Abstract
Proinsulin C-peptide is internalized into cells, but a function of its intracellular localization has not been established. We now demonstrate that, upon cellular entry, C-peptide is localized to the nucleoli, where it promotes transcription of genes encoding for ribosomal RNA. We find that C-peptide binds to histones and enhances acetylation of lysine residue 16 of histone H4 at the promoter region of genes for ribosomal RNA. In agreement with synchrony of ribosomal RNA synthesis and cell proliferation, we show that C-peptide stimulates proliferation in chondrocytes and HEK-293 cells. This regulation of ribosomal RNA provides a mechanism by which C-peptide can exert transcriptional effects and implies that the peptide has growth factor activity.
Keywords: Chromatin/Epigenetics, Growth Factors, Histones/Modification, Hormones/Peptide, RNA/Ribosomal RNA, RNA/Synthesis, Transcription/Regulation
Introduction
C-peptide is synthesized as a prohormone and, after processing, is released as a 31-amino acid peptide from pancreatic β-cells to the blood in amounts equimolar to those of insulin (1, 2). Type-1 diabetic patients therefore have a deficiency of C-peptide in addition to that of insulin (3) and suffer early from several conditions, including microalbuminuria and glomerular hyperfiltration, that can be improved by C-peptide administration (4–8). C-peptide binds specifically to cell membranes, which has been ascribed to a pertussis toxin-sensitive receptor (9). Both free and rhodamine-labeled C-peptide (Rh-C-peptide)2 is internalized into mouse fibroblast Swiss-3T3 and human embryonic kidney 293 (HEK-293) cells within 1 h in an active, pertussis toxin-sensitive fashion, implying C-peptide to be an intracrine factor (10). The internalization has been verified recently in other cell types and also shown to be mediated via endocytosis (11). Previous studies have investigated extracellular actions of C-peptide (12), as well as several effects presumably mediated through signaling pathways originating from the surface binding (13, 14; cf. 7). In contrast, intracrine factors concern peptides that exert effects within the cell of synthesis or a target cell (15), and several intracrine factors, including basic fibroblast growth factor and angiotensin II, regulate gene transcription upon nuclear binding (16). C-peptide lacks a nuclear localization signal and deviates from many intracrine factors by its very low pI, ∼3.5. Previous studies have implicated acidic peptides in transcriptional regulation (17, 18).
In the current study, we show that C-peptide upon nuclear entry is localized to nucleoli, where ribosomal DNA (rDNA) generates ribosomal RNA (rRNA) precursors. In the human genome, there are >400 copies of RNA-encoding genes, and epigenetic control mechanisms regulate to which extent they are transcribed (19). In active rRNA genes, promoters are unmethylated and associated with histones that are acetylated (20); in silent genes, the pattern is the opposite. Acetylated lysine residue 16 of histone 4 (H4K16Ac) has been shown to increase gene transcription (21, 22) and to be an epigenetic marker for actively transcribed rRNA genes (19, 23, 24). We now investigated C-peptide effects on rRNA synthesis and H4K16 acetylation, as well as interactions of C-peptide with histone proteins. We further investigated whether the ability of C-peptide to stimulate rRNA expression is accompanied by proliferation in chondrocytes, a type-1 diabetes relevant model system.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
HEK-293 and Swiss-3T3 cells were cultured as described (10). Human chondrosarcoma (HCS-2/8) cells were maintained in Dulbecco's modified Eagle's medium/F12 (Invitrogen) medium supplemented with 20% fetal bovine serum and 20 μg/ml gentamycin. Treatment with C-peptide was performed post-serum starvation with 1 μm concentrations for 24 h, unless otherwise described. Human C-peptide was used throughout the study.
Immunofluorescence and Confocal Microscopy Imaging
Cells were seeded on coverslips, allowed to settle, and serum-starved overnight. Swiss-3T3 cells were stimulated at 37 °C for 30–120 min with 1–5 μm Rh-C-peptide. HEK-293 cells were stimulated at 37 °C for 30–240 min with 0.1–5 μm C-peptide and probed with a polyclonal rabbit anti-acetyl-H4K16 antibody (Upstate Technology). Preparation of samples was performed as described (10). Cells were costained with Hoechst 33342 and SYTO RNASelect green fluorescent cell stain (Molecular Probes, Invitrogen) according to the manufacturer's protocol.
Luciferase Gene Reporter Assays
Transfections were performed in 24-well plates with Lipofectamine 2000 (Invitrogen) and 100 ng of both a luciferase reporter (pHrD-IRES-Luc) containing an internal ribosome entry site (IRES) downstream of the human rDNA promoter and pCMX-β-galactosidase reference plasmid per well. Four h post-transfection, cells were treated with C-peptide. Twenty four h after treatment, extracts were assayed for luciferase and β-galactosidase activity in a microplate luminometer/photometer reader (Orion Microplate Luminometer; Berthold detection systems).
