Summary
Two-dimensional patterning of the follicular epithelium in Drosophila oogenesis is required for the formation of three-dimensional eggshell structures. Our analysis of a large number of published gene expression patterns in the follicle cells suggested that they follow a simple combinatorial code, based on six spatial building blocks and the operations of union, difference, intersection, and addition. The building blocks are related to the distribution of the inductive signals, provided by the highly conserved EGFR and DPP pathways. We demonstrated the validity of the code by testing it against a set of newly identified expression patterns, obtained in a large-scale transcriptional profiling experiment. Using the proposed code, we distinguished 36 distinct patterns for 81 genes expressed in the follicular epithelium and characterized their joint dynamics over four stages of oogenesis. This work provides the first systematic analysis of the diversity and dynamics of two-dimensional gene expression patterns in a developing tissue.
Introduction
Drosophila eggshell is an elaborate three-dimensional structure that is derived from the follicular epithelium in the developing egg chamber (Figure 1A-C). The dorsal-anterior structures of the eggshell, including the dorsal appendages and operculum, are formed by the follicle cells that are patterned by Gurken (GRK), a TGFα-like ligand secreted by the oocyte, and Decapentaplegic (DPP), a BMP2/4-type ligand secreted by the follicle cells stretched over the nurse cells, reviewed in (Dobens and Raftery, 2000; Berg, 2005). GRK and DPP control the expression of multiple genes in the follicular epithelium. Under their action, the expression of a Zn-finger transcription factor Broad (BR) evolves into a pattern with two patches on either side of the dorsal midline (Deng and Bownes, 1997; Yakoby et al., 2008). The BR-expressing cells form the roof (upper part) of the dorsal appendages (James and Berg, 2003; Dorman et al., 2004; Ward and Berg, 2005). Adjacent to the BR-expressing cells are two stripes of cells that express rhomboid (rho), a gene that is directly repressed by BR and encodes ligand-processing protease in the EGFR pathway (Ruohola-Baker et al., 1993; Sapir et al., 1998; Lee et al., 2001; Ward and Berg, 2005). These cells form the floor (lower part) of the appendages (James and Berg, 2003; Dorman et al., 2004; Berg, 2005).
Figure 1. Patterning of the follicle cells and dorsal appendage morphogenesis.
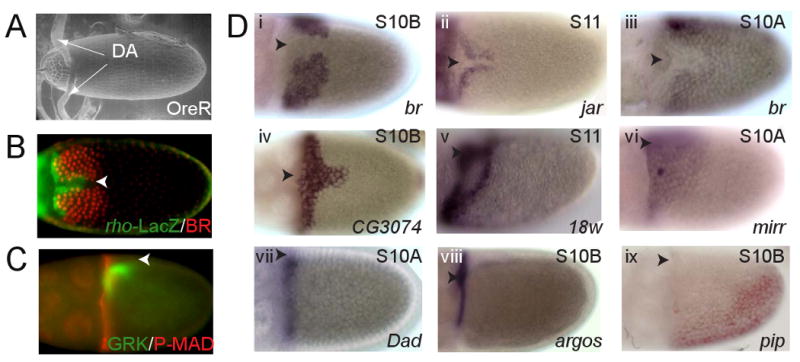
(A) ESEM image of the Drosophila eggshell (dorsal view, anterior to the left). The dorsal appendages (DA) are tubular structures located on the dorsal side of the eggshell.
(B) Dorsal view of a stage 12 egg chamber. Each of the appendages is formed by the two adjacent groups of cells within the follicular epithelium. The cells expressing a Zn-finger transcription factor Broad (BR, red) form the roof of the future appendage. The floor of the appendage is formed by the cells that express rho (green), a protease in the Drosophila EGFR pathway.
(C) Lateral view of a stage 10B egg chamber. Patterning of the dorsal eggshell structures depends on the localized activation of the EGFR and DPP pathways in the follicular epithelium. The activation of the EGFR pathway is initiated by GRK (green). The early pattern of EGFR activation is distributed as a broad dorsoventral gradient. The DPP pathway is activated by DPP, a BMP2/4-type ligand secreted by the stretch cells and at the anterior border of the follicle cells associated with the oocyte. This generates an anterior-posterior pattern of MAD phosphorylation (P-MAD, red).
(D) Examples of gene expression patterns in the follicular epithelium during the stages of oogenesis corresponding to the dorsoventral patterning of the eggshell. Gene expression is visualized by whole-mount in situ hybridization. Images i-iv show dorsal views, vi-ix show lateral views.
The patterns of genes expressed during the stages of oogenesis that correspond to the formation of dorsal eggshell structures are very diverse (Figure 1D). At the same time, inspection of a large number of published patterns suggests that they can be “constructed” from a small number of building blocks. For instance, the T-shaped pattern of CG3074 is similar to the domain “missing” in the early pattern of br (Figure 1D,iv,iii), while the two patches in the late pattern of br appear to correspond to the two “holes” in the expression of 18w (Figure 1D,i,v). Based on a number of similar observations, we hypothesized that all of the published patterns could be constructed from just six basic shapes, or primitives, which reflect the anatomy of the egg chamber and the spatial structure of the patterning signals (Figure 2).
Figure 2. Building blocks and spatial operations in the proposed combinatorial code.
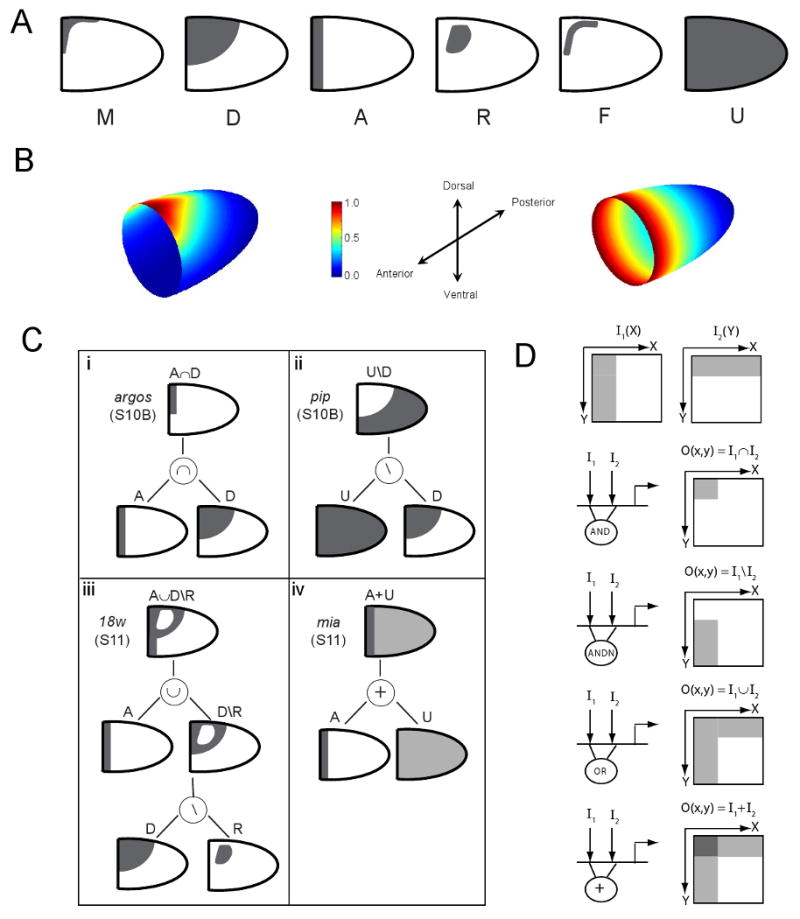
(A) Lateral views of the six geometric building blocks (primitives) used to describe two-dimensional gene expression in the follicular epithelium.
(B) The first three building blocks are connected to the spatial structure of the patterning inputs. The spatial pattern of GRK protein in the oocyte (not shown), which is a proxy for the spatial pattern of ligand secretion, has a concave boundary and potentially explains the origin of the M primitive. The D primitive originates from the convex level-sets of the spatial distribution of secreted GRK in the lateral region of the egg chamber, computed using a biophysical model of GRK secretion, diffusion, binding, and internalization. The A primitive reflects the AP gradient of DPP sinaling, computed using a biophysical model of DPP secretion, diffusion, binding and internalization. (C) Complex patterns are constructed from primitives and the operations of (i) intersection (∩), (ii) difference (\), (iii) union (∪), and (iv) addition (+). The four examples show the construction of the patterns for argos, pip, 18w, and mia.
(D) Schematic showing how the four spatial operations could be realized by the Boolean and arithmetic operations at the regulatory regions of target genes (see text for details). The diagram shows the two hypothetical spatial inputs that are locally processed by the AND, ANDN (AND NOT), OR, and addition operations. I/O refer to input/output, respectively.
In computer graphics, representation of geometrical objects in terms of a small number of building blocks is known under the name of Constructive Solid Geometry (CSG), which provides a way do describe complex shapes in terms of just a few parameters – the types of the building blocks, such as cylinders, spheres, and cubes, their sizes, and operations, such as difference, union, and intersection (Requicha and Voelcker, 1982; Foley et al., 1992). Thus, information about a large number of structures can be stored in a compact form of statements that contain information about the types of the building blocks and the operations from which these structures were assembled. Here, we describe a similar approach for two-dimensional patterns and demonstrate how it enables the synthesis, comparison, and analysis of gene expression at the tissue scale.
