Abstract
The mitochondrial genome of Physarum polycephalum encodes five tRNAs, four of which are edited by nucleotide insertion. Two of these tRNAs, tRNAmet1 and tRNAmet2, contain predicted mismatches at the beginning (proximal end) of the acceptor stem. In addition, the putative 5′ end of tRNAmet2 overlaps the 3′ end of a small, abundant, noncoding RNA, which we term ppoRNA. These anomalies led us to hypothesize that these two Physarum mitochondrial tRNAs undergo additional editing events. Here, we show that tRNAmet1 and tRNAmet2 each has a nonencoded G at its 5′ end. In contrast to the other nucleotides that are added to Physarum mitochondrial RNAs, these extra G residues are likely added post-transcriptionally based on (1) the absence of added G in precursor transcripts containing inserted C and AA residues, (2) the presence of potential intermediates characteristic of 5′ replacement editing, and (3) preferential incorporation of GTP into tRNA molecules under conditions that do not support transcription. This is the first report of both post-transcriptional nucleotide insertions and the addition of single Gs in P. polycephalum mitochondrial transcripts. We postulate that tRNAmet1 and tRNAmet2 are acted upon by an activity similar to that present in the mitochondria of certain other amoebozoons and chytrid fungi, suggesting that enzymes that repair the 5′ end of tRNAs may be widespread.
Keywords: RNA editing, tRNA, nucleotide substitution
INTRODUCTION
RNAs in the mitochondria of Physarum polycephalum undergo maturation through an array of RNA editing events, including the insertion of nonencoded nucleotides (Mahendran et al. 1991; Gott et al. 1993; Takano et al. 2001), the deletion of encoded nucleotides (Gott et al. 2005), and C-to-U substitutions (Gott et al. 1993). The vast majority of such changes involve the specific insertion of 1–2 nucleotides (nt) (C, U, AA, UU, GC, CU, GU, or UA) at defined sites. Thus far, nearly 500 insertion sites have been identified, with another approximately 500 insertion sites predicted to occur in mitochondrial RNAs that remain to be characterized (Beargie et al. 2008). Editing creates open reading frames within mRNAs and contributes to the secondary and tertiary structures of mitochondrial tRNAs and rRNAs. On average, inserted nucleotides comprise ∼4% of the total residues of mRNAs and ∼2% of the residues of mature rRNAs and tRNAs (Miller et al. 1993; Gott and Rhee 2007).
A variety of tRNA editing events have been reported in mitochondrial tRNAs (Price and Gray 1998), including C-to-U edits in plants (Maréchal-Drouard et al. 1993, 1996a,b; Binder et al. 1994), marsupials (Janke and Pääbo 1993), and trypanosomatid protozoa (Alfonzo et al. 1999); 5′-replacement editing in amoebozoan protists (Lonergan and Gray 1993a,b) and chytridomycete fungi (Laforest et al. 1997); 3′-replacement editing in various metazoan animals (Yokobori and Pääbo 1995a,b, 1997; Tomita et al. 1996) and the jakobid flagellate Seculamonas ecuadoriensis (Leigh and Lang 2004); and insertion editing in slime molds (Antes et al. 1998). First documented in the amoeboid protist Acanthamoeba castellanii (Lonergan and Gray 1993a), 5′-replacement editing of mitochondrial tRNAs has been inferred on the basis of mitochondrial DNA sequence in other amoebozoons such as Dictyostelium discodieum and Polysphondylium pallidum, and edited mitochondrial tRNAs have been characterized in the latter protist (E Schindel and MW Gray, unpubl.). The biochemical characteristics of 5′ tRNA editing in A. castellanii (Price and Gray 1999a) and in a phylogenetically unrelated chytrid fungus, Spizellomyces punctatus (Bullerwell and Gray 2005), are strikingly similar, suggesting very similar mechanisms in the two cases. These editing activities may be evolutionarily related to those involved in 5′ end maturation of nuclear histidine tRNAs (Cooley et al. 1982), although the histidine guanylyltransferases add a G at position −1 rather than +1.