C-peptide Interactions with Histone Proteins
For Biacore analysis, biotinylated C-peptide was immobilized on streptavidin-coated sensor chips (10). Histone extracts were prepared from Swiss-3T3 cells (Abcam), resuspended in Biacore running buffer (0.01 m Tris-HCl, pH 7.4, 3 mm EDTA, 0.005% surfactant P20, 0.15 m NaCl) and added at a flow rate of 5 μl/min.
For affinity precipitation, biotinylated C-peptide was immobilized on streptavidin beads according to the manufacturer's protocol (Dynabeads, Invitrogen). Histone extracts prepared from Swiss-3T3 cells were added for 60 min, after which beads were washed three times with buffer (150 mm sodium phosphate, 150 mm NaCl, pH 7.0) and eluted in sample loading buffer. Samples were separated on an SDS-PAGE gel and transferred to polyvinylidene difluoride membranes that were probed with an anti-acetyl-H4K16 antibody.
Mass Spectrometry Analysis of C-peptide Interactions
Protein bands were destained and digested with trypsin in a Massprep robotic system (Waters Corp.) (25). Digests were concentrated by evaporating solvents under a stream of nitrogen and analyzed by liquid chromatography tandem mass spectrometry, using Waters CapLC and Q-Tof Ultima API instruments. Data processing was made using Protein Lynx global server 2.3, and data base matching was made using Phenyx (PhenyxOnline, GeneBio) with a fragment tolerance of 0.1 Da. Uniprot 1.0 was used as the data bank.
rRNA Analysis
HEK-293 cells were treated with C-peptide for 90 min-24 h, and total RNA was extracted using phenol/chloroform/isoamyl alcohol. Samples were resolved on a 1% agarose gel. The 28 S and 18 S rRNA bands were visualized on a UV transilluminator.
RNA Isolation and PCR Analysis
Following treatment with C-peptide, RNA from both the control and treated cells was isolated using RNA-Bee (AMS Biotechnology Ltd.) or a RNeasy kit (Qiagen). cDNA synthesis was performed using 1 μg RNA with Superscript II reverse transcriptase (Invitrogen), according to the manufacturer's protocol. PCR amplifications of the 47 S ribosomal gene (forward 5′-CCT GTC GTC GGA GAG GTT GG-3′ and reverse 5′-ACC CCA CGC CTT CCC ACA C-3′) and the G3PDH housekeeping gene (forward 5′-ATG GCC TTC CGT GTC CCC ACT G-3′ and reverse 5′-TGA GTG TGG CAG GGA CTC CCC A-3′) were performed at an annealing temperature of 50 °C, 45 cycles, with reagents from Invitrogen, according to the manufacturer's protocol.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed with a ChIP assay kit (Upstate Biotechnology) according to the manufacturer's protocol. Chromatin was immunoprecipitated using the antibodies indicated. PCR amplification of the 47 S ribosomal promoter region (forward 5′-GTT TCC GAG ATC CCC GTG GGG AGC-3′ and reverse 5′-GAC AGC GTG TCA GCA ATA ACC CGG-3′) was performed at an annealing temperature of 55 °C, 45 cycles.
Cell Count Analysis
HEK-293 cells were seeded at a density of 100,000 cells/ml. One day later, cells were counted (0 h), and 0.3 μm C-peptide was added. Cell counts were then performed 24, 48, and 72 h post-treatment using a hemacytometer.
Cell Cycle Distribution Analysis by Flow Cytometry
The distribution of cells in the G1, S, and G2/M cell cycle phases was determined by DNA flow cytometry (26). Cells were fixed in 1% paraformaldehyde and subsequently frozen overnight in 95% ethanol. Thirty min prior to analysis, cells were resuspended in phosphate-buffered saline with 50 μg/ml propidium iodide and 5 μg/ml RNase A. Samples were analyzed on a fluorescence-activated cell sorting (FACS) Calibur flow cytometer, using Cell Quest software (BD Biosciences).
Flow Cytometric Analysis of Bromodeoxyuridine Incorporation
Cells were subjected to 10 μm bromodeoxyuridine (BrdUrd) for 15 min, harvested, and prepared according to a protocol for BrdUrd incorporation (BD Pharmingen). Samples were analyzed on a FACS-Calibur flow cytometer, using Cell Quest software (BD Biosciences).
Cell Death and Proliferation Analysis
Cells were seeded in 96- well plates 72 h prior to experiments and then treated with C-peptide for 72 h. Cell proliferation was analyzed by the WST-1 kit and BrdUrd incorporation enzyme-linked immunosorbent assay (ELISA) (Roche Diagnostics) as described by the manufacturer. Cell death was analyzed by a cell death detection ELISA kit (Roche Diagnostics) and Hoechst 33342 staining, according to the manufacturer's protocol.