Results
Building blocks for two-dimensional patterns
The six building blocks used in our annotation system can be related to the structure of the egg chamber and the spatial distribution of the EGFR and DPP signals (Figure 2A,B). The first primitive, M (for “midline”), is related to the EGFR signal. It reflects high levels of EGFR activation and has a concave boundary, which can be related to the spatial pattern of GRK secretion from the oocyte (Peri et al., 1999; Queenan et al., 1999; Nakamura and Matsuno, 2003). The second primitive, denoted by D (for “dorsal”), reflects the intermediate levels of EGFR signaling during the early phase of EGFR activation by GRK, and is defined as a region of the follicular epithelium that is bounded by a level set (line of constant value) of the DV profile of EGFR activation. The boundary of this shape is convex and can be extracted from the experimentally validated computational model of the GRK gradient (Goentoro et al., 2006a; Chang et al., 2008). The third primitive, denoted by A (for “anterior”), is an anterior stripe which is obtained from a level set of the early pattern of DPP signaling in the follicular epithelium (Lembong et al., 2008). This pattern is uniform along the DV axis, as visualized by the spatial pattern of phosphorylated MAD (P-MAD) (Jekely and Rorth, 2003; Shravage et al., 2007). Thus, the D, M, and A primitives represent the spatial distribution of the inductive signals at the stage of eggshell patterning when the EGFR and DPP pathways act as independent AP and DV gradients.
Each of the next two primitives, denoted R (for “roof”) and F (for “floor”), is composed of two identical regions, shaped as the respective expression domains of br and rho (Figure 1B, (Ruohola-Baker et al., 1993; Deng and Bownes, 1997)), and reflect spatial and temporal integration of the EGFR and DPP pathways in later stages of eggshell patterning (Peri et al., 1999; Astigarraga et al., 2007; Yakoby et al., 2008). The mechanisms responsible for the emergence of the F and R domains are not fully understood. It was shown that the R domain is established as a result of sequential action of the feedforward and feedback loops within the EGFR and DPP pathways (Yakoby et al., 2008). The formation of the F domain requires the activating EGFR signal and repressive BR signal, expressed in the R domain (Peri et al., 1999; Ward and Berg, 2005; Ward et al., 2006). Thus, at the current level of understanding, the R and F domains should be viewed as just two of the shapes that are commonly seen in the two-dimensional expression patterns in the follicular epithelium (Figure 1D). The sixth primitive, U (for “uniform”), is spatially uniform and will be used in combination with other primitives to generate more complex patterns.
Combinatorial construction of complex patterns
While a number of patterns, such as those of jar and Dad (Figure 1D,ii,vii), can be described with just a single primitive, more complex patterns are constructed combinatorially, using the operations of intersection (∩), difference (\), and union (∪). E.g., the dorsal anterior stripe of argos expression (Figure 1D,viii) is obtained as an intersection of the A and D primitives (A∩D, Figure 2C,i). The ventral pattern of pip (Figure 1D,ix) is obtained as a difference of the U and D primitives (U\D, Figure 2C,ii). The pattern of 18w (Figure 1D,v) is constructed from the A, D, and R primitives, joined by the operations of union and difference (A∪D\R, Figure 2C,iii). For a small number of published patterns, our annotations reflect the experimentally demonstrated regulatory connections. E.g., the U\D annotation for pip, reflects that actual repression of pip by the dorsal gradient of EGFR activation (Pai et al., 2000; James et al., 2002; Peri et al., 2002). For a majority of genes, our annotations should be viewed as a way to schematically represent a two-dimensional pattern and as a hypothetical description of regulation.
The geometric operations of intersection, difference, and union can be implemented by the Boolean operations performed at the regulatory regions of individual genes (Davidson, 2005), (Figure 2D). Boolean operations evaluate expression at each point and assign a value of zero (off) or one (on). As an example, consider a regulatory module, hypothesized for argos (Figure 2Ci), that performs a logical AND operation on two inputs: the output of the module is one only when both inputs are present. When both of the inputs are spatially distributed, the output is nonzero only in those regions of space where both inputs are present, leading to an output that corresponds to the intersection of the two inputs. Similarly, a spatial difference of the two inputs can be realized by a regulatory module that performs the ANDN (ANDNOT) operation. This is the case for pip, repressed by the DV gradient of Gurken signaling and activated by a still unknown uniform signal (Figure 2Cii)(Sen et al., 1998; Pai et al., 2000; James et al., 2002). Finally, a regulatory module that performs an OR operation is non-zero when at least one of the inputs is nonzero. When the inputs are spatially distributed, the output is their spatial union (illustrated for 18w, Figure 2C,iii).
Boolean operations on primitives lead to patterns with just two levels of expression (the gene is either expressed or not). In addition to Boolean logic, developmental cis-regulatory modules and systems for postranscriptional control of gene expression can perform analog operations, leading to multiple nonzero levels of output (Yuh et al., 1998; Davidson, 2001; Buchler et al., 2003; Longabaugh et al., 2005; Istrail et al., 2007; Cory and Perkins, 2008). Consider a module that adds the two binary inputs, shaped as the primitives (Figure 2D). The output is nonzero in the domain shaped as the union of the two primitives, but is characterized by two nonzero levels of expression. We reserve this type of annotation only for those cases where the application of Boolean operations would lead to a loss of the spatial structure of the pattern (such as the A+U expression pattern of mia at stage 11 of oogenesis, Figure 2C,iv). For example, the union of the A and U primitives is a U primitive, whereas the sum of these primitives is an anterior band superimposed on top of a spatially uniform background (Figures 2C,iv and 3E,iv).
Figure 3. Large-scale test of the annotation and analysis of the spatial diversity of gene expression patterns in the follicular epithelium.
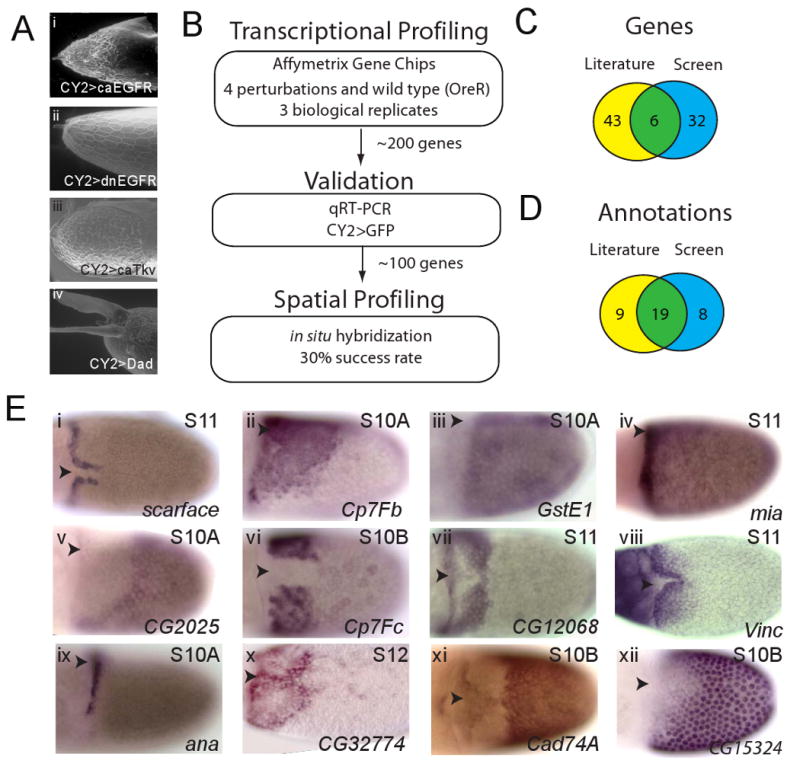
(A) ESEM images of eggshell morphologies induced by the perturbations of EGFR and DPP signaling in the follicular epithelium used in the microarray screen: (i) uniform EGFR activation (CY2>caEGFR), (ii) uniform EGFR downregulation (CY2>dnEGFR), (iii) uniform DPP signaling (CY2>caTkv), (iv) uniform repression of DPP signaling (CY2>Dad).
(B) Summary of the transcriptional profiling approach (see text for details). The screen identified 91 new patterns, corresponding to 32 genes expressed in stages 10A through 12.
(C) Venn diagram comparing the sets of genes described in the literature and identified in the transcriptional profiling screen.
(D) Venn diagram comparing the sets of the annotations that were described in the literature and identified in the screen.
(E) In situ hybridization images showing twelve examples of new gene expression patterns identified in a microarray-based screen (see text for details). The arrows mark the location of the dorsal midline.
Testing the combinatorial code
As a first test of our combinatorial code, we used it to describe the two-dimensional patterns of the 49 genes previously shown to be expressed between stages 10A and 12 of oogenesis. The results of the annotion of this group of genes, that have been studied for approximately two decades, are shown in Table 1 (see also Table S1 for references to the original publications). For ∼50% of these genes we relied on published in situ hybridization images. For the other genes (26/49), we have either verified, corrected the published patterns, or completed the temporal profile in our own in situ hybridization experiments. This expanded set of 118 gene expression patterns, all of which could be successfully annotated using our system of six primitives and four operations. The entire dataset required only 28 logical statements (Table 1). This analysis of the previously published and verified data supports the feasibility of our approach.