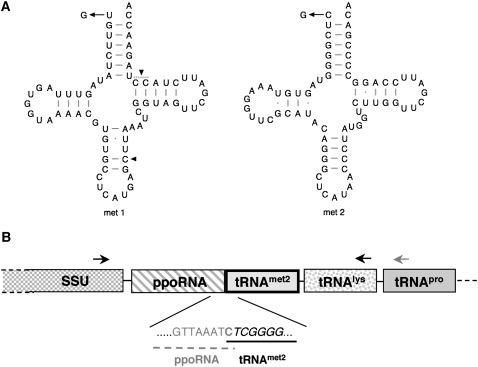
The Physarum mitochondrial genome encodes five tRNA genes, and four of the five tRNA products have been shown to contain extra, nonencoded nucleotides (Antes et al. 1998). Curiously, two of the five tRNAs encoded in the Physarum mitochondrial genome are predicted to contain mismatches at the beginning (proximal end) of the acceptor stem: a UxC in tRNAmet1 and a CxC in tRNAmet2 (Fig. 1A; Antes et al. 1998). In addition, the predicted tRNAmet2 product appears to overlap by a single nucleotide a small, abundant, noncoding RNA (ppoRNA) (Fig. 1B; C Bullerwell, G Burger, J Gott, O Kourennaia, M Schnare, and M Gray, unpubl.). Based on these observations, we hypothesized that the production of the mature forms of tRNAmet1 and tRNAmet2 requires an additional RNA editing event at their 5′ ends that would create standard G-C base-pairs.
FIGURE 1.
5′ RNA editing is predicted for two of the five tRNAs encoded in the mitochondrial genome of Physarum polycephalum. (A) Mitochondrial tRNAmet1 and tRNAmet2 each contain a mismatch at the beginning of the acceptor stem (Antes et al. 1998). Sites predicted to contain a G residue in the mature tRNA products are indicated with arrows; sites of internal C insertions identified by Antes et al. (1998) are indicated with arrowheads. (B) Apparent overlap of the 5′ end of tRNAmet2 with the 3′ end of ppoRNA. The approximate positions of primers used for reverse transcription (gray) and PCR (black) of nascent transcripts are shown.
Two of the known forms of tRNA editing could operate to generate base-paired acceptor stems in Physarum mitochondrial tRNAmet1 and tRNAmet2, which are matured from longer, polycistronic transcripts (Antes et al. 1998; Byrne and Gott 2004). In Physarum mitochondria, the insertion of nonencoded nucleotides is a cotranscriptional process in which extra residues are added at specific sites within nascent transcripts (Cheng et al. 2001). It is therefore possible that the predicted editing events occur via cotranscriptional insertion of a G at the 5′ end of each tRNA, followed by endonucleolytic cleavage of the primary transcript, presumably by RNase P. Alternatively, these tRNAs could undergo an additional form of RNA editing similar to the post-transcriptional 5′ replacement editing observed in the mitochondria of A. castellanii (Lonergan and Gray 1993a,b) and S. punctatus (Laforest et al. 1997).
Here, we demonstrate that tRNAmet1 and tRNAmet2 contain, at their 5′ ends, the predicted G residues that would create a standard 5′ G:C 3′ base-pair at the beginning of the acceptor stem of each tRNA. We also provide strong evidence that these G residues are added by a post-transcriptional 5′ replacement mechanism rather than the cotranscriptional mechanism responsible for nucleotide additions in internal regions of tRNAs, rRNAs, and mRNAs. Thus, maturation of tRNAs in Physarum mitochondria involves two separate forms of editing involving nonencoded nucleotides: cotranscriptional insertion within internal regions and post-transcriptional changes at 5′ termini. These data add to the already impressive number of distinct editing mechanisms operating in Physarum mitochondria. The implications of these findings are discussed.
RESULTS AND DISCUSSION
Internal editing sites within the tRNAs encoded in the Physarum mitochondrial genome have been reported previously (Antes et al. 1998). Interestingly, two of the tRNAs, met1 and met2, were each predicted to have a mismatch at the first base pair of the acceptor stem (positions 1:72, standard numbering) (Fig. 1A). Lack of pairing at these positions is a characteristic feature of initiator tRNAmet in prokaryotes and organelles (Jühling et al. 2009); but given the existence of RNA editing in Physarum mitochondria, we suspected that these apparent mismatches might be “corrected” in the mature tRNA products.
The Physarum mitochondrial tRNAmet2 is synthesized as part of a longer transcript that minimally includes the small subunit (SSU) rRNA, ppoRNA, tRNAmet2, tRNAlys, and tRNApro (Fig. 1B; Antes et al. 1998; Byrne and Gott 2004). Characterization of the abundant ppoRNA, which is encoded immediately upstream of the tRNAmet2 gene, indicated that about 73% of these molecules have a C at their 3′ end, with the remaining molecules ending in a U (C Bullerwell, G Burger, J Gott, O Kourennaia, M Schnare, and M Gray, unpubl.). Given that there is only a single encoded C at this position and that it was previously suggested this C is the 5′ end of tRNAmet2 (Antes et al. 1998), it is possible that an extra residue is added at this position in the primary transcript. We therefore decided to characterize both the mature tRNA species and the nascent transcript.
