Abstract
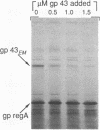
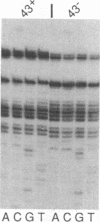
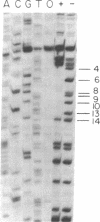
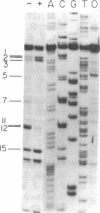
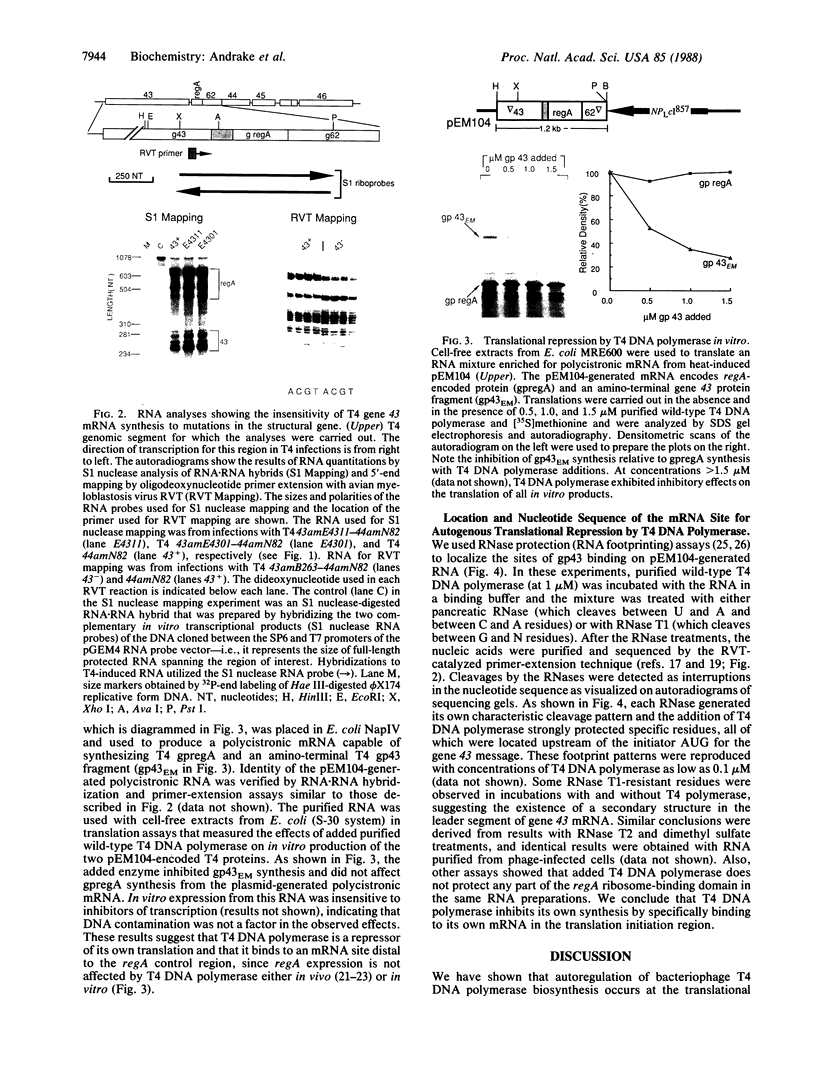
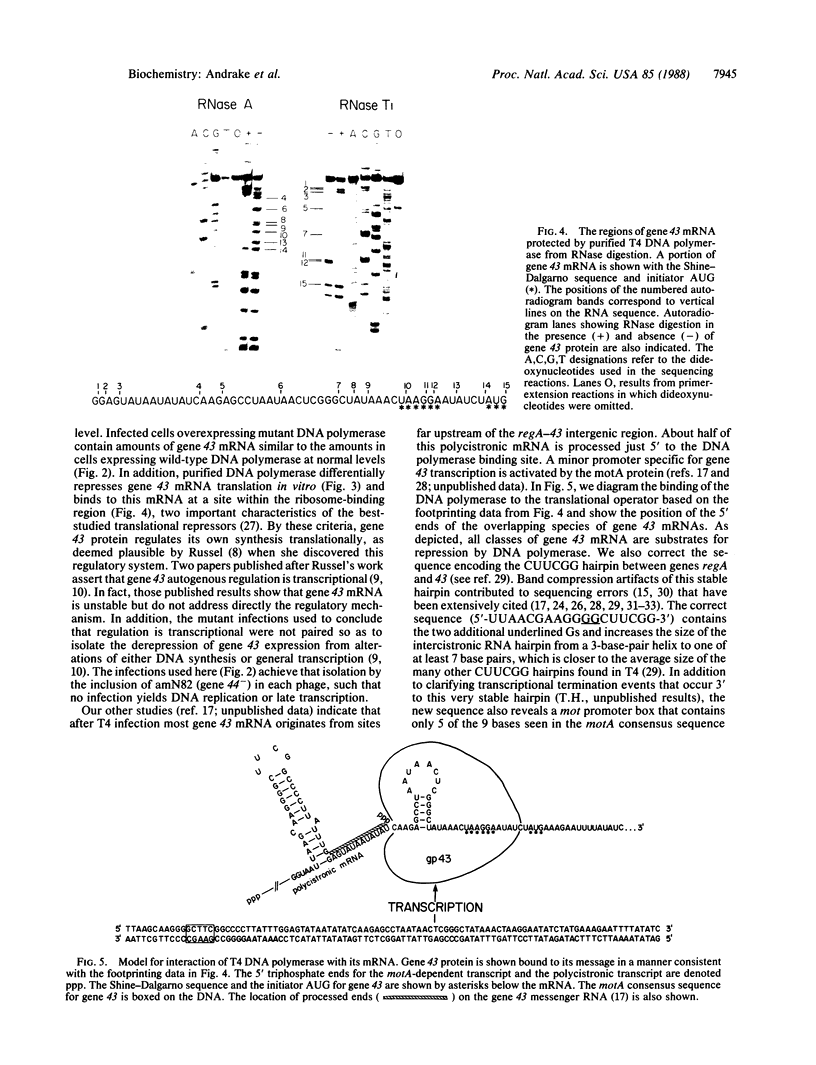
In bacteriophage T4 the protein product of gene 43 (gp43) is a multifunctional DNA polymerase that is essential for replication of the phage genome. The protein harbors DNA-binding, deoxyribonucleotide-binding, DNA-synthesizing (polymerase) and 3'-exonucleolytic (editing) activities as well as a capacity to interact with several other T4-induced replication enzymes. In addition, the T4 gp43 is a repressor of its own synthesis in vivo. We show here that this protein is an autogenous repressor of translation, and we have localized its RNA-binding sequence (translational operator) to the translation initiation domain of gene 43 mRNA. This mechanism for regulation of T4 DNA polymerase expression underscores the ubiquity of translational repression in the control of T4 DNA replication. Many T4 DNA polymerase accessory proteins and nucleotide biosynthesis enzymes are regulated by the phage-induced translational repressor regA, while the T4 single-stranded DNA-binding protein (T4 gp32) is, like gp43, autogenously regulated at the translational level.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adari H. Y., Rose K., Williams K. R., Konigsberg W. H., Lin T. C., Spicer E. K. Cloning, nucleotide sequence, and overexpression of the bacteriophage T4 regA gene. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1901–1905. doi: 10.1073/pnas.82.7.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi A., Spahr P. F. Nucleotide sequence at the binding site for coat protein on RNA of bacteriophage R17. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3033–3037. doi: 10.1073/pnas.69.10.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher M. S. Direct translation of a circular messenger DNA. Nature. 1968 Dec 14;220(5172):1088–1091. doi: 10.1038/2201088a0. [DOI] [PubMed] [Google Scholar]
- Calame K., Ihler G. Physical location of ribosome binding sites on lambda DNA. J Mol Biol. 1977 Nov;116(4):841–853. doi: 10.1016/0022-2836(77)90274-1. [DOI] [PubMed] [Google Scholar]