Statistical Analysis
In gene reporter assays, statistical analysis was performed using two-tailed, paired student's t test. For proliferation and cell death assay analysis, differences between the groups were tested by one-way analysis of variance (the Holm-Sidak method). Results are presented as mean values ± S.E. *, p < 0.05, **, p < 0.01, and ***, p < 0.001.
RESULTS
C-peptide Is Localized to the Nucleolus
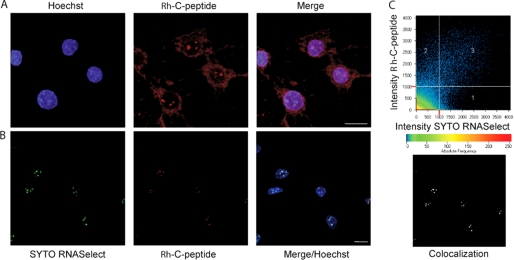
To gain insight into the intracellular localization of C-peptide, we monitored Rh-C-peptide (red, Fig. 1A) by confocal microscopy and found that it accumulates in a distinct nuclear compartment as illustrated by Hoechst nuclear staining (blue). Simultaneous SYTO RNASelect staining (green) for nucleoli established colocalization with Rh-C-peptide (colocalized pixels pseudo-colored white with the Zeiss software, Fig. 1, B and C).
FIGURE 1.
C-peptide is localized to nucleoli. A, confocal images of Rh-C-peptide (red) and nuclear Hoechst staining (blue) in Swiss-3T3 cells upon C-peptide exposure (1 h). B, colocalization of Rh-C-peptide (red) and the selective nucleolar marker SYTO RNASelect (green) shown by confocal fluorescence analysis as in A. C, colocalization of Rh-C-peptide (y-axe) and SYTO RNASelect (x-axe) staining pattern analyzed using the Zeiss Software (colocalized pixels are pseudo-colored white and are depicted in the lower panel). Scale bar, 20 μm.
C-peptide Rapidly Stimulates rRNA Synthesis
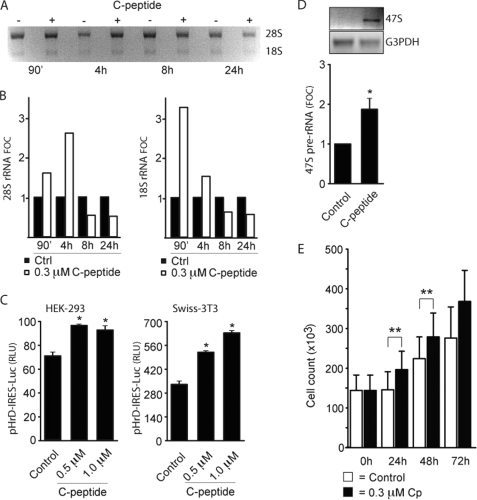
Our observation of C-peptide in the nucleoli raised the question whether the peptide can exert stimulatory effects on the production of rRNA. To address this, an equal number of serum-starved HEK-293 cells were either treated with C-peptide or untreated (90 min-24 h), and total RNA extracted was examined by RNA gel analysis. A marked increase in the levels of mature rRNAs (18 S and 28 S rRNA) was observed already within 90 min in cells treated with C-peptide versus those untreated (Fig. 2A and B). The rRNA increase after C-peptide treatment was confirmed with ribosomal precursor RNA (47 S) versus glyceraldehyde-3-phosphate dehydrogenase-specific primers by reverse transcriptase (RT-)PCR (Fig. 2D).
FIGURE 2.
C-peptide induces transcription of rDNA. A and B, equivalent samples of HEK-293 cells (∼25 × 106) were exposed to 0.3 μm C-peptide. After 90 min to 24 h, total RNA was extracted and the levels of 28 S and 18 S rRNA expression were examined by RNA gel analysis (∼4 μg/sample). Quantification of mature rRNA expression in treated relative to control samples as obtained by densitometric analysis (ImageJ software). The image shown is representative of three similar experiments. C, HEK-293 and Swiss-3T3 cells were cotransfected with β-galactosidase reference plasmid and pHrD-IRES-Luc, a luciferase reporter containing an IRES downstream of the human rDNA promoter, and treated with C-peptide. Cells were harvested after 24 h, and cell extracts were assayed for luciferase and β-galactosidase activity. Relative light units (RLU) were computed after normalization to β-galactosidase activity. D, RT-PCR analysis of pre-rRNA 47 S in HEK-293 cells (∼25 × 106) treated for 24 h with 0.3 μm C-peptide. Quantification of pre-rRNA expression relative to the untreated sample is presented, using the ImageJ software. E, HEK-293 cells (1 × 105/ml) were exposed to 0.3 μm C-peptide for 0–72 h, with cell counts performed at 24, 48, and 72 h post-treatment using a hemacytometer. Error bars represent S.E. (n = 3) and fold over control (FOC) was computed after normalization to intensity (treated versus control samples).