A G residue is present at the 5′ end of tRNAmet1 and tRNAmet2
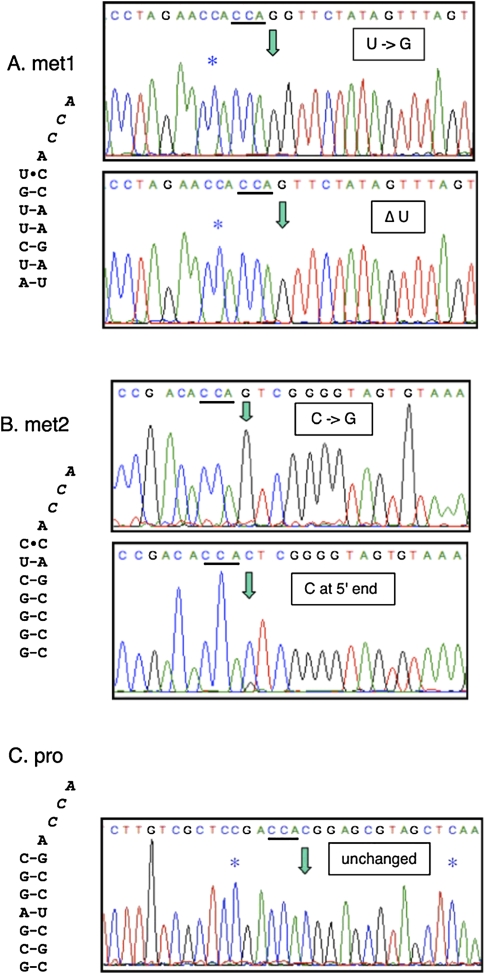
To determine the nucleotide present at the 5′ end of tRNAmet1 and tRNAmet2, we used the strategy employed by Price and Gray (1999b) to characterize mature mitochondrial tRNAs in A. castellanii. In this approach, tRNAs are circularized with RNA ligase prior to RT-PCR, cloning, and sequencing (Lohan and Gray 2004, 2007). As a control, using the same strategy, we also examined tRNApro, which is inferred not to undergo 5′ editing. As we predicted, many tRNAmet1 and tRNAmet2 molecules had a G rather than the encoded C (in the case of met2) or U (met1) at their 5′ ends, with all tRNAmet1 clones also containing the expected internal C insertion (Fig. 2). Similar experiments using primers specific for circularized tRNApro indicated that all tRNApro clones contained both of the internally added C residues (Fig. 2, blue asterisks) but had the encoded C at their 5′ ends; i.e., no evidence of 5′ editing was seen for this tRNA. All clones derived from circularized tRNA contained a 3′-CCA end, but no inferences can be drawn from this observation due to the possibility of preferential ligation of tRNAs having this structure. From these experiments, we conclude that some form of 5′-replacement editing occurs during the maturation of tRNAs having a mismatch at the beginning of the acceptor stem.
FIGURE 2.
Editing at the 5′ ends of tRNAmet1 and tRNAmet2. Sequence of representative cloned RT-PCR products derived from circularized tRNAs; traces of the region between the primers used for PCR amplification are shown. The nonencoded CCA ends (italics) are underlined, and the 5′ ends of the mature tRNAs are indicated by green arrows in the traces. Internal C insertion sites are indicated with blue asterisks. (A) The acceptor stem of tRNAmet1 and traces from clones with a G at its 5′ end (edited, top) or missing the 5′ nucleotide (potential editing intermediate, bottom). Both contain the extra C that is added in this region. (B) Acceptor stem of tRNAmet2 and traces from tRNAmet2 clones having a 5′ G (edited, top) or C (unedited, bottom). (C) Clone from circularized tRNApro containing the encoded C at its 5′ end and internal C insertions.
Presence of potential editing intermediates
Interestingly, none of the cloned RT-PCR products derived from circularized tRNAmet1 contained the encoded 5′ U, but more than half were missing the 5′ nucleotide entirely (Fig. 2A). Since tRNAmet1 occupies the 5′ end of the tRNAmet1-tRNAglu2 precursor RNA (Antes et al. 1998), such molecules could potentially be generated via either initiation of transcription at position +2 or cleavage at the position of the mismatched U. As was the case with the completely edited molecules, all of the 5′ -truncated tRNAmet1 clones contained both the internal C insertion and a 3′-CCA end (Fig. 2A), suggesting that these tRNAs could potentially be intermediates in a 5′ editing reaction. Circularized tRNAmet2 clones lacking the 5′ nucleotide were not observed, but one contained a C at its 5′ end (Fig. 2B). This clone must be derived from processed RNA based on the added CCA at the circularization junction, and is likely a result of processing between the ppoRNA and tRNAmet2 (Fig. 1), since about 25% of ppoRNA molecules have a U at their 3′ ends (C Bullerwell, G Burger, J Gott, O Kourennaia, M Schnare, and M Gray, unpubl.). The presence of tRNAmet2 having a C at its 5′ end is not consistent with a mechanism involving cotranscriptional G insertion, although the possibility of partial editing at this site cannot be ruled out. Antes et al. (1998) have reported partial editing of unprocessed tRNA precursors, but in all cases cDNAs derived from processed tRNAs were fully edited.