Table.
| Flybase ID | Symbol | Stage 10A | Stage 10B | Stage 11 | Stage 12 | |
|---|---|---|---|---|---|---|
| 1 | FBgn0004364 | 18w | A+U | A∪(D\R) | A∪(D\R) | A∪M |
| 2 | FBgn0004569 | argos | A∩D | M | M | |
| 3 | FBgn0023407 | B4 | A | |||
| 4 | FBgn0001090 | bnb | A∩D | |||
| 5 | FBgn0000210 | br | U\M | (A\D)∪R | ||
| 6 | FBgn0024250 | brk | U\A | U\A | ||
| 7 | FBgn0031150 | bves | U\D | U\D | ||
| 8 | FBgn0039709 | Cad99C | U | A | A∪(D\R) | A\D |
| 9 | FBgn0041342 | Cct1 | A | A | R | |
| 10 | FBgn0034709 | CG3074 | A | A∪M | F | F |
| 11 | FBgn0038469 | CG4009 | D\M | D\M | D+U | |
| 12 | FBgn0033631 | CG9027 | A∩D | |||
| 13 | FBgn0032126 | CG13113 | F | |||
| 14 | FBgn0032120 | CG33298 | D | D | A | |
| 15 | FBgn0000359 | Cp36 | A+D | A+D | ||
| 16 | FBgn0020493 | Dad | A | A | ||
| 17 | FBgn0000490 | dpp | A | A | A | A |
| 18 | FBgn0000568 | Eip75B | D | |||
| 19 | FBgn0004865 | Eip78C | D | |||
| 20 | FBgn0000575 | emc | A | A | A∪R | A∪R |
| 21 | FBgn0010470 | Fkbp13 | A+U | (A∪D)+U | (A∪D)+U | (A∪D)+U |
| 22 | FBgn0011591 | fng | U\D | U\D | ||
| 23 | FBgn0001987 | Gli | F | |||
| 24 | FBgn0001257 | ImpL2 | A | M | F∪R | F |
| 25 | FBgn0011225 | jar | U | A+U | F | F∪R |
| 26 | FBgn0027339 | jim-2 | U\(A∪D) | U\D | ||
| 27 | FBgn0001297 | kay-RB | U | F | A\D | |
| 28 | FBgn0015399 | kek1 | M | M | F | |
| 29 | FBgn0020278 | loco-c2 | D | D | D | |
| 30 | FBgn0024211 | mfas | U\D | |||
| 31 | FBgn0014343 | mirr | A∪D | A∪R | F | |
| 32 | FBgn0002778 | mnd | A∪(U\D) | |||
| 33 | FBgn0003089 | pip-RA | A∪(U\D) | U\D | ||
| 34 | FBgn0003118 | pnt-P1 | A∪M | A∪M | F | |
| 35 | FBgn0003118 | pnt-P2 | M | R | R | |
| 36 | FBgn0243512 | puc | A | A∪D | ||
| 37 | FBgn0004635 | rho | D\M | F | F | |
| 38 | FBgn0010851 | sgl | F | F | M | |
| 39 | FBgn0003388 | shd | A | |||
| 40 | FBgn0005638 | slbo | A | A | ||
| 41 | FBgn0023423 | slmb | F | R | R | |
| 42 | FBgn0085450 | Snoo | (A∩D)∪(D\D) | D\D | ||
| 43 | FBgn0003463 | sog | A∩D | A∩D | ||
| 44 | FBgn0014388 | sty | M | A∪M | (A\D)∪F | |
| 45 | FBgn0003716 | tkv | (A\D)∪R | (A\D)∪R | ||
| 46 | FBgn0014076 | Vm32E | U\(A∪D) | U\A | ||
| 47 | FBgn0003984 | vn | F | |||
| 48 | FBgn0041709 | yellow-g | D\R | |||
| 49 | FBgn0035328 | yellow-g2 | A\D | M∪(U\D) | M∪(U\D) | |
| 50 | FBgn0011746 | ana | A∩D | A∩D | ||
| 51 | FBgn0036715 | Cad74A | U\R | U\R | U\R | |
| 52 | FBgn0039905 | CG2052 | U\D | U\D | ||
| 53 | FBgn0036612 | CG4998 | A | A∪(D\(M∪R)) | A∪(D\(M∪R)) | A∪(D\(M∪R)) |
| 54 | FBgn0029568 | CG11381 | M∪(U\D) | M∪(U\D) | M∪(U\D) | M∪(U\D) |
| 55 | FBgn0039637 | CG11880 | A | A | A∪D | |
| 56 | FBgn0039695 | CG12068 | U | (A∪D)\M | A∪(D\F) | (A∪D)\M |
| 57 | FBgn0029966 | CG15324 | U\M | U | U | |
| 58 | FBgn0051522 | CG31522 | U | A+U | A | A |
| 59 | FBgn0051900 | CG31900 | A∪(U\D) | A∪(U\D) | ||
| 60 | FBgn0052774 | CG32774 | D\R | D\R | ||
| 61 | FBgn0053099 | CG33099 | U | (A\D)∪(U\A) | ||
| 62 | FBgn0085446 | CG34417 | U\M | (U\M)+R | (U\M)+R | R |
| 63 | FBgn0000355 | Cp15 | U | A+U | U | U |
| 64 | FBgn0000360 | Cp38 | D\M | (A+U)\M | U | |
| 65 | FBgn0014465 | Cp7Fb | A∪D | A∪R | U\F | U\R |
| 66 | FBgn0014466 | Cp7Fc | (A∪D)\M | (A+U)\M | (A+U)\M | |
| 67 | FBgn0011577 | dally | A+U | A+U | A\D | |
| 68 | FBgn0004638 | drk/Grb2 | U | U | M | M |
| 69 | FBgn0034335 | GstE1 | U\A | U\M | R | F |
| 70 | FBgn0001230 | Hsp68 | A | A+U | D\R | |
| 71 | FBgn0023001 | melt | D | D\M | ||
| 72 | FBgn0014342 | mia | A∩D | A+U | A | |
| 73 | FBgn0011754 | PhKγ | A | A\D | ||
| 74 | FBgn0003205 | Ras85D | D | F | F | F |
| 75 | FBgn0033033 | scarface | F | F | F | |
| 76 | FBgn0015296 | Shc | U | F | F | F |
| 77 | FBgn0003507 | srp | D\M | D | ||
| 78 | FBgn0003865 | tsg | A∩D | |||
| 79 | FBgn0004397 | Vinc | F | F∪R | F∪R | |
| 80 | FBgn0016075 | vkg | A | A | D | |
| 81 | FBgn0024179 | wit | A | A\D |
As a more rigorous test of the proposed code, we applied it to a large set of newly identified genes and patterns. We reasoned that new eggshell patterning genes could be discovered by screening for targets of the EGFR and DPP signals. Based on this, we used the GAL4/UAS system to perturb the EGFR and DPP signals at the stages 9-10 of oogenesis in a manner that induced clear perturbations of the dorsal eggshell structures (Figure 3A) (Brand and Perrimon, 1993; Twombly et al., 1996; Queenan et al., 1997; Yakoby et al., 2008). Specifically, ectopic activation of EGFR signaling abolishes the dorsoventral polarity of the eggshell, completely eliminates the dorsal appendages, and generates an operculum-like material at the anterior of the eggshell (Figure 3A,i). Uniform inhibition of EGFR signaling in the follicle cells abolishes the dorsal eggshell structures (Figure 3A,ii). Uniform activation of DPP signaling also leads to the loss of dorsal appendages and greatly expands the operculum (Figure 3A,iii). Finally, uniform expression of the intracellular inhibitor of DPP signaling leads to eggshells with a smaller operculum and deformed appendages (Figure 3A,iv).
At the next step, we used Affymetrix Gene Chip microarrays to identify ∼100 genes that changed in abundance in stage 9-10 egg chambers, when compared to the wild type (Figure 3B). The details of transcriptional profiling experiments, their statistical analysis, and validation are described in the Supplemental Figures and Text. Briefly, we selected ∼200 genes that responded to perturbations in both the EGFR and DPP pathways to identify potential targets of the EGFR and DPP signal integration. We then used a large scale qRT-PCR transcriptional profiling approach to validate all of these targets and eliminated ones that were likely induced due to stress, reducing the number of the candidate genes to ∼100. Using in situ hybridization, we found that ∼1/3 of these genes are expressed in the wild type ovary during the stages of oogenesis relevant for eggshell patterning (Figure 3C, Table 1). Some of the identified genes were known to be expressed before, while others are new (Figure 3C-E). In addition to identifying new genes, we identified a number of novel spatial patterns, e.g. the mask-like pattern of the putative cell adhesion gene Cad74A (Figure 3C,ix) (Zartman et al., 2008). Importantly, the proposed code describes all identified patterns, demonstrating that it provides an adequate language for gene expression in the follicular epithelium.