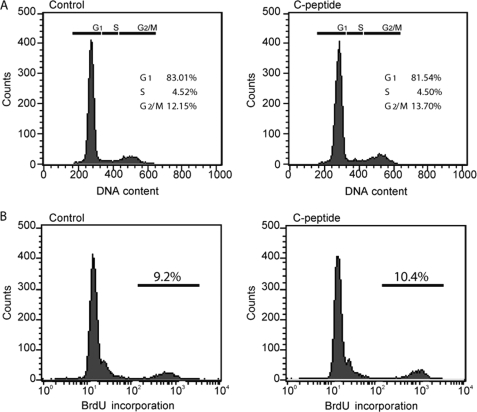
In addition, we investigated whether the early stimulation of rRNA synthesis observed in HEK-293 cells could be indirectly due to an early effect of C-peptide on the cell cycle. Cell cycle distribution was investigated by BrdUrd and propidium iodide staining, and no significant effect on the distribution between the different phases of the cell cycle was observed at early times (Fig. 3, A and B). These results indicate that C-peptide-induced rRNA production is not an indirect consequence of a modified cell cycle progression but instead suggested a direct transcriptional effect as the cause.
FIGURE 3.
Absence of early effects of C-peptide on cell proliferation. Swiss-3T3 cells were treated for 24 h with 1 μm C-peptide. A, distribution of the cells in the different phases of the cell cycle was determined by their DNA content upon propidium iodide staining and FACS analysis. B, cells were pulsed with BrdUrd for 15 min, and the proportion of cells in S phase (BrdUrd-incorporating cells) was monitored by FACS analysis. Similar results were obtained with HEK-293 cells.
As 18 S and 28 S rRNA are products of the 47 S transcription unit, we wanted to establish whether C-peptide regulates transcription of rDNA. HEK-293 and Swiss-3T3 cells were transfected with pHrD-IRES-Luc, a luciferase reporter construct that contains an IRES downstream of the human rDNA promoter (27). After 4 h of transfection, cells were treated with C-peptide for an additional 24 h. C-peptide treatment increased the luciferase expression from pHrD-IRES-Luc compared with that from untreated cells (Fig. 2C). The stimulatory effect of C-peptide on rDNA transcription was further confirmed in intact cells, by an increase of 47 S rRNA as measured by RT-PCR after 24 h (Fig. 2D). Hence, exposure of cells to C-peptide results in increased expression of rRNA already within 90 min, as shown both for pre-rRNA and mature RNA (18 and 28 S), establishing that C-peptide activates ribosomal gene expression. This activation was found to have late stage proliferative effects on HEK-293 cells as measured by cell counting (Fig. 2E).
C-peptide Regulates rRNA Synthesis via Epigenetic Stimulation
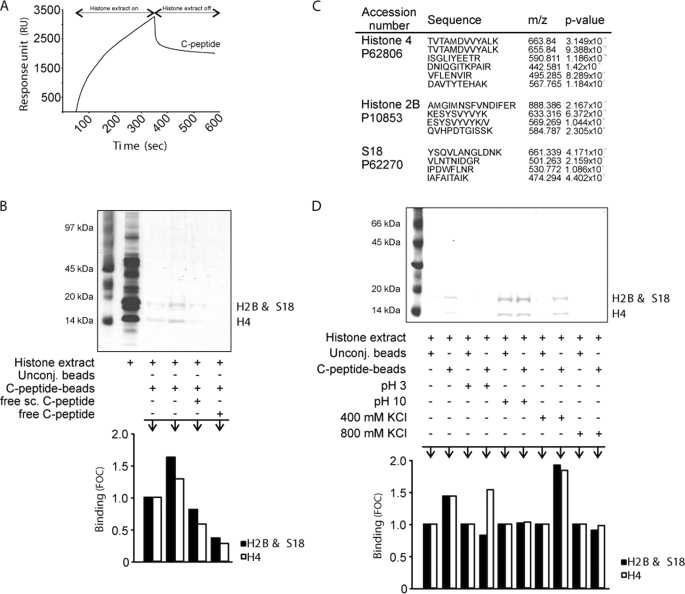
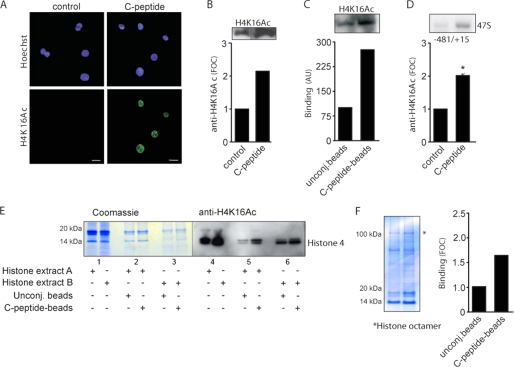
Because C-peptide localizes to nucleoli and increases transcription of 47 S rRNA, we next examined whether transcriptional activation is coordinated with epigenetic modifications of histones. We first investigated the binding of C-peptide to histone extracts from Swiss-3T3 cells using surface plasmon resonance, revealing that histone extracts bind to C-peptide (Fig. 4A). To identify the individual components binding to C-peptide, we performed a similar experiment, affinity-precipitating histone extracts using magnetic beads with immobilized C-peptide and subsequent SDS-PAGE analysis (Fig. 4B). Proteins were identified by mass spectrometry (liquid chromatography tandem mass spectrometry), and histone H4, histone H2B, and ribosomal protein S 18 were found as binding components (Fig. 4C). These interactions were found to be specific, in particular regarding H4, as preincubation of the histone extract with scrambled C-peptide did not reduce the binding of the extract to the same extent as with C-peptide (Fig. 4B). The interaction of C-peptide with H4 was pH-sensitive (Fig. 4D), and all three interactions were augmented at 400 mm KCl (Fig. 3D). Hence, C-peptide interacts specifically with histone proteins, in particular H4. Interactions with the histone octamer was also observed (Fig. 5F).