- Calame K., Ihler G. Visualization of ribosome--single-stranded DNA complexes in the electron microscope. Biochemistry. 1977 Mar 8;16(5):964–971. doi: 10.1021/bi00624a024. [DOI] [PubMed] [Google Scholar]
- Chao J., Leach M., Karam J. In vivo functional interaction between DNA polymerase and dCMP-hydroxymethylase of bacteriophage T4. J Virol. 1977 Nov;24(2):557–563. doi: 10.1128/jvi.24.2.557-563.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Waard A., Paul A. V., Lehman I. R. The structural gene for deoxyribonucleic acid polymerase in bacteriophages T4 and T5. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1241–1248. doi: 10.1073/pnas.54.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Burke R. L., Alberts B. M. Affinity purification of bacteriophage T4 proteins essential for DNA replication and genetic recombination. Proc Natl Acad Sci U S A. 1983 May;80(9):2442–2446. doi: 10.1073/pnas.80.9.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulford W., Model P. Specificity of translational regulation by two DNA-binding proteins. J Mol Biol. 1984 Feb 25;173(2):211–226. doi: 10.1016/0022-2836(84)90190-6. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Goulian M., Lucas Z. J., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXV. Purification and properties of deoxyribonucleic acid polymerase induced by infection with phage T4. J Biol Chem. 1968 Feb 10;243(3):627–638. [PubMed] [Google Scholar]
- Guild N., Gayle M., Sweeney R., Hollingsworth T., Modeer T., Gold L. Transcriptional activation of bacteriophage T4 middle promoters by the motA protein. J Mol Biol. 1988 Jan 20;199(2):241–258. doi: 10.1016/0022-2836(88)90311-7. [DOI] [PubMed] [Google Scholar]
- Hsu T., Wei R. X., Dawson M., Karam J. D. Identification of two new bacteriophage T4 genes that may have roles in transcription and DNA replication. J Virol. 1987 Feb;61(2):366–374. doi: 10.1128/jvi.61.2.366-374.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. M., Ao S. Z., Casjens S., Orlandi R., Zeikus R., Weiss R., Winge D., Fang M. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science. 1988 Feb 26;239(4843):1005–1012. doi: 10.1126/science.2830666. [DOI] [PubMed] [Google Scholar]
