1466-GSTROM. [DOI] [PubMed] [Google Scholar]
- 4.Tworek JA, Appelman HD, Singleton TP, Greenson JK. Stromal tumors of the jejunum and ileum. Mod Pathol. 1997;10:200–9. [PubMed] [Google Scholar]
- 5.Miettinen M, Kopczynski J, Makhlouf HR, et al. Gastrointestinal stromal tumors, intramural leiomyomas, and leiomyosarcomas in the duodenum: a clinicopathologic, immunohistochemical, and molecular genetic study of 167 cases. The American journal of surgical pathology. 2003;27:625–41. doi: 10.1097/00000478-200305000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Tworek JA, Goldblum JR, Weiss SW, Greenson JK, Appelman HD. Stromal tumors of the anorectum: a clinicopathologic study of 22 cases. The American journal of surgical pathology. 1999;23:946–54. doi: 10.1097/00000478-199908000-00013. [DOI] [PubMed] [Google Scholar]
- 7.Miettinen M, Monihan JM, Sarlomo-Rikala M, et al. Gastrointestinal stromal tumors/smooth muscle tumors (GISTs) primary in the omentum and mesentery: clinicopathologic and immunohistochemical study of 26 cases. The American journal of surgical pathology. 1999;23:1109–18. doi: 10.1097/00000478-199909000-00015. [DOI] [PubMed] [Google Scholar]
- 8.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22:3813–25. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 9.Mazur MT, Clark HB. Gastric stromal tumors. Reappraisal of histogenesis. The American journal of surgical pathology. 1983;7:507–19. doi: 10.1097/00000478-198309000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Graadt van Roggen JF, van Velthuysen ML, Hogendoorn PC. The histopathological differential diagnosis of gastrointestinal stromal tumours. Journal of clinical pathology. 2001;54:96–102. doi: 10.1136/jcp.54.2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinrich MC, Corless CL. Gastric GI stromal tumors (GISTs): the role of surgery in the era of targeted therapy. J Surg Oncol. 2005;90:195–207. doi: 10.1002/jso.20230. discussion. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian S, West RB, Corless CL, et al. Gastrointestinal stromal tumors (GISTs) with KIT and PDGFRA mutations have distinct gene expression profiles. Oncogene. 2004;23:7780–90. doi: 10.1038/sj.onc.1208056. [DOI] [PubMed] [Google Scholar]
- 13.Tarn C, Merkel E, Canutescu AA, et al. Analysis of KIT mutations in sporadic and familial gastrointestinal stromal tumors: therapeutic implications through protein modeling. Clin Cancer Res. 2005;11:3668–77. doi: 10.1158/1078-0432.CCR-04-2515. [DOI] [PubMed] [Google Scholar]
- 14.Debiec-Rychter M, Sciot R, Le Cesne A, et al. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer. 2006;42:1093–103. doi: 10.1016/j.ejca.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 15.Eisenberg BL, Judson I. Surgery and imatinib in the management of GIST: emerging approaches to adjuvant and neoadjuvant therapy. Annals of surgical oncology. 2004;11:465–75. doi: 10.1245/ASO.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 16.DeMatteo RP, Lewis JJ, Leung D, Mudan SS, Woodruff JM, Brennan MF. Two hundred gastrointestinal stromal tumors: recurrence patterns and prognostic factors for survival. Annals of surgery. 2000;231:51–8. doi: 10.1097/00000658-200001000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–80. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 18.Verweij J, Casali PG, Zalcberg J, et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet. 2004;364:1127–34. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 19.Van den Abbeele AD, Badawi RD. Use of positron emission tomography in oncology and its potential role to assess response to imatinib mesylate therapy in gastrointestinal stromal tumors (GISTs) Eur J Cancer. 2002;38(Suppl 5):S60–5. doi: 10.1016/s0959-8049(02)80604-9. [DOI] [PubMed] [Google Scholar]
- 20.Zalcberg JR, Verweij J, Casali PG, et al. Outcome of patients with advanced gastro-intestinal stromal tumours crossing over to a daily imatinib dose of 800 mg after progression on 400 mg. Eur J Cancer. 2005;41:1751–7. doi: 10.1016/j.ejca.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 21.Prenen H, Cools J, Mentens N, et al. Efficacy of the kinase inhibitor SU11248 against gastrointestinal stromal tumor mutants refractory to imatinib mesylate. Clin Cancer Res. 2006;12:2622–7. doi: 10.1158/1078-0432.CCR-05-2275. [DOI] [PubMed] [Google Scholar]