FIGURE 4.
Specific binding of C-peptide to histone H4. A, sensorgram showing binding of C-peptide to Swiss-3T3 histone extracts assessed by surface plasmon resonance technology. B, Swiss-3T3 histone extracts (∼2 μg) were affinity-precipitated with C-peptide-conjugated beads and control beads in the presence of additives as indicated, separated by SDS-PAGE, and stained with Coomassie Brilliant Blue. C, protein identification of tryptic digests from binding experiments. M denotes oxidized methionine. D, Swiss-3T3 histone extracts (∼2 μg) were affinity-precipitated with C-peptide-conjugated beads and control beads at pH 3 and 10, and in 400 and 800 mm KCl, analyzed by SDS-PAGE, and Coomassie staining. Quantification of band staining intensity with the ImageJ software and FOC was computed as described in Fig. 2. Unconj., unconjugated beads; sc., scrambled.
FIGURE 5.
C-peptide stimulates acetylation of H4K16. A, HEK-293 cells were treated for 1 h with 1 μm C-peptide, fixed, and submitted to immunofluorescence detection using antibodies directed against mono-acetylated H4K16 (green). Nuclei were costained with Hoechst 33342 (blue). Scale bar, 20 μm. B, HEK-293 cells were treated for 1 h with 1 μm C-peptide, and cell lysates were separated on an SDS-PAGE gel and transferred to polyvinylidene difluoride membranes that were probed with an anti-acetyl-H4K16 antibody. Quantification of H4K16Ac staining is shown. C, Swiss-3T3 histone extracts were affinity-precipitated with C-peptide-conjugated beads and control beads and analyzed by immunoblotting using H4K16Ac antibodies. Quantification of H4K16Ac in complexes is shown. Relative arbitrary units (AU) were computed after normalization to intensity (treated versus control samples). D, enrichment of H4K16Ac at the promoter region of rDNA was determined by ChIP analysis using chromatin prepared from control and 90 min C-peptide-treated HEK-293 cells (∼25 × 106). Samples were precipitated using H4K16Ac antibodies. Quantification of DNA band intensity is shown. E, Swiss-3T3 histone extracts A (normal) and B (from cells overexpressing SIRT1) were affinity-precipitated with C-peptide-conjugated beads and control beads and separated by SDS-PAGE. Samples were analyzed by Coomassie staining and transferred to polyvinylidene difluoride membranes that were probed with an anti-acetyl-H4K16 antibody. Quantification of band staining intensity is shown. F, Swiss-3T3 histone extracts (∼2 μg) were affinity-precipitated with C-peptide-conjugated beads and control beads in the presence of additives as indicated, separated by SDS-PAGE, and stained with Coomassie Brilliant Blue. Quantification of band staining intensity with the ImageJ software and fold over control (FOC) was computed as described in Fig. 2. Unconj., unconjugated.
Acetylation of H4K16 has been shown to be an epigenetic marker for actively transcribed rRNA genes (19, 23, 24). We therefore investigated whether C-peptide can promote this specific modification. Serum-starved HEK-293 cells were treated with C-peptide for 1 h, and cells were processed for immunofluorescence using antibodies directed against monoacetylated lysine 16 on H4. By confocal imaging analysis, we detected a pronounced increase in H4K16Ac staining intensity in HEK-293 cells treated with C-peptide (Fig. 5A). The increased H4K16 acetylation upon C-peptide exposure was further confirmed by immunoblotting of both control and treated cell extracts (Fig. 5B).