Diversity of spatial patterns
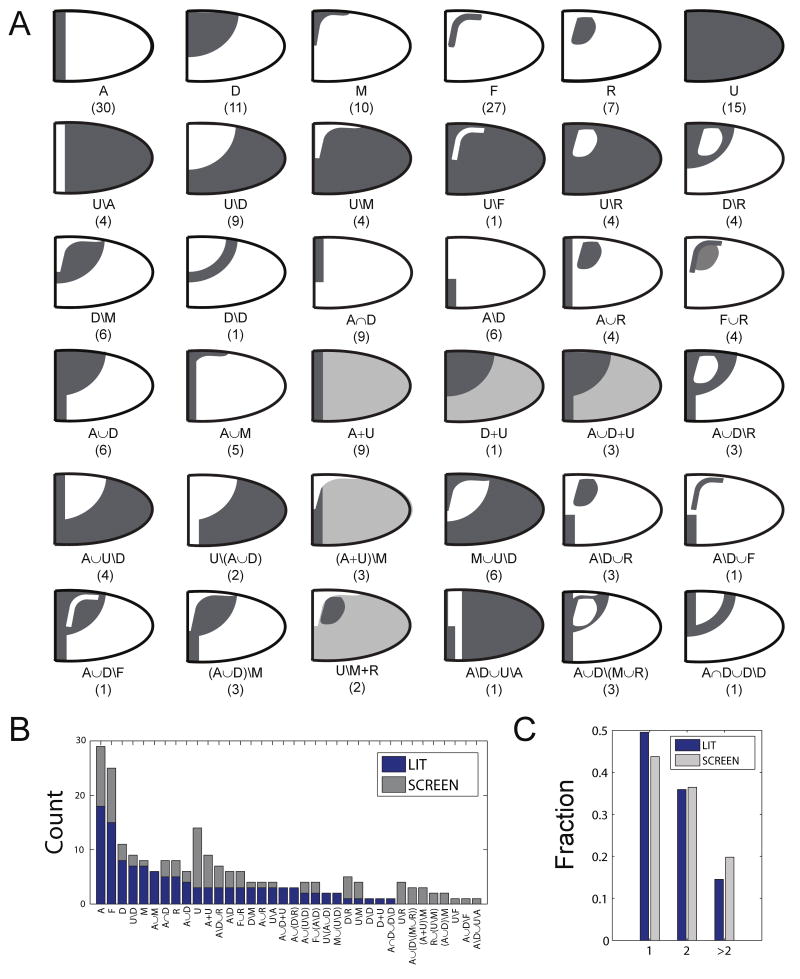
Our experiments have essentially doubled the number of reported expression patterns in the follicle cells. In combination with the published data, we have collected 211 two-dimensional spatial patterns for 81 genes at four consecutive stages of oogenesis (stages 10A, 10B, 11, and 12). This set of data forms the basis for our analysis of the diversity of the spatial gene expression patterns in the follicular epithelium. We assigned each of the 211 images in our database to 36 distinct patterns (Figure 4A-C). Out of these, six are the primitives themselves, 16 are binary terms (e.g, A∩D for argos at stage 10A), while the remaining 14 are built from three or four terms (e.g., 18w at stage 10B). Approximately two thirds of the patterns can be described with just four primitives that are directly related to the early spatial patterns of the EGFR and DPP pathway activation (D, M, A, and U). The number of distinct patterns is low when compared to the number of hypothetical patterns that can be generated within the framework of our combinatorial system. For example, if we consider only patterns that potentially could be built from two primitives and three operations, we can generate 6*6*3=108 patterns, compared to the 16 observed patterns described by only 2 primitives. This is a consequence of the fact that the primitives in our system are not completely independent and are downstream of a smaller number (most likely, just two) inductive signals.
Figure 4. Diversity of transcriptional patterns in stages 10A-12 of oogenesis.
(A) Qualitatively different classes of two-dimensional patterns based on the annotation of 211 expression patterns from the literature and our own experiments. The numbers indicate the number of times a given pattern is observed in the database.
(B) Counts for the number of genes in a given pattern annotation in the published data (LIT) and our experiments (SCREEN).
(C) Distribution of published (LIT) and new (SCREEN) patterns by the number of primitives required to describe them. ∼80% are described with one or two primitives.
Our annotation describes the shape of the boundary of the spatial pattern, but not the precise location of this boundary or the quantitative expression level within the domain. For example, the ventral patterns of bves, jim, fng, and pip, are all annotated with the same expression (U\D), even though the expression domains of these genes overlap only partially (Sen et al., 1998; Doerflinger et al., 1999; Jordan et al., 2000; Lin et al., 2007). Qualitative similiarity of spatial patterns for a group of genes, which corresponds to the exact match of our annotations, can be used as a proxy for a similar gene regulatory strategy. Analyzing the statistics of such events (see Experimental Procedures), we found that an exact match of the statements used to describe the real patterns, is a rare event. For example, the probability that any three genes, selected at random from our database, are co-expressed in the same pattern in at least one stage of oogenesis, is <1%.
Sharing the same annotation is not only rare, but also a transient event (Figure 5A). We found that most of the groups of genes with the same annotation at any given point either converge to the similar pattern from diverse patterns in the past, or diverge from a common annotation to different patterns at later stages. For example, given that a pair of genes is expressed in the same pattern at some point of oogenesis, the probability that this pair will still be co-expressed at some other time point is estimated to be only ∼14%. The numbers are much lower for triplets and quadruplets of co-expressed genes (∼4% and 1%, respectively; see Experimental Procedures for details). In other words, the fact that any group of genes is expressed in a similar pattern is not predictive of common expression patterns at other time points. Thus, patterns are dynamically reassembled from a small number of building blocks at every stage of oogenesis.
Figure 5. Dynamics of two-dimensional gene expression patterns in the follicle cells.
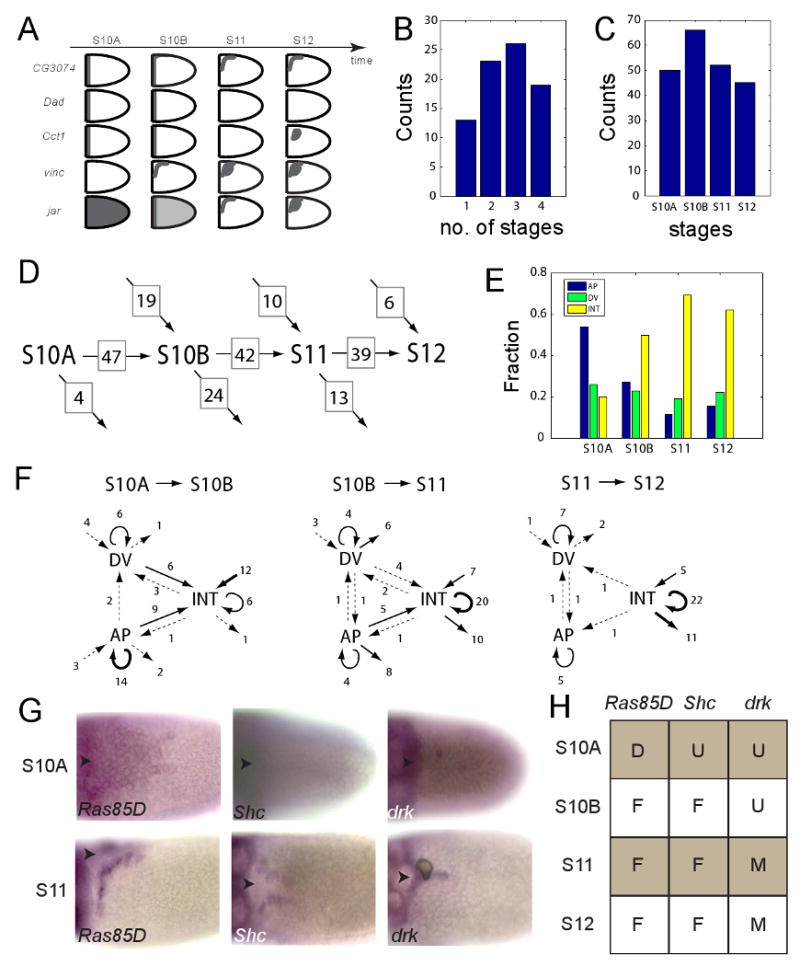
(A) Convergence and divergence of co-expression in annotated patterns at different stages. For example, the patterns of CG3074, Dad, and Cct1 coincide at stage 10A, but separate at later stages of oogenesis. On the other hand, vinc and jar, which are differentially expressed at stage 10A, are co-expressed during stage 12. Genes expressed in primitive patterns can transition both into other primitives and into composite patterns (described by 2 or more primitives).
(B) Most of the genes are expressed in more than one stage of oogenesis. 15 genes are expressed only once between stages 10A-12, 22 genes are expressed twice, 24 genes are expressed in three time points, and 20 genes are expressed in all four stages.
(C) Numbers of genes expressed in each of the four analyzed stages of oogenesis.
(D) Dynamic flow chart for the numbers of genes expressed during different stages of oogenesis, including the number of genes appearing at or dissappearing after a given stage (arrows pointing toward or away from a given stage) and the number of genes that continue to the next stage (arrows pointing toward the next stage).
(E) Dynamics of the fractions of the patterns assigned AP, DV, and INT classes, see text for details.
(F) Network-like representation of the AP, DV, and INT classes of genes at four consecutive stages of oogenesis. The number of genes that transition between the classes are indicated. Arrow thickness highlights the frequency of a given transition (thick: ≥10 genes, medium: 4-9 genes, dashed: 1-3 genes).