Absence of added G residue in primary transcripts
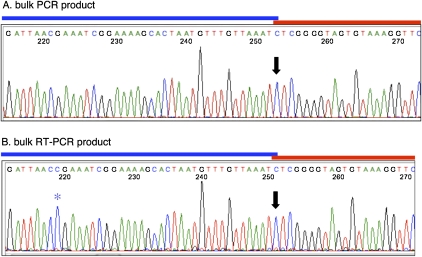
To determine whether a G is added during transcription of the tRNAmet2 precursor, we generated RT-PCR products derived from the nascent transcript using primers specific for genes lying upstream (SSU rRNA) and downstream (tRNAlys) of the tRNAmet2 gene. The same primers were also used to amplify the genomic sequence. Sequence traces from the bulk PCR and RT-PCR products are shown in Figure 3. The precursor RNA clearly does not have an added G (or any other added nucleotide) at the 5′ end of tRNAmet2, despite the fact that all other nonencoded nucleotides both upstream and downstream are present. This finding is consistent with previous results with cloned RT-PCR products derived from this region. In those experiments, all RT-PCR clones representing nascent (unprocessed) transcripts made in vivo were completely edited by nucleotide insertion at known insertion sites, but none contained an extra G at the 5′ end of tRNAmet2 (Byrne and Gott 2004). Likewise, none of 16 independent partially edited clones made during run-on transcription in vitro had an inserted G at this position despite high levels of editing at other sites (Byrne and Gott 2004). Thus, we conclude that the G at the 5′ end of the mature tRNAmet2 is not derived from a G inserted into the nascent transcript.
FIGURE 3.
No extra G is present at the ppoRNA-tRNAmet2 boundary in RNA precursors containing added nucleotides at other positions. Sequence traces of bulk PCR and RT-PCR products derived from mitochondrial DNA (A) and nascent transcripts (B) containing the 3′ end of the mitochondrial small subunit rRNA (SSU), ppoRNA, tRNAmet2, and tRNAlys. The region encoding the 3′ portion of ppoRNA (blue line) and the 5′ portion of tRNAmet2 (red line) is enlarged here. The traces from which these are extracted are shown in Supplemental Figure S1. The site of potential overlap of the ppoRNA and tRNAmet2 is indicated by a black arrow, and the nonencoded C inserted in this portion of the ppoRNA is marked with a blue asterisk.
Detection of a G addition activity in isolated Physarum mitochondria
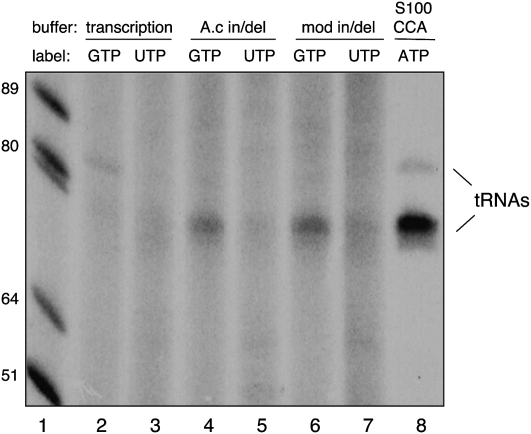
A partially purified activity from A. castellanii (Price and Gray 1999a) and S. punctatus (Bullerwell and Gray 2005) mitochondria is capable of removing nucleotides from the 5′ end of tRNA substrates and replacing them in a 3′-to-5′ direction using the 3′ portion of the acceptor stem as template. To investigate whether a similar activity might be present in Physarum mitochondria, we carried out labeling experiments with isolated organelles. These experiments were done in three different buffers: (1) our standard mitochondrial transcription buffer, (2) the buffer used to characterize the A. castellanii 5′ replacement activity, and (3) a modified version of the A. castellanii buffer containing a lower level of Mg+2 (see Materials and Methods). Mitochondria were isolated and preincubated in buffer to reduce endogenous nucleotide pools prior to the addition of [α-32P]-UTP or [α-32P]-GTP. After a 30-min incubation, nucleic acids were isolated and subjected to gel electrophoresis alongside mitochondrial tRNAs labeled by the endogenous CCA-adding activity (Fig. 4, lane 8).
FIGURE 4.