We then assessed whether C-peptide can promote this modification by acting directly at the promoter of 47 S rRNA. First, Swiss-3T3 histone extracts were affinity-precipitated with C-peptide conjugated beads, and the immunoblotted complexes were probed with H4K16Ac antibodies. This analysis showed that C-peptide interacts with Lys16-acetylated H4 (Fig. 5C). To assess whether the C-peptide induced acetylation of H4K16 occurs at the 47 S ribosomal promoter region, we performed ChIP assays. This revealed a pronounced C-peptide-responsive enrichment of acetylated H4K16 at the 47 S ribosomal promoter region (Fig. 5D). We conclude from these experiments that binding of C-peptide to H4 and the induction of H4K16 acetylation increases the expression of both pre-rRNA and mature RNA.
Since we also have shown binding of C-peptide to H4, we investigated whether the presence of Lys16-acetylation affects binding. Histone extracts from normal HEK-293 cells (extract A) and HEK-293 cells overexpressing sirtuin 1 (SIRT1) (extract B), a histone/protein deacetylase that can deacetylate histone 4 at Lys16 (28), were analyzed with magnetic beads as described above followed by SDS-PAGE analysis. Samples were analyzed both by Coomassie staining and Western blot analysis (anti-H4K16Ac) (Fig. 5E), revealing that C-peptide interacts more with nonacetylated than acetylated H4K16 (Fig. 5E, lanes 6 and 4).
C-peptide Regulates Proliferation via Stimulation of rRNA Synthesis
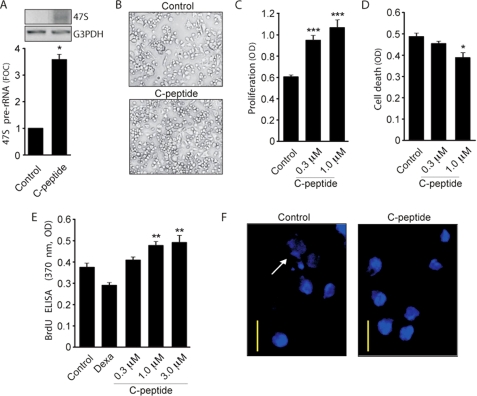
As the transcription of the rRNA-encoding gene is coupled to the potential of a cell to proliferate, we investigated whether the ability of C-peptide to stimulate rRNA gene expression is accompanied by proliferation. To study proliferation as an effect of increased rDNA transcription, we used HCS-2/8 chondrocytes derived from a human chondrosarcoma, a cell line previously studied as a model for chondrocyte differentiation (29, 30). We investigated whether C-peptide promotes rRNA gene expression. Chondrocytes were exposed to C-peptide for 72 h, total RNA was extracted, and pre-rRNA expression levels were measured by RT-PCR. C-peptide was found to induce expression of 47 S (Fig. 6A), indicating that the effect of C-peptide is not limited to Swiss-3T3 and HEK-293 cells but also occurs in chondrocytes. The stimulatory effect of C-peptide on rRNA synthesis suggests that the peptide can have proliferative effects. After 72 h of exposure to C-peptide, cell counting under a phase contrast microscope (Fig. 6B) and measurement with a cell proliferation kit (WST-1) and BrdUrd staining (Fig. 6, C and E) established that C-peptide exerts proliferative effects on chondrocytes. The incidence of cell death was monitored using a cell death detection ELISA kit and Hoechst staining, and C-peptide was found to significantly reduce spontaneous chondrocyte cell death (Fig. 6, D and F).
FIGURE 6.
C-peptide induces proliferation of chondrocytes. Human chondrocyte HCS-2/8 cells were exposed to C-peptide (0.3, 1, and 3 μm) for 72 h. A, total RNA was extracted from control and treated cells (∼1 × 106) and 47 S pre-rRNA expression analyzed by RT-PCR. Quantification of DNA band staining intensity with the ImageJ software is shown, and FOC was computed as described in Fig. 2. B, cells were observed under contrast phase microscopy and photographed. C, after treatment, the cell proliferation reagent WST-1 was added and incubation continued for 1 h at 37 °C. The amount of the formazan dye formed, as measured by ELISA, correlated with the number of metabolically active cells. D, cell death was assayed using the Cell Death Detection ELISA kit. The relative quantities of histone-complexed DNA fragments (mono- and oligonucleosomes) were measured by ELISA and correlate to the number of dead cells. Experiments were performed on cells that were serum-starved overnight. Error bars represent S.E. (n = 3). E, effect of different concentrations of C-peptide after a 72-h stimulation on BrdUrd incorporation in chondrocytes. Dexamethasone (25 μm) was used as a positive control. n = 5, p < 0.01. In B–E, ∼20 × 103 cells/sample were used. F, frequency of apoptotic cells (arrows) in control versus C-peptide treated cells as observed after staining with Hoechst. Scale bar, 20 μm.