(G) Early and late expression stages for Ras85D, Shc, and drk (the arrowheads mark the location of the dorsal midline).
(H) Summary of expression patterns Ras85D, Shc, and drk at four consecutive stages of oogenesis.
Dynamics of pattern formation
Based on our database, each stage is characterized by the expression of ∼50 genes, most of which are expressed in multiple stages of oogenesis (Figures 5A-C, Tables S2,3). The sets of genes expressed at two consecutive stages show a considerable overlap, and the differences between the two sets constitute the genes that either stop being expressed or are expressed de novo (Figure 5D). Most of the genes are expressed in more than one stage of oogenesis and their expression patterns are very dynamic (Figure 5C). In general, the probability that a gene, which is expressed in multiple stages of oogenesis, will be expressed in different patterns is 74±10%.
To characterize dynamics at the scale of dozens of genes, we grouped their expression patterns into three classes. The first class, called AP, is composed of four patterns constructed from A and U primitives, is related to the AP patterning signals alone, and likely reflects regulation by the DPP pathway (A, U, U\A, and A+U). The second class, called DV, is composed of eight patterns that could be assigned to the DV patterning signal alone. This class is composed of patterns constructed from the D, M, and U primitives and their combinations (D, M, U\D, D\D, D\M, D+U, U\M, and M∪(U\D)). Thus, the AP and DV classes can be viewed as simple responses to the EGFR and DPP signals during the early stages of eggshell patterning. The remaining 24 annotations form the third class, called INT (for ‘integration’). This class is constructed from combinations of purely AP and purely DV patterns and/or the R and F primitives.
Next, we analyzed how the sets of genes that belong to each of these classes change over four consequtive time points (Figure 5E). Purely AP or DV patterns dominate the expression patterns in stage 10A egg chambers. Over time, however, most of the expressed genes appear as complex patterns, as can be expected based on the change in the spatial distribution of the EGFR and BMP signaling (Wasserman and Freeman, 1998; Peri et al., 1999; Yakoby et al., 2008). Complex patterns (members of the INT class) appear in two different ways. First, a gene can evolve into a more complex pattern from a simpler one. The change of the rho pattern, from D\M to F annotation, is one example (Peri et al., 1999). Second, a gene can first appear as a complex pattern, which is the case for the F pattern of Vinc. We found that newly appearing genes (after stage 10) are more likely to belong to the complex (INT) class. The set memberships of the simple, AP and DV, classes are more transient than that of the INT class, which tends to be more maintained over time (Figure 5F), potentially reflecting the reinforcing action of the feedback loops in the EGFR and DPP systems.
A qualitative change in the spatial expression pattern of a gene can reflect the use of a different regulatory region and/or the dynamics of the inductive signal. In the follicle cells, the dynamics of patterns and convergence into the INT category likely reflects the reinforcing action of the feedforward and feedback loops that split the spatial profiles of the EGFR and DPP signaling along the dorsal midline (Peri et al., 1999; Yakoby et al., 2008). For the EGFR system, this was attributed to the action of the midline-expressed EGFR inhibitor and a switch in the activating ligand: from the oocyte-derived GRK to SPI, secreted by the rho-expressing follicle cells. Thus, in both the early and late stages of oogenesis, EGFR activation is thought to be generated by a locally produced ligand that acts through a uniformly expressed receptor.
Interestingly, one of the genes identified in our experiments is Ras85D, which is essential for signal transmission from activated EGFR. During the patterning of the follicular epithelium, Ras is expressed in a dynamic pattern, which can be described as the U→ →F→F→F, sequence (stages 9 through 12, respectively), indicating that it is regulated by EGFR signaling (Figure 5G). This suggests a new layer of regulation, which depends on the ability of the follicle cells to transduce signals downstream of EGFR. At least two other components of the EGFR pathway, Shc and drk/Grb2, are expressed in dynamic patterns (Figure 5G,H). One function of this highly coordinated patterning of multiple pathway components is to localize EGFR signaling to the F domain and to prevent the spreading of a traveling wave of the EGFR activation across the entire follicular epithelium (Pribyl et al., 2003).
Pattern annotations and connections to inductive signals
In addition to enabling the analysis of patterns at the tissue scale, our annotations guide mechanistic studies of individual genes and gene groups. As an example, the A∩D annotation for the stage 10A expression of ana, a secreted glycoprotein identified in our transcriptional profiling experiments (Figure 3E,ix; 6A), suggests that it is generated by a local AND gate that responds to the anterior DPP and dorsoventral GRK gradients. Thus, removal of either of these signals should erase the pattern. In agreement with this prediction, the pattern of ana is abolished in response to uniform inhibition of DPP signaling (Figure 6A,iv) and uniform inhibition of the EGFR signaling (not shown). Thus, experiments with complete inhibition of the EGFR and DPP inputs support the model whereby the A∩D pattern is established by a locally acting AND gate. At the same time, partial reduction of either of the inputs is predicted to reduce the size of the stripe of the A∩D pattern. This prediction is supported by the fact that the size of the ana pattern is reduced in flies homozygous for the weak allele of Ras (Figure 6A,iii)(Schnorr et al., 2001).
Figure 6. Pattern annotations and connection to inductive signals.
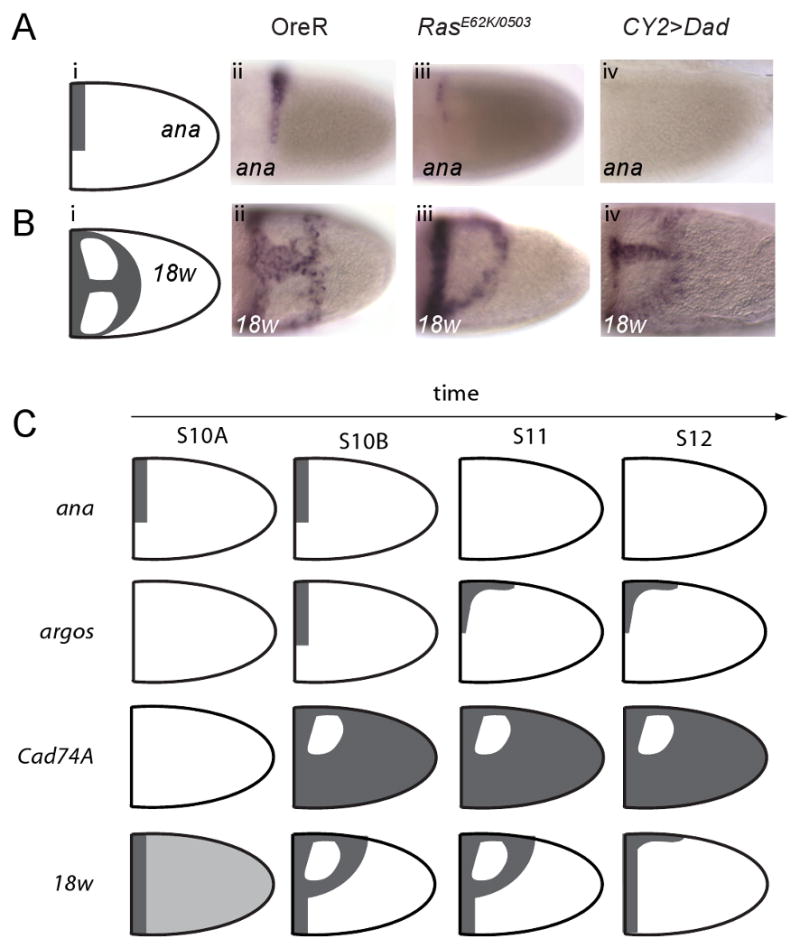
(A) Schematic representation (i) and in situ hybridization image (ii) of ana expression at S10A, lateral view. Changes in the ana pattern in flies with reduced level of EGFR signaling (iii) and DPP signaling (iv). The pattern is greatly reduced in flies with the weak allele of Ras (iii) and is completely obliterated in flies overexpressing Dad, an inhibitor of DPP signaling (iv).
(B) Schematic representation (i) and in situ hybridization image (ii) of 18w expression at S10A, dorsal view. Changes in the 18w pattern in flies with reduced level of EGFR signaling (iii) and DPP signaling (iv). The midline part of the pattern is missing in flies with the weak allele of Ras (iii). The anterior part of the pattern is greatly reduced in flies overexpressing Dad (iv). The observed changes are highly correlated with the corresponding changes in the pattern of BR, expressed in the R domain.
(C) Summary of the expression dynamics of ana, argos, Cad74A, and 18w at four consecutive stages of oogenesis.
We predict that these changes in the spatial pattern of ana will be also observed for other genes with the A∩D annotation. One of these genes is argos, a negative feedback inhibitor induced by high levels of EGFR signaling (Figures 1D,viii;2C,I;6C) (Wasserman and Freeman, 1998; Mantrova et al., 1999). The AND-gate model for argos is supported by the fact that the uniform activation of EGFR generates ectopic argos expression only in the anterior part of the follicular epithelium, suggesting a necessary role of the anterior DPP signal (Queenan et al., 1997).