- Hughes M. B., Yee A. M., Dawson M., Karam J. Genetic mapping of the amino-terminal domain of bacteriophage T4 DNA polymerase. Genetics. 1987 Mar;115(3):393–403. doi: 10.1093/genetics/115.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam J. D., Bowles M. G. Mutation to overproduction of bacteriophage T4 gene products. J Virol. 1974 Feb;13(2):428–438. doi: 10.1128/jvi.13.2.428-438.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam J., Bowles M., Leach M. Expression of bacteriophage T4 genes 45, 44, and 62. I. Discoordinate synthesis of the T4 45- and 44-proteins. Virology. 1979 Apr 15;94(1):192–203. doi: 10.1016/0042-6822(79)90449-5. [DOI] [PubMed] [Google Scholar]
- Karam J., McCulley C., Leach M. Genetic control of mRNA decay in T4 phage-infected Escherichia coli. Virology. 1977 Feb;76(2):685–700. doi: 10.1016/0042-6822(77)90251-3. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Engman H. W., Yap W. Y. Tertiary initiation of replication in bacteriophage T4. Deletion of the overlapping uvsY promoter/replication origin from the phage genome. J Biol Chem. 1988 Aug 15;263(23):11348–11357. [PubMed] [Google Scholar]
- Krisch H. M., Van Houwe G., Belin D., Gibbs W., Epstein R. H. Regulation of the expression of bacteriophage T4 genes 32 and 43. Virology. 1977 May 1;78(1):87–98. doi: 10.1016/0042-6822(77)90080-0. [DOI] [PubMed] [Google Scholar]
- McPheeters D. S., Christensen A., Young E. T., Stormo G., Gold L. Translational regulation of expression of the bacteriophage T4 lysozyme gene. Nucleic Acids Res. 1986 Jul 25;14(14):5813–5826. doi: 10.1093/nar/14.14.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters D. S., Stormo G. D., Gold L. Autogenous regulatory site on the bacteriophage T4 gene 32 messenger RNA. J Mol Biol. 1988 Jun 5;201(3):517–535. doi: 10.1016/0022-2836(88)90634-1. [DOI] [PubMed] [Google Scholar]
- Menkens A. E., Kreuzer K. N. Deletion analysis of bacteriophage T4 tertiary origins. A promoter sequence is required for a rifampicin-resistant replication origin. J Biol Chem. 1988 Aug 15;263(23):11358–11365. [PubMed] [Google Scholar]
- Miller E. S., Karam J., Dawson M., Trojanowska M., Gauss P., Gold L. Translational repression: biological activity of plasmid-encoded bacteriophage T4 RegA protein. J Mol Biol. 1987 Apr 5;194(3):397–410. doi: 10.1016/0022-2836(87)90670-x. [DOI] [PubMed] [Google Scholar]