DISCUSSION
Our previous work on C-peptide internalization, together with our finding of nucleolar C-peptide localization in this study, made us investigate whether C-peptide stimulates rDNA transcription. Insulin has been reported to stimulate rRNA production (31) but has not been detected in nucleoli. Indeed, both analysis of mature (18 and 28 S) rRNA and rRNA precursor (47 S) levels reveal that C-peptide rapidly stimulates rRNA synthesis, congruent with the reported internalization rate of C-peptide (10).
C-peptide differs from most biologically active peptides by being very acidic, with a pI ∼3.5. Other studies have implicated acidic peptides in transcriptional regulation (17, 18). One of them, prothymosin α, has been found to interact with free histone proteins (18) and to modulate the interaction of histone H1 with chromatin (32). To study how C-peptide can stimulate rDNA transcription, we first investigated whether C-peptide interacts with histone extracts and observed that it had sequence-specific interactions with predominantly H4, but also H2B and S 18. The interaction with S18 further confirmed the nucleolar localization of C-peptide, as ribosome assembly occurs in the nucleolus. The observed H4 interaction made us investigate whether C-peptide stimulates rRNA synthesis via epigenetic modification of H4, and we found that C-peptide can induce this modification already within 1 h.
To assess whether this modification is involved in the observed C-peptide-dependent stimulation of rRNA synthesis, we performed ChIP analysis and found increased amounts of acetylated H4K16 at the 47 S ribosomal promoter region in response to C-peptide treatment. As we show both binding of C-peptide to H4 and stimulation of acetylation, we investigated whether acetylation of H4K16 occurs at the 47 S promoter or whether C-peptide relocates already acetylated H4K16 to the 47 S promoter. Binding of C-peptide to H4 in a normal histone extract compared with that in a histone extract from cells overexpressing SIRT1, a histone deacetylase specific for H4K16 acetylation (28), revealed more interaction with nonacetylated than with acetylated H4K16. This could suggest that C-peptide interacts with core H4 and induces its acetylation at the promoter region of 47 S. Cell cycle analysis revealed that the increased rDNA transcription was not a general effect on cell cycle regulation, concurrent with the observation that cell number and total RNA content were different in control and treated cells only at late time points where rDNA stimulatory effects were no longer observed. Together, these results show a mechanism whereby C-peptide controls transcription of the rRNA-encoding gene, and suggests that C-peptide binding to H4 at the 47 S promoter region induces acetylation of Lys16.
Because long term complications of type-1 diabetes frequently develop despite insulin therapy and include reduced renal function ameliorated by C-peptide administration (cf. Ref. 7), we further studied our mechanistic findings of rDNA-associated proliferation with C-peptide also in chondrocytes, the principal cells in the cartilage portion of the growth plate. Chondrocytes constitute a type-1 diabetes relevant system because decreased bone strength and bone healing, indicative of a reduced number of proliferating chondrocytes, have been shown in type-1 diabetes patients and in rodent models of type-1 diabetes (33, 34). Indeed, C-peptide was now also in the chondrocyte system shown to stimulate rDNA transcription as well as cell proliferation. Suppression of cell death was observed, although that effect was less pronounced than on cell proliferation.
In conclusion, we report that proinsulin C-peptide regulates rRNA expression in several cell lines. We show that C-peptide localizes to the nucleoli and binds specifically to nucleolar proteins, including S 18 (35). Furthermore, we demonstrate that C-peptide binds to H4 and increases the acetylation of H4K16, an epigenetic marker for actively transcribed rRNA-encoding genes. In human chondrocytes, we further demonstrate that the rRNA regulatory effect of C-peptide stimulates proliferation. Hence, our data on regulation of rRNA expression by C-peptide provide a mechanism whereby C-peptide can act as a growth factor.
Acknowledgment
We thank Dr. S. T. Jacob (Ohio State University) for providing the pHrD-IRES-Luc construct.
This work was supported by the Swedish Research Council, the Ramón Areces Foundation, the Swedish Cancer Society, the Swedish Medical Society, the Swedish Children's Cancer Foundation, Karolinska Institutet, and the Knut and Alice Wallenberg Foundation.
- Rh-C-peptide
- rhodamine-labeled C-peptide
- Luc
- luciferase
- IRES
- internal ribosome entry site
- ChIP
- chromatin immunoprecipitation
- FACS
- fluorescence-activated cell sorting
- BrdUrd
- bromodeoxyuridine
- ELISA
- enzyme-linked immunosorbent assay
- RT
- reverse transcriptase
- Ac
- acetylated.