Another example of the connection between spatial patterns, as described by our annotations, and regulatory signals is provided by the A∪D\R pattern of a cell adhesion molecule 18w (Figure 2B,i,ii). The D\R part of the annotation suggests that 18w is repressed by BR, a transcription factor expressed in the R domain. Thus, in flies with reduced level of EGFR signaling (Schnorr et al., 2001), the change in the BR domain from the two-domain R pattern to a single domain pattern should lead to the midline repression of 18w. This is exactly what is observed experimentally: the wild type A∪D\R pattern is converted into a A∪D\D pattern (Figure 6B,iii). Similarly, uniform inhibition of DPP signaling, which generates ectopic BR in the anterior cells, leads to the loss of 18w expression from a part of the A domain, as predicted by its wild type annotation (Figure 6B,iv). We observed similar transitions in the spatial pattern of Cad74A, a cell adhesion molecule with the U\R annotation (Zartman et al., 2008). In this case, BR is sufficient for repressing Cad74A in the R domain, illustrating the predictive power of our annotations.
In a clear demonstration of a general trend revealed by the analysis of pattern dynamics, the highly correlated initial patterns of ana and argos and those of 18w and Cad74A, follow different dynamics (Figure 6C). At stage 11 ana expression disappears, whereas argos evolves into a midline-type pattern. These differences might reflect differences in the quantitative parameters of the locally acting regulatory modules. One possibility is that the AND gates, hypothesized to establish the wild type A∩D patterns, are activated at different levels of the EGFR and DPP signals and respond in qualitatively different ways to changes in these signals at later stages of oogenesis.
Discussion
A combinatorial code for two-dimensional patterns
Signaling pathways guide organogenesis through the spatial and temporal control of gene expression. While the identities of genes controlled by any given signal can be identified using a combination of genetic and transcriptional profiling techniques, systematic analysis of the diversity of induced patterns requires a formal approach for pattern quantification, categorization and comparison (Leptin, 2005; Dequéant et al., 2006; Brady et al., 2007). Such approaches are only beginning to be developed (Kumar et al., 2002; Megason and Fraser, 2007; Fisher et al., 2008; Fowlkes et al., 2008). Multiplex detection of gene expression, which has a potential to convert images of the spatial distribution of transcripts into a vector format preferred by a majority of statistical methods, is currently feasible only for a small number of genes and systems with simple anatomies (Kosman et al., 2004; Luengo Hendriks et al., 2007). We presented an alternative approach, based on the combinatorial construction of patterns from simple building blocks.
In general, the building blocks can be identified as shapes that are overrepresented in a large set of experimentally collected gene expression patterns. This approach can be potentially pursued in systems where mechanisms of pattern formation are yet to be explored. At the same time, in well-studied systems, the building blocks can be linked to identified patterning mechanisms. We chose six primitives, based on the features that are commonly observed in real patterns and related to the structure of the tissue as well as the spatial distribution of the inductive signals. A similar approach will be useful whenever a two-dimensional cellular layer is patterned by a small number of signals, when cells can convert smoothly varying signals into spatial patterns with sharp boundaries, and when the regulatory regions of target genes have the ability to combinatorially process the inductive signals. One system where this approach could be feasible is the wing imaginal disk which is patterned by the spatially orthogonal WG and DPP morphogens (Cadigan, 2002; Butler et al., 2003; Jacobsen et al., 2006).
We have shown that six primitives are sufficient to describe the experimentally observed patterns during stages 10-12 ooogenesis. A natural question is whether it is possible to accomplish this with a smaller number of primitives. Two of our primitives, R and F, could be potentially constructed from the D, M, and A primitives, which are related to the patterns EGFR and DPP activation during the earlier stages of eggshell patterning. Specifically, recent studies of br regulation suggest that the R domain is formed as a difference of the D, A, and M patterns (Yakoby et al., 2008). Furthermore, the formation of the F domain requires repressive action in the adjacent R domain (Ward et al., 2006). With the R and F domains related to the other four primitives, the size of our spatial alphabet will be reduced even further (from 6 to 4), but at the expense of increasing the complexity of the expressions used to describe various spatial patterns.
Systems biology of epithelial patterning
Previously, the question of the diversity of the spatial patterns has been adressed only in one-dimensional systems. For example, transcriptional responses to the Dorsal morphogen gradient in the early Drosophila embryo, give rise to three-types of patterns in the form of the dorsal, lateral, and ventral bands (Markstein et al., 2004). Our work provides the first attempt to characterize the diversity and dynamics of two-dimensional patterns. We identified 36 qualitatively different patterns and proposed that each of them can be constructed using a compact combinatorial code. The sizes of the datasets from the literature, and our own transcriptional profiling experiments are approximately the same (118 and 93 patterns, respectively; Figure 3B,C). Based on this, we expect that newly discovered patterns will be readily described using our annotation system.
We found that a gene expressed in more than one stage of oogenesis, is more likely to appear in different patterns, and that groups of genes sharing the same pattern at one time point are more likely to scatter in the future than to stay together. More detailed understanding of the dynamics of the spatial patterns of the EGFR and DPP pathway activation is crucial for explaining these trends and the two observed scenarios for the emergence of complex patterns. A gene that makes its first appearance as a complex pattern, such as the A∩D pattern of argos at stage 10B, can be a direct target of the EGFR and DPP signal integration. In contrast, a gene such as Cct1, which changes from the A to R patterns, can be a dedicated target of DPP signaling alone, and changes as a consequence of change in the spatial pattern of DPP signaling (Gupta and Schupbach, 2003; Lembong et al., 2008; Yakoby et al., 2008). Future tests of such hypotheses require analysis of cis-regulatory modules responsible for gene regulation in the follicular epithelium. While only a few enhancers have been identified at this time (Tolias et al., 1993; Andrenacci et al., 2000; Ward et al., 2006), our categorization of patterns should accelerate the identification of enhancers for a large number of genes.
Proposed for the spatial patterns of transcripts, our annotations can also describe patterns of protein expression, modification, and subcellular localization patterns. For example, the stage 10A patterns of MAD phosphorylation and CIC nuclear localization can be accurately described using the A and U\D annotations, respectively. The ultimate challenge is to use the information about the patterning of the follicular epithelium to explore how it is transformed into the three-dimensional eggshell. A number of genes in the assembled database encode cytoskeleton and cell adhesion molecules, suggesting that they provide a link between patterning and morphogenesis (Ward and Berg, 2005; Kleve et al., 2006; Laplante and Nilson, 2006; Zartman et al., 2008). We hypothesize that the highly correlated expression patterns of these genes give rise to the spatial patterns of force generation and mechanical properties of cells that eventually convert the epithelium into a target three-dimensional structure.
Experimental Procedures
Fly stocks and genetics
Wild type: OreR, UAS lines: UAS-Dad (Tsuneizumi et al., 1997), UAS-tkv* (caTKV), (Lecuit et al., 1996; Nellen et al., 1996), UAS-dnEGFR (a gift from A. Michelson), UAS-λ-top 4.2 (caEGFR) (Queenan et al., 1997), and UAS-2EGFP-AH3 (Halfon et al., 2002). The UAS lines were crossed to the CY2-GAL4 driver (Queenan et al., 1997; Goentoro et al., 2006b). Flies were grown on agar cornmeal medium; all crosses were done at 23°C, except for the caTKV, where the cross was done at 18C, and adult progeny were transferred to 23°C prior to dissection.
Dissection and RNA extraction
Previous transcriptional profiling studies of oogenesis used either the entire ovary or sorted subpopulations of the follicle cells (Bryant et al., 1999; Jordan et al., 2005). We hand-dissected stage 9-10 egg chambers. Flies were transferred to a fresh cornmeal vial with yeast and incubated at 23°C for 24h prior to dissection. Ovaries were hand-dissected in cold PBS and egg chambers at stages 9–10 (defined in Spradling, 1993) were separated from older and younger stages and divided into three groups of equal number egg chambers. To prevent RNA degradation, egg chambers were collected in groups of 20 in the RNA stabilizing buffer (RNeasy Mini Kit, Qiagen, Valencia, CA), over 5 rounds to accumulate a total of 100 egg chambers for each biological replicate (a total of 300 egg chambers from each genetic background). Details of RNA extraction, microarray experiments, qRT-PCR validation, and data analysis are described in the Supplementary Material.
In situ hybridization, immunohistochemistry, microscopy
Digoxigenin-labeled antisense RNA probes were synthesized using cDNA clones obtained from the Drosophila Gene Collection (http://www.fruitfly.org/DGC/index.html). For genes with no available cDNA, gene-specific PCR primers were designed to obtain 900-1200 bp products and cloned using StrataClone PCR cloning kit (Stratagene). All clones were sequenced (GeneWiz) and BLASTed against the D. melanogaster genome. In situ hybridization was carried out as described elsewhere (Wang et al., 2006), but without the RNase digestion step, which, as we found, does not allow detection of patterns in the main body follicle cells. Immunohistochemistry procedures are described elsewhere (Yakoby et al., 2008).