Processed tRNAs are preferentially labeled with [α32P]GTP in isolated mitochondria. Isolated mitochondria were incubated in transcription buffer (lanes 2,3), the optimal buffer for the A. castellanii 5′ editing activity (lanes 4,5), or a modified version of the A. castellanii buffer (lanes 6,7), in the presence of [α-32P]GTP (lanes 2,4,6) or [α-32P]UTP (lanes 3,5,7) as described in Materials and Methods. Lane 8 shows the position of mature tRNAs labeled with [α-32P]ATP by the CCA-adding activity present in S100 extracts derived from Physarum mitochondria. End-labeled, denatured DNA size markers are shown in lane 1.
Under the run-on transcription conditions used here, very little incorporation of label into mature tRNA species would be expected due to (1) the limited amount of transcription observed under severely limiting nucleotide concentrations (Visomirski-Robic and Gott 1997) and (2) the relatively short incubation times, which might not be sufficient for complete processing of the very large precursor. Indeed, only trace amounts of labeled nucleotides were incorporated into mature tRNA species when mitochondria were incubated in transcription buffer (Fig. 4, lanes 2,3). Strikingly, however, under conditions optimal for the A. castellanii tRNA replacement activity, significant incorporation of labeled nucleotide into endogenous tRNA was observed when [α-32P]-GTP (Fig. 4, lanes 4,6) but not [α-32P]-UTP (Fig. 4, lanes 5,7) was present. This result cannot be explained by differences in endogenous nucleotide pools, since GTP levels are significantly higher than UTP levels in isolated mitochondria (L Visomirski-Robic, unpubl.). These data indicate that there is an activity present in Physarum mitochondria that is capable of post-transcriptional addition of GTP into RNAs that comigrate with mature tRNAs (Fig. 4, lane 8).
Thirteen of the 16 tRNAs encoded in the A. castellanii mitochondrial genome contain mismatches in their acceptor stems (Lonergan and Gray 1993a,b, Price and Gray 1999b). However, unlike the tRNAs studied here, the mismatches in A. castellanii tRNAs can occur anywhere within the proximal three positions of the acceptor stem, with some tRNAs containing two or even three mismatches (Lonergan and Gray 1993a). The A. castellanii tRNA editing activity is capable of deleting up to 3 nt on the 5′ side of the acceptor stem and filling in the recessed end (Price and Gray 1999a). Interestingly, Physarum mitochondrial tRNAs were not labeled with UTP under replacement conditions (Fig. 4, lanes 5,7), despite the fact that tRNAmet1 has a U at position 3 and tRNAmet2 has a U at position 2 that could potentially be replaced if a putative deletion/addition activity similar to the A. castellanii enzyme were present in P. polycephalum mitochondria. Given that the mismatches in the two Physarum tRNAs are at the beginning of the acceptor stem, it may be that the putative editing activity in Physarum mitochondria is only capable of deleting a single nucleotide from the 5′ end of tRNAs. Alternatively, base-pairing may simply inhibit nucleolytic removal of additional nucleotides, resulting in a much less robust signal.
The work presented here demonstrates that tRNAmet1 and tRNAmet2 are edited at their 5′ ends by replacement of U or C, respectively, by G. Although G insertion has been observed previously as part of internal GU or GC dinucleotide insertions, this is the first report of single G additions in Physarum mitochondria. Three major findings support our conclusion that editing at the 5′ end of these tRNAs is likely to occur via a post-transcriptional mechanism that is distinct from previously identified forms of editing in this organism. First, the existence of potential editing intermediates in processed tRNAs containing 3′-CCA ends is consistent with a post-transcriptional mechanism, but not with cotranscriptional nucleotide insertion. Second, the absence of an added nucleotide at the 5′ end of tRNAmet2 in a polycistronic precursor that contains inserted nucleotides both upstream and downstream argues against cotranscriptional G insertion. Third, Physarum mitochondria contain an activity that specifically incorporates GTP into the 5′ end of mature tRNAs. tRNAmet1 is also subject to editing by internal nucleotide insertions (Fig. 2; Antes et al. 1998); thus, this tRNA requires two different forms of RNA editing for its maturation. To our knowledge this is the first example of a tRNA that is edited both co- and post-transcriptionally. Although we cannot rule out the existence of an entirely novel editing activity, it seems more likely (based on the fact that the GTP incorporation occurs under conditions optimal for the 5′ editing activity in A. castellanii mitochondria) that the 5′ tRNA editing activity in P. polycephalum mitochondria is related to the enzymes present in A. castellanii and S. punctatus. CCA-adding enzymes are present even in organisms having tRNAs with encoded CCA ends (Deutscher 1990), and we suggest that RNA repair activities that act specifically at the 5′ ends of tRNAs may actually be quite widespread in nature.
MATERIALS AND METHODS
RNA isolation and circularization reactions
Mitochondria were isolated in buffered sucrose (BSS) as described previously (Visomirski-Robic and Gott 1995) and lysed in 20 mM Tris-HCl (pH 7.7), 1 mM EDTA, and 0.1% SDS. Total mitochondrial nucleic acids were deproteinized via sequential extractions with phenol:chloroform:isoamyl alcohol (25:24:1 [v:v:v]) and chloroform:isoamyl alcohol (24:1 [v:v]), followed by ethanol precipitation. After treatment with DNase I (Roche), mitochondrial RNAs were again deproteinized and precipitated. Enrichment for salt-soluble RNAs (sRNA) and circularization reactions were carried out essentially according to the method as described by Price and Gray (1999b). Briefly, 60 μg of total mitochondrial RNA were brought to 1.2 M NaCl, incubated overnight at 4°C, and centrifuged 10 min at 12,000g at 4°C. RNAs present in the supernatant were precipitated with ethanol, resuspended, deproteinized, and again precipitated with ethanol. Five micrograms of sRNA were heated for 5 min to 90°C, quickly cooled on ice, and then incubated overnight at 37°C in 50 mM HEPES (pH 7.5), 15 mM MgCl2, 3.3 mM DTT, 10% DMSO, 0.01 μg/μL BSA, and 80 μM ATP with 10 units T4 RNA ligase (Promega). Ligated RNAs were deproteinized, precipitated with ethanol, and resuspended in 10 μL of water.
RT-PCR and PCR reactions
For cDNA synthesis from circularized tRNAs, 1 μg ligated sRNA was mixed with 1 pmol of primer, heated for 2 min to 90°C, cooled to room temperature over ∼20 min, and incubated on ice for 15 min. Annealed primer templates were then incubated in 50 mM Tris (pH 8.3, at 42°C), 50 mM KCl, 10 mM MgCl2, 10 mM DTT, 0.5 mM spermidine, and 60 μM each dNTP with 10 units of AMV reverse transcriptase (Life Sciences) for 45 min at 42°C. Primers used for cDNA synthesis were as follows: cirRTmet1.3 (5′-GAGGCACACGTTTTACC-3′), cirRTmet2.1 (5′-AGCCCTGTATGCGAACC-3′), and cirRTpro1 (5′-AACCGAATGCTCTACCAG-3′). One-tenth of the appropriate cDNA was used as the template in subsequent PCR reactions using Taq polymerase (Roche) under conditions suggested by the supplier. Primers were phosphorylated by incubation with polynucleotide kinase prior to PCR reactions containing cirRTmet1.3 and cirmet1.4 (5′-TCGGTAGTTCGATTCTAC-3′), cirRTmet2.1 and cirmet2.2 (5′-TGGTTCGATTCCAGGCC-3′), or cirRTpro1 and cirpro2 (5′-GGGACCGAAAGGTTGC-3′). The resulting RT-PCR products were gel purified and ligated to pBSM13+ (Stratagene) that had been digested with SmaI (New England Biolabs) and treated with calf intestine alkaline phosphatase (Roche).
For cDNA synthesis from nascent transcripts, 2 μg of total mitochondrial RNA in 10 mM Tris (pH 8.3, at 42°C) and 250 mM KCl were mixed with primer cirRTpro1, heated for 3 min to 95°C, incubated for 10 min at 65°C, and then held on ice. Reverse transcription was carried out in 25 mM Tris (pH 8.3, at 42°C), 125 mM KCl, 15 mM MgCl2, 7.5 mM DTT, and 380 μM each dNTP with 5 units of AMV reverse transcriptase (Life Sciences) for 1 h at 42°C. One-tenth of the cDNA product was used in a PCR reaction using primers 1ssu (5′-TCACGTACAGACCGCCC-3′) and tRNAK3 (5′-TGGTTGGCTCCACAGGACTTGC-3′) and Taq polymerase (New England BioLabs) under recommended conditions. A parallel reaction containing 100 ng total mitochondrial nucleic acid was carried out to amplify the genomic sequence. The RT-PCR and PCR products were gel purified and sequenced directly by Biotic Solutions.
Labeling experiments
Mitochondria were isolated as described above and preincubated in the appropriate buffer (see below) for 5 min at 35°C prior to the addition of nucleotide. Mitochondria at a final concentration of 0.75 mg/mL were then incubated with 0.3 μM [α-32P]UTP or [α-32P]GTP for 30 min at 35°C. Reactions were stopped by the addition of 4 volumes of stop mix (20 mM Tris-HCl [pH 7.5], 15 mM EDTA, 0.25% SDS) followed by sequential extractions with phenol:chloroform:isoamyl alcohol (25:24:1) and chloroform:isoamyl alcohol (24:1). Nucleic acids were precipitated with ethanol, resuspended in 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA, and electrophoresed on an 8% denaturing acrylamide gel. The composition of the incubation buffers is as follows: Transcription buffer: 20 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 10 mM KCl, 2 mM DTT; A. castellanii deletion/insertion buffer: 40 mM HEPES (pH 7), 7.5 mM MgCl2, 5 mM KCl, 1 mM DTT; Modified A. castellanii buffer: 40 mM HEPES (pH 7), 5 mM MgCl2, 5 mM KCl, 1 mM DTT.
The tRNA marker in Figure 4, lane 8, was generated by the endogenous CCA-adding activity. An S100 extract from Physarum mitochondria was preincubated in 15 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 5 mM KCl, 1.5 mM DTT, and 5% glycerol for 5 min at 35°C to deplete nucleotide pools and then incubated in the presence of [α-32P]ATP and CTP for an additional 30 min at 35°C prior to nucleic acid isolation and gel electrophoresis as described above.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Neeta Parimi and Angela Stout for early contributions to this project, and Murray N. Schnare for generating the tRNA secondary structure diagrams. Funding for this work was provided by NIH grant GM54663 to J.M.G. and CIHR grant MOP-4124 to M.W.G.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1958810.
REFERENCES
- Alfonzo JD, Blanc V, Estévez AM, Rubio MA, Simpson L. C to U editing of the anticodon of imported mitochondrial tRNATrp allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 1999;18:7056–7062. doi: 10.1093/emboj/18.24.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antes T, Costandy H, Mahendran R, Spottswood M, Miller D. Insertional editing of mitochondrial tRNAs of Physarum polycephalum and Didymium nigripes. Mol Cell Biol. 1998;18:7521–7527. doi: 10.1128/mcb.18.12.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beargie C, Liu T, Corriveau M, Lee HY, Gott J, Bundschuh R. Genome annotation in the presence of insertional RNA editing. Bioinformatics. 2008;24:2571–2578. doi: 10.1093/bioinformatics/btn487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S, Marchfelder A, Brennicke A. RNA editing of tRNAPhe and tRNACys in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol Gen Genet. 1994;244:67–74. doi: 10.1007/BF00280188. [DOI] [PubMed] [Google Scholar]
- Bullerwell CE, Gray MW. In vitro characterization of a tRNA editing activity in the mitochondria of Spizellomyces punctatus, a chytridiomycete fungus. J Biol Chem. 2005;280:2463–2470. doi: 10.1074/jbc.M411273200. [DOI] [PubMed] [Google Scholar]
- Byrne EM, Gott JM. Unexpectedly complex editing patterns at dinucleotide insertion sites in Physarum mitochondria. Mol Cell Biol. 2004;24:7821–7828. doi: 10.1128/MCB.24.18.7821-7828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YW, Visomirski-Robic LM, Gott JM. Non-templated addition of nucleotides to the 3′ end of nascent RNA during RNA editing in Physarum. EMBO J. 2001;20:1405–1414. doi: 10.1093/emboj/20.6.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Appel B, Söll D. Post-transcriptional nucleotide addition is responsible for the formation of the 5′ terminus of histidine tRNA. Proc Natl Acad Sci. 1982;70:6475–6479. doi: 10.1073/pnas.79.21.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Ribonucleases, tRNA nucleotidyltransferase, and the 3′ processing of tRNA. Prog Nucleic Acid Res Mol Biol. 1990;39:209–240. doi: 10.1016/s0079-6603(08)60628-5. [DOI] [PubMed] [Google Scholar]
- Gott JM, Rhee AC. Insertion/deletion editing in Physarum polycephalum. In: Goringer HU, editor. RNA editing. Springer-Verlag; Berlin: 2007. pp. 85–104. [Google Scholar]
- Gott JM, Visomirski LM, Hunter JL. Substitutional and insertional RNA editing of the cytochrome c oxidase subunit 1 mRNA of Physarum polycephalum. J Biol Chem. 1993;268:25483–25486. [PubMed] [Google Scholar]
- Gott JM, Parimi N, Bundschuh R. Discovery of new genes and deletion editing in Physarum mitochondria enabled by a novel algorithm for finding edited mRNAs. Nucleic Acids Res. 2005;33:5063–5072. doi: 10.1093/nar/gki820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke A, Pääbo S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 1993;21:1523–1525. doi: 10.1093/nar/21.7.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest M-J, Roewer I, Lang BF. Mitochondrial tRNAs in the lower fungus Spizellomyces punctatus: tRNA editing and UAG “stop” codons recognized as leucine. Nucleic Acids Res. 1997;25:626–632. doi: 10.1093/nar/25.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J, Lang BF. Mitochondrial 3′ tRNA editing in the jakobid Seculamonas ecuadoriensis: A novel mechanism and implications for tRNA processing. RNA. 2004;10:615–621. doi: 10.1261/rna.5195504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohan AJ, Gray MW. Methods for analysis of tRNA editing in Acanthamoeba castellanii. Methods Mol Biol. 2004;265:315–332. doi: 10.1385/1-59259-775-0:315. [DOI] [PubMed] [Google Scholar]
- Lohan AJ, Gray MW. Analysis of 5′- or 3′-terminal tRNA editing: Mitochondrial 5′ tRNA editing in Acanthamoeba castellanii as the exemplar. Methods Enzymol. 2007;424:221–242. doi: 10.1016/S0076-6879(07)24010-8. [DOI] [PubMed] [Google Scholar]
- Lonergan KM, Gray MW. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science. 1993a;259:812–816. doi: 10.1126/science.8430334. [DOI] [PubMed] [Google Scholar]
- Lonergan KM, Gray MW. Predicted editing of additional transfer RNAs in Acanthamoeba castellanii mitochondria. Nucleic Acids Res. 1993b;21:4402. doi: 10.1093/nar/21.18.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendran R, Spottswood MR, Miller DL. RNA editing by cytidine insertion in mitochondria of Physarum polycephalum. Nature. 1991;349:434–438. doi: 10.1038/349434a0. [DOI] [PubMed] [Google Scholar]
- Maréchal-Drouard L, Ramamonjisoa D, Cossett A, Weil JH, Dietrich A. Editing corrects mispairing in the acceptor stem of bean and potato mitochondrial phenylalanine transfer RNAs. Nucleic Acids Res. 1993;21:4909–4914. doi: 10.1093/nar/21.21.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal-Drouard L, Kumar R, Remacle C, Small I. RNA editing of larch mitochondrial tRNAHis precursors is a prerequisite for processing. Nucleic Acids Res. 1996a;24:3229–3234. doi: 10.1093/nar/24.16.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal-Drouard L, Cosset A, Remacle C, Ramamonjisoa D, Dietrich A. A single editing event is a prerequisite for efficient processing of potato mitochondrial phenylalanine tRNA. Mol Cell Biol. 1996b;16:3504–3510. doi: 10.1128/mcb.16.7.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D, Mahendran R, Spottswood M, Costandy H, Wang S, Ling ML, Yang N. Insertional editing in mitochondria of Physarum. Semin Cell Biol. 1993;4:261–266. doi: 10.1006/scel.1993.1031. [DOI] [PubMed] [Google Scholar]
- Price DH, Gray MW. Editing of transfer RNA. In: Grosjean H, Benne R, editors. Modification and editing of RNA: The alteration of RNA structure and function. ASM Press; Washington, DC: 1998. pp. 289–305. [Google Scholar]
- Price DH, Gray MW. A novel nucleotide incorporation activity implicated in the editing of mitochondrial transfer RNAs in Acanthamoeba castellanii. RNA. 1999a;5:302–317. doi: 10.1017/s1355838299981840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH, Gray MW. Confirmation of predicted edits and demonstration of unpredicted edits in Acanthamoeba castellanii mitochondrial tRNAs. Curr Genet. 1999b;35:23–29. doi: 10.1007/s002940050428. [DOI] [PubMed] [Google Scholar]
- Takano H, Abe T, Sakurai R, Moriyama Y, Miyazawa Y, Nozaki H, Kawano S, Sasaki N, Kuroiwa T. The complete DNA sequence of the mitochondrial genome of Physarum polycephalum. Mol Gen Genet. 2001;264:539–545. doi: 10.1007/s004380000357. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ueda T, Watanabe K. RNA editing in the acceptor stem of squid mitochondrial tRNATyr. Nucleic Acids Res. 1996;24:4987–4991. doi: 10.1093/nar/24.24.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visomirski-Robic LM, Gott JM. Accurate and efficient insertional RNA editing in isolated Physarum mitochondria. RNA. 1995;1:681–691. [PMC free article] [PubMed] [Google Scholar]
- Visomirski-Robic LM, Gott JM. Insertional editing in isolated Physarum mitochondria is linked to RNA synthesis. RNA. 1997;3:821–837. [PMC free article] [PubMed] [Google Scholar]
- Yokobori S-I, Pääbo S. Transfer RNA editing in land snail mitochondria. Proc Natl Acad Sci. 1995a;92:10432–10435. doi: 10.1073/pnas.92.22.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori S-I, Pääbo S. tRNA editing in metazoans. Nature. 1995b;377:490. doi: 10.1038/377490a0. [DOI] [PubMed] [Google Scholar]
- Yokobori S, Pääbo S. Polyadenylation creates the discriminator nucleotide of chicken mitochondrial tRNATyr. J Mol Biol. 1997;265:95–99. doi: 10.1006/jmbi.1996.0728. [DOI] [PubMed] [Google Scholar]