REFERENCES
- 1.Rubenstein A. H., Clark J. L., Melani F., Steiner D. F. (1969) Nature 224, 697–699 [Google Scholar]
- 2.Steiner D. F. (1967) Trans. N Y Acad. Sci. 30, 60–68 [DOI] [PubMed] [Google Scholar]
- 3.Cahill G. F., Jr. (1973) N. Engl. J. Med. 288, 1181–1182 [DOI] [PubMed] [Google Scholar]
- 4.Johansson B. L., Borg K., Fernqvist-Forbes E., Kernell A., Odergren T., Wahren J. (2000) Diabet. Med. 17, 181–189 [DOI] [PubMed] [Google Scholar]
- 5.Rebsomen L., Pitel S., Boubred F., Buffat C., Feuerstein J. M., Raccah D., Vague P., Tsimaratos M. (2006) Diabetes Metab. 32, 223–228 [DOI] [PubMed] [Google Scholar]
- 6.Sima A. A., Zhang W., Grunberger G. (2004) Exp. Diabesity Res. 5, 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wahren J., Ekberg K., Jörnvall H. (2007) Diabetologia 50, 503–509 [DOI] [PubMed] [Google Scholar]
- 8.Ekberg K., Brismar T., Johansson B. L., Lindström P., Juntti-Berggren L., Norrby A., Berne C., Arnqvist H. J., Bolinder J., Wahren J. (2007) Diabetes Care 30, 71–76 [DOI] [PubMed] [Google Scholar]
- 9.Rigler R., Pramanik A., Jonasson P., Kratz G., Jansson O. T., Nygren P., Stâhl S., Ekberg K., Johansson B., Uhlén S., Uhlén M., Jörnvall H., Wahren J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindahl E., Nyman U., Melles E., Sigmundsson K., Ståhlberg M., Wahren J., Obrink B., Shafqat J., Joseph B., Jörnvall H. (2007) Cell. Mol. Life Sci. 64, 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luppi P., Geng X., Cifarelli V., Drain P., Trucco M. (2009) Diabetologia 52, 2218–2228 [DOI] [PubMed] [Google Scholar]
- 12.Shafqat J., Melles E., Sigmundsson K., Johansson B. L., Ekberg K., Alvelius G., Henriksson M., Johansson J., Wahren J., Jörnvall H. (2006) Cell. Mol. Life Sci. 63, 1805–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohtomo Y., Aperia A., Sahlgren B., Johansson B. L., Wahren J. (1996) Diabetologia 39, 199–205 [DOI] [PubMed] [Google Scholar]
- 14.Li Z. G., Zhang W., Sima A. A. (2003) Diabetes Metab. Res. Rev. 19, 375–385 [DOI] [PubMed] [Google Scholar]
- 15.Re R. N. (1989) J. Mol. Cell. Cardiol. 21, 63–69 [DOI] [PubMed] [Google Scholar]
- 16.Re R. N., Cook J. L. (2007) Nat. Clin. Pract. Cardiovasc. Med. 4, 549–557 [DOI] [PubMed] [Google Scholar]
- 17.Calzuola I., Giavarini F., Sassi P., De Angelis L., Gianfranceschi G. L., Marsili V. (2005) Peptides 26, 2074–2085 [DOI] [PubMed] [Google Scholar]
- 18.Covelo G., Sarandeses C. S., Díaz-Jullien C., Freire M. (2006) J. Biochem. 140, 627–637 [DOI] [PubMed] [Google Scholar]
- 19.Grummt I., Pikaard C. S. (2003) Nat. Rev. Mol. Cell Biol. 4, 641–649 [DOI] [PubMed] [Google Scholar]
- 20.Santoro R., Li J., Grummt I. (2002) Nat. Genet. 32, 393–396 [DOI] [PubMed] [Google Scholar]
- 21.Akhtar A., Becker P. B. (2000) Mol. Cell 5, 367–375 [DOI] [PubMed] [Google Scholar]
- 22.Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. (2006) Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 23.Hirschler-Laszkiewicz I., Cavanaugh A., Hu Q., Catania J., Avantaggiati M. L., Rothblum L. I. (2001) Nucleic Acids Res. 29, 4114–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence R. J., Earley K., Pontes O., Silva M., Chen Z. J., Neves N., Viegas W., Pikaard C. S. (2004) Mol. Cell 13, 599–609 [DOI] [PubMed] [Google Scholar]
- 25.Jägerbrink T., Lexander H., Palmberg C., Shafqat J., Sharoyko V., Berggren P. O., Efendic S., Zaitsev S., Jörnvall H. (2007) Cell. Mol. Life Sci. 64, 1310–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph B., Marchetti