Annotation of in situ hybridizaton images
In situ hybridization images were annotated using 6 blocks and 4 operations (see the main text). Annotation of all of the patterns was done independently by four different scientists. In deciding on the annotation for every gene and time point, we examined 10-20 images collected from multiple focal planes from multiple egg chambers at different orientations on the microscope slide (see Figures S3-5 for examples of representative datasets). Since fixed and stained egg chambers are frequently connected to each other within the ovariole and are found in a wide range of orientations, standard image registration and normalization approches, similar to those used in the computer-assisted annotations of in situ hybridization images of gene expression in the Drosophila embryo (Kumar et al., 2002; Gurunathan et al., 2004; Peng et al., 2007), were not applicable. Furthermore, the color reactions at the final step of in situ hybridization were developed for different periods of time for different genes (the development times ranged from 10 minutes to 3 hours). As a consequence, images for different genes have different levels of background, which makes the segmentation of images, required for automated extraction of gene expression patterns, impossible at this stage. Given these limitations and a reasonably small size of the images in our database (100's, as opposed to 1000's in the fly embryo dataset) we used human annotation to summarize the information contained in multiple images. Similar approaches have been used in analyzing in situ hybridization images in Drosophila and other organisms (Tomancak et al., 2002; Sprague et al., 2006; Darnell et al., 2007; Visel et al., 2007).
Analysis of pattern diversity and dynamics
To characterize the frequency of co-expression, defined as the exact match of annotations, we first generated lists of all nonredundant k-tuples (pairs, triples, quadruplets) of genes in the list of 81 genes in our database. We then identified those k-tuples that share the annotation at least once in one of the four analyzed stages of oogenesis. The ratio of the number of k-tuples that are co-expressed in at least one stage serves as the estimate for the frequency of co-expression. For the gene pairs, triplets, and quadruplets, these estimates and 95% confidence intervals (computed using the Bernoulli model (Wasserman, 2003)) are: 0.10±0.01, 0.0099±0.0007, and 0.0011±0.0001, respectively. Similar analysis was done to estimate the probabilities of pattern convergence and scatterring.
Accession numbers
The Affymetrix Gene Chip data generated for this study are available in the GEO database (GSE12477).
Supplementary Material
Acknowledgments
This work was supported by the P50 GM071508 and R01GM078079 grants from the NIH. TS is supported by the Howard Hughes Medical Institute. CAB was supported by the BWF Graduate Training Program in Biological Dynamics, the NSF-IGERT PICASso Program, and by the Sigma-Xi Grant in Aid of Research. We thank Denise Montell and Xiaobo Wang for the in situ hybridization protocol. We are grateful to Pam Carroll, Hong Xaio, Aiqing He for help with the statistical design and quality control for transcriptional profiling assays, and to Donna Storton for help with the microarray facility. We thank Tiffany Vora and Stefano De Renzis for their help with the microarray experiments and Jack Leatherbarrow for assistance in generating the in situ hybridization probes. We also thank Thomas Funkhouser, Sudhir Kumar, Dannie Durand, Olga Troyanskaya, Stuart Newfeld, Mathieu Coppey, and Gregory Reeves for helpful discussions during the course of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrenacci D, Cavaliere V, Cernilogar FM, Gargiulo G. Spatial activation and repression of the drosophila vitelline membrane gene VM32E are switched by a complex cis-regulatory system. Dev Dyn. 2000;218:499–506. doi: 10.1002/1097-0177(200007)218:3<499::AID-DVDY1006>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Astigarraga S, Grossman R, Diaz-Delfin J, Caelles C, Paroush Z, Jimenez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. Embo Journal. 2007;26:668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg CA. The Drosophila shell game: patterning genes and morphological change. Trends Genet. 2005;21:346–355. doi: 10.1016/j.tig.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bryant Z, Subrahmanyan L, Tworoger M, LaTray L, Liu CR, Li MJ, van den Engh G, Ruohola-Baker H. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proc Natl Acad Sci. 1999;96:5559–5564. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc Natl Acad Sci. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MJ, Jacobsen TL, Cain DM, Jarman MG, Hubank M, Whittle JR, Phillips R, Simcox A. Discovery of genes with highly restricted expression patterns in the Drosophila wing disc using DNA oligonucleotide microarrays. Development. 2003;130:659–670. doi: 10.1242/dev.00293. [DOI] [PubMed] [Google Scholar]
- Cadigan KM. Regulating morphogen gradients in the Drosophila wing. Semin Cell Dev Biol. 2002;13:83–90. doi: 10.1016/s1084-9521(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Chang WL, Liou W, Pen HC, Chou HY, Chang YW, Li WH, Chiang W, Pai LM. The gradient of Gurken, a long-range morphogen, is directly regulated by Cbl-mediated endocytosis. Development. 2008;135:1923–1933. doi: 10.1242/dev.017103. [DOI] [PubMed] [Google Scholar]
- Cory SM, Perkins TJ. Implementing arithmetic and other analytic operations by transcriptional regulation. PLoS Comput Biol. 2008;4:e1000064. doi: 10.1371/journal.pcbi.1000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Davey S, Konieczka JH, Yatskievych TA, Antin PB. GEISHA: an in situ hybridization gene expression resource for the chicken embryo. Cytogenetic and Genome Research. 2007;117:30–35. doi: 10.1159/000103162. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Genomic regulatory systems. San Diego: Academic Press; 2001. [Google Scholar]
- Davidson EH. The Regulatory Genome: Gene Regulatory Networks In Development And Evolution. Academic Press; 2005. [Google Scholar]
- Deng WM, Bownes M. Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development. 1997;124:4639–4647. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- Dequéant ML, Glynn E, Gaudenz K, Wahl M, Chen J, Mushegian A, Pourquié O. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- Dobens LL, Raftery LA. Integration of epithelial patterning and morphogenesis in Drosophila oogenesis. Dev Dyn. 2000;218:80–93. doi: 10.1002/(SICI)1097-0177(200005)218:1<80::AID-DVDY7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Doerflinger H, Lepesant JA, Yanicostas C. Differential expression of the Drosophila zinc finger gene jim in the follicular epithelium. Mech Dev. 1999;86:177–182. doi: 10.1016/s0925-4773(99)00109-4. [DOI] [PubMed] [Google Scholar]
- Dorman JB, James KE, Fraser SE, Kiehart DP, Berg CA. bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Developmental Biology. 2004;267:320–341. doi: 10.1016/j.ydbio.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Fisher ME, Clelland AK, Bain A, Baldock RA, Murphy P, Downie H, Tickle C, Davidson DR, Buckland RA. Integrating technologies for comparing 3D gene expression domains in the developing chick limb. Dev Biol. 2008;317:13–23. doi: 10.1016/j.ydbio.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JD, van Dam A, Feiner SK, Hughes JF. Computer Graphics: principles and practice. 2. Addison-Wesley; 1992. [Google Scholar]
- Fowlkes CC, Hendriks CL, Keränen SV, Weber GH, Rübel O, Huang MY, Chatoor S, DePace AH, Simirenko L, Henriquez C, et al. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- Goentoro LA, Reeves GT, Kowal CP, Martinelli L, Schupbach T, Shvartsman SY. Quantifying the gurken morphogen gradient in Drosophila oogenesis. Dev Cell. 2006a;11:263–272. doi: 10.1016/j.devcel.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goentoro LA, Yakoby N, Goodhouse J, Schupbach T, Shvartsman SY. Quantitative analysis of the GAL4/UAS system in Drosophila oogenesis. Genesis. 2006b;44:66–74. doi: 10.1002/gene.20184. [DOI] [PubMed] [Google Scholar]
- Gupta T, Schupbach T. Cct1, a phosphatidylcholine biosynthesis enzyme, is required for Drosophila oogenesis and ovarian morphogenesis. Development. 2003;130:6075–6087. doi: 10.1242/dev.00817. [DOI] [PubMed] [Google Scholar]
- Gurunathan R, Van Emden B, Panchanathan S, Kumar S. Identifying spatially similar gene expression patterns in early stage fruit fly embryo images: binary feature versus invariant moment digital representations. Bmc Bioinformatics. 2004;5 doi: 10.1186/1471-2105-5-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon MS, Gisselbrecht S, Lu J, Estrada B, Keshishian H, Michelson AM. New fluorescent protein reporters for use with the Drosophila Gal4 expression system and for vital detection of balancer chromosomes. Genesis. 2002;34:135–138. doi: 10.1002/gene.10136. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Bilder D. Mass transit: Epithelial morphogenesis in the Drosophila egg chamber. Developmental Dynamics. 2005;232:559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- Istrail S, De-Leon SB, Davidson EH. The regulatory genome and the computer. Dev Biol. 2007;310:187–195. doi: 10.1016/j.ydbio.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Jacobsen TL, Cain D, Paul L, Justiniano S, Alli A, Mullins JS, Wang CP, Butchar JP, Simcox A. Functional analysis of genes differentially expressed in the Drosophila wing disc: role of transcripts enriched in the wing region. Genetics. 2006;174:1973–1982. doi: 10.1534/genetics.106.056788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James KE, Berg CA. Temporal comparison of Broad-Complex expression during eggshell-appendage patterning and morphogenesis in two Drosophila species with different eggshell-appendage numbers. Gene Expr Patterns. 2003;3:629–634. doi: 10.1016/s1567-133x(03)00136-4. [DOI] [PubMed] [Google Scholar]