- 22.Weisberg E, Wright RD, Jiang J, et al. Effects of PKC412, nilotinib, and imatinib against GIST-associated PDGFRA mutants with differential imatinib sensitivity. Gastroenterology. 2006;131:1734–42. doi: 10.1053/j.gastro.2006.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schittenhelm MM, Shiraga S, Schroeder A, et al. Dasatinib (BMS-354825), a dual SRC/ABL kinase inhibitor, inhibits the kinase activity of wild-type, juxtamembrane, and activation loop mutant KIT isoforms associated with human malignancies. Cancer research. 2006;66:473–81. doi: 10.1158/0008-5472.CAN-05-2050. [DOI] [PubMed] [Google Scholar]
- 24.Bauer S, Yu LK, Demetri GD, Fletcher JA. Heat shock protein 90 inhibition in imatinib-resistant gastrointestinal stromal tumor. Cancer research. 2006;66:9153–61. doi: 10.1158/0008-5472.CAN-06-0165. [DOI] [PubMed] [Google Scholar]
- 25.Godwin AK, Vanderveer L, Schultz DC, et al. A common region of deletion on chromosome 17q in both sporadic and familial epithelial ovarian tumors distal to BRCA1. American journal of human genetics. 1994;55:666–77. [PMC free article] [PubMed] [Google Scholar]
- 26.Corless CL, McGreevey L, Haley A, Town A, Heinrich MC. KIT mutations are common in incidental gastrointestinal stromal tumors one centimeter or less in size. The American journal of pathology. 2002;160:1567–72. doi: 10.1016/S0002-9440(10)61103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant JD, Somers LA, Zhang Y, Manion FJ, Bidaut G, Ochs MF. FGDP: functional genomics data pipeline for automated, multiple microarray data analyses. Bioinformatics (Oxford, England) 2004;20:282–3. doi: 10.1093/bioinformatics/btg407. [DOI] [PubMed] [Google Scholar]
- 28.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Arciero CA, Wang C, Broccoli D, Godwin AK. BRCC36 is essential for ionizing radiation-induced BRCA1 phosphorylation and nuclear foci formation. Cancer research. 2006;66:5039–46. doi: 10.1158/0008-5472.CAN-05-4194. [DOI] [PubMed] [Google Scholar]
- 31.Heinrich MC, Corless CL, Blanke CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006;24:4764–74. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton AT, Huntley S, Tran-Gyamfi M, Baggott DM, Gordon L, Stubbs L. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome research. 2006;16:584–94. doi: 10.1101/gr.4843906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mark C, Abrink M, Hellman L. Comparative analysis of KRAB zinc finger proteins in rodents and man: evidence for several evolutionarily distinct subfamilies of KRAB zinc finger genes. DNA and cell biology. 1999;18:381–96. doi: 10.1089/104454999315277. [DOI] [PubMed] [Google Scholar]
- 34.Lovering R, Trowsdale J. A gene encoding 22 highly related zinc fingers is expressed in lymphoid cell lines. Nucleic acids research. 1991;19:2921–8. doi: 10.1093/nar/19.11.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unoki M, Okutsu J, Nakamura Y. Identification of a novel human gene, ZFP91, involved in acute myelogenous leukemia. International journal of oncology. 2003;22:1217–23. [PubMed] [Google Scholar]
- 36.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. (New York, NY. [DOI] [PubMed] [Google Scholar]
- 37.Chung YJ, Kim TM, Kim DW, et al. Gene expression signatures associated with the resistance to imatinib. Leukemia. 2006;20:1542–50. doi: 10.1038/sj.leu.2404310. [DOI] [PubMed] [Google Scholar]
- 38.van Wijnen AJ, Massung RF, Stein JL, Stein GS. Human H1 histone gene promoter CCAAT box binding protein HiNF-B is a mosaic factor. Biochemistry. 1988;27:6534–41. doi: 10.1021/bi00417a051. [DOI] [PubMed] [Google Scholar]
- 39.Vogel MJ, Guelen L, de Wit E, et al. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome research. 2006;16:1493–504. doi: 10.1101/gr.5391806. [DOI] [PMC free article] [PubMed] [Google Scholar]