- Miller R. C., Jr, Young E. T., 2nd, Epstein R. H., Krisch H. M., Mattson T., Bolle A. Regulation of the synthesis of the T4 DNA polymerase (gene 43). Virology. 1981 Apr 15;110(1):98–112. doi: 10.1016/0042-6822(81)90011-8. [DOI] [PubMed] [Google Scholar]
- Nossal N. G. Prokaryotic DNA replication systems. Annu Rev Biochem. 1983;52:581–615. doi: 10.1146/annurev.bi.52.070183.003053. [DOI] [PubMed] [Google Scholar]
- Shub D. A., Gott J. M., Xu M. Q., Lang B. F., Michel F., Tomaschewski J., Pedersen-Lane J., Belfort M. Structural conservation among three homologous introns of bacteriophage T4 and the group I introns of eukaryotes. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1151–1155. doi: 10.1073/pnas.85.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer E. K., Rush J., Fung C., Reha-Krantz L. J., Karam J. D., Konigsberg W. H. Primary structure of T4 DNA polymerase. Evolutionary relatedness to eucaryotic and other procaryotic DNA polymerases. J Biol Chem. 1988 Jun 5;263(16):7478–7486. [PubMed] [Google Scholar]
- Trimble R. B., Maley Level of specific prereplicative mRNA's during bacteriophage T4 regA-, 43- and T4 43- infection of Escherichia coli B. J Virol. 1976 Feb;17(2):538–549. doi: 10.1128/jvi.17.2.538-549.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowska M., Miller E. S., Karam J., Stormo G., Gold L. The bacteriophage T4 regA gene: primary sequence of a translational repressor. Nucleic Acids Res. 1984 Aug 10;12(15):5979–5993. doi: 10.1093/nar/12.15.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M., Nossal N. G. Bacteriophage T4 gene 44/62 and gene 45 polymerase accessory proteins stimulate hydrolysis of duplex DNA by T4 DNA polymerase. J Biol Chem. 1982 Oct 25;257(20):12435–12443. [PubMed] [Google Scholar]
- Warner H. R., Barnes J. E. Deoxyribonucleic acid synthesis in Escherichia coli infected with some deoxyribonucleic acid polymerase-less mutants of bacteriophage T4. Virology. 1966 Jan;28(1):100–107. doi: 10.1016/0042-6822(66)90310-2. [DOI] [PubMed] [Google Scholar]
- Winter R. B., Morrissey L., Gauss P., Gold L., Hsu T., Karam J. Bacteriophage T4 regA protein binds to mRNAs and prevents translation initiation. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7822–7826. doi: 10.1073/pnas.84.22.7822. [DOI] [PMC free article] [PubMed] [Google Scholar]