- James KE, Dorman JB, Berg CA. Mosaic analyses reveal the function of Drosophila Ras in embryonic dorsoventral patterning and dorsal follicle cell morphogenesis. Development. 2002;129:2209–2222. doi: 10.1242/dev.129.9.2209. [DOI] [PubMed] [Google Scholar]
- Jekely G, Rorth P. Hrs mediates downregulation of multiple signalling receptors in Drosophila. Embo Reports. 2003;4:1163–1168. doi: 10.1038/sj.embor.7400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan KC, Clegg NJ, Blasi JA, Morimoto AM, Sen J, Stein D, McNeill H, Deng WM, Tworoger M, Ruohola-Baker H. The homeobox gene mirror links EGF signalling to embryonic dorso-ventral axis formation through notch activation. Nat Genet. 2000;24:429–433. doi: 10.1038/74294. [DOI] [PubMed] [Google Scholar]
- Jordan KC, Hatfield SD, Tworoger M, Ward EJ, Fischer KA, Bowers S, Ruohola-Baker H. Genome wide analysis of transcript levels after perturbation of the EGFR pathway in the Drosophila ovary. Dev Dyn. 2005;232:709–724. doi: 10.1002/dvdy.20318. [DOI] [PubMed] [Google Scholar]
- Kleve CD, Siler DA, Syed SK, Eldon ED. Expression of 18-wheeler in the follicle cell epithelium affects cell migration and egg morphology in Drosophila. Developmental Dynamics. 2006;235:1953–1961. doi: 10.1002/dvdy.20820. [DOI] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Kumar S, Jayaraman K, Panchanathan S, Gurunathan R, Marti-Subirana A, Newfeld SJ. BEST: A novel computational approach for comparing gene expression patterns from early stages of Drosophila melanogaster development. Genetics. 2002;162:2037–2047. doi: 10.1093/genetics/162.4.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante C, Nilson LA. Differential expression of the adhesion molecule Echinoid drives epithelial morphogenesis in Drosophila. Development. 2006;133:3255–3264. doi: 10.1242/dev.02492. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis regulate egf receptor activation in Drosophila. Cell. 2001;107:161–171. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- Lembong J, Yakoby N, Shvartsman SY. Spatial regulation of BMP signaling by patterned receptor expression. Tissue Eng Part A. 2008;14:1479–1477. doi: 10.1089/ten.tea.2008.0098. [DOI] [PubMed] [Google Scholar]
- Leptin M. Gastrulation movements: the logic and the nuts and bolts. Developmental Cell. 2005;8:305–320. doi: 10.1016/j.devcel.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Lin S, Zhao D, Bownes M. Blood vessel/epicardial substance (bves) expression, essential for embryonic development, is down regulated by Grk/EFGR signalling. Int J Dev Biol. 2007;51:37–44. doi: 10.1387/ijdb.052108sl. [DOI] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Luengo Hendriks CL, Keränen SV, Fowlkes CC, Simirenko L, Weber GH, DePace AH, Henriquez C, Kaszuba DW, Hamann B, Eisen MB, et al. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: data acquisition pipeline. Genome Biol. 2007;7:R123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantrova EY, Schulz RA, Hsu T. Oogenic function of the myogenic factor D-MEF2: Negative regulation of the Decapentaplegic receptor gene thick veins. Proceedings Of The National Academy Of Sciences Of The United States Of America. 1999;96:11889–11894. doi: 10.1073/pnas.96.21.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–2394. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- Megason SG, Fraser S. Imaging in systems biology. Cell. 2007;130:784–795. doi: 10.1016/j.cell.2007.08.031. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Matsuno K. Species-specific activation of EGF receptor signaling underlies evolutionary diversity in the dorsal appendage number of the genus Drosophila eggshells. Mech Dev. 2003;120:897–907. doi: 10.1016/s0925-4773(03)00164-3. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- Pai L, Barcelo G, Schupbach T. D-cbl, negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell. 2000;103:51–61. doi: 10.1016/s0092-8674(00)00104-5. [DOI] [PubMed] [Google Scholar]
- Peng HC, Long FF, Zhou J, Leung G, Eisen MB, Myers EW. Automatic image analysis for gene expression patterns of fly embryos. Bmc Cell Biology. 2007;8 doi: 10.1186/1471-2121-8-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri F, Bokel C, Roth S. Local Gurken signaling and dynamic MAPK activation during Drosophila oogenesis. Mech Dev. 1999;81:75–88. doi: 10.1016/s0925-4773(98)00228-7. [DOI] [PubMed] [Google Scholar]
- Peri F, Technau M, Roth S. Mechanisms of Gurken-dependent pipe regulation and the robustness of dorsoventral patterning in Drosophila. Development. 2002;129:2965–2975. doi: 10.1242/dev.129.12.2965. [DOI] [PubMed] [Google Scholar]
- Pribyl M, Muratov CB, Shvartsman SY. Discrete models of autocrine signaling in epithelial layers. Biophys J. 2003;84:3624–3635. doi: 10.1016/S0006-3495(03)75093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queenan AM, Barcelo G, VanBuskirk C, Schupbach T. The transmembrane region of Gurken is not required for biological activity, but is necessary for transport to the oocyte membrane in Drosophila. Mech Dev. 1999;89:35–42. doi: 10.1016/s0925-4773(99)00196-3. [DOI] [PubMed] [Google Scholar]
- Queenan AM, Ghabrial A, Schupbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- Requicha AAG, Voelcker HB. Solid Modeling - a Historical Summary and Contemporary Assessment. Ieee Computer Graphics and Applications. 1982;2:9–&. [Google Scholar]
- Ruohola-Baker H, Grell E, Chou TB, Baker D, Jan L, Jan YN. Spatially localized rhomboid is required for establishing the dorsal-ventral axis in Drosophila oogenesis. Cell. 1993;73:953–965. doi: 10.1016/0092-8674(93)90273-s. [DOI] [PubMed] [Google Scholar]
- Sapir A, Schweitzer R, Shilo BZ. Sequential activation of the EGF receptor pathway during Drosophila oogenesis establishes the dorsoventral axis. Development. 1998;125:191–200. doi: 10.1242/dev.125.2.191. [DOI] [PubMed] [Google Scholar]
- Schnorr JD, Holdcraft R, Chevalier B, Berg CA. Ras1 interacts with multiple new signaling and cytoskeletal loci in Drosophila eggshell patterning and morphogenesis. Genetics. 2001;129:2209–2222. doi: 10.1093/genetics/159.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen J, Goltz JS, Stevens L, Stein D. Spatially restricted expression of pipe in the Drosophila egg chamber defines embryonic dorsal-ventral polarity. Cell. 1998;95:471–481. doi: 10.1016/s0092-8674(00)81615-3. [DOI] [PubMed] [Google Scholar]
- Shravage BV, Altmann G, Technau M, Roth S. The role of Dpp and its inhibitors during eggshell patterning in Drosophila. Development. 2007;134:2261–2271. doi: 10.1242/dev.02856. [DOI] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, et al. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;1 doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias PP, Konsolaki M, Halfon MS, Stroumbakis ND, Kafatos FC. Elements controlling follicular expression of the s36 chorion gene during Drosophila oogenesis. Mol Cell Biol. 1993;13:5898–5906. doi: 10.1128/mcb.13.9.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3:RESEARCH0088. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneizumi K, Nakayama T, Kamoshida Y, Kornberg TB, Christian JL, Tabata T. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997;389:627–631. doi: 10.1038/39362. [DOI] [PubMed] [Google Scholar]
- Twombly V, Blackman RK, Jin H, Graff JM, Padgett RW, Gelbart WM. The TGF-beta signaling pathway is essential for Drosophila oogenesis. Development. 1996;122:1555–1565. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- Visel A, Carson J, Oldekamp J, Warnecke M, Jakubcakova V, Zhou X, Shaw CA, Alvarez-Bolado G, Eichele G. Regulatory pathway analysis by high-throughput in situ hybridization. PloS Genet. 2007;3:1867–1883. doi: 10.1371/journal.pgen.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Bo JY, Bridges T, Dugan KD, Pan TC, Chodosh LA, Montell DJ. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Developmental Cell. 2006;10:483–495. doi: 10.1016/j.devcel.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Ward EJ, Berg CA. Juxtaposition between two cell types is necessary for dorsal appendage tube formation. Mechanisms of Development. 2005;122:241–255. doi: 10.1016/j.mod.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Ward EJ, Zhou XF, Riddiford LM, Berg CA, Ruohola-Baker H. Border of Notch activity establishes a boundary between the two dorsal appendage tube cell types. Developmental Biology. 2006;297:461–470. doi: 10.1016/j.ydbio.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Wasserman JD, Freeman M. An autoregulatory cascade of EGF receptor signaling patterns the Drosophila egg. Cell. 1998;95:355–364. doi: 10.1016/s0092-8674(00)81767-5. [DOI] [PubMed] [Google Scholar]
- Wasserman L. All of statistics: A Concise Course in Statistical Inference. Springer; 2003. [Google Scholar]
- Yakoby N, Lembong J, Schupbach T, Shvartsman SY. Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development. 2008;135:343–351. doi: 10.1242/dev.008920. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Bolouri H, Davidson EH. Genomic Cis-Regulatory Logic: Experimental and Computational Analysis of a Sea Urchin Gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- Zartman JJ, Yakoby N, Bristow CA, Zhou X, Schlichting X, Dahmann C, Shvartsman SY. Cad74A is regulated by Br and is required for robust dorsal appendage formation in Drosophila oogenesis. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.07.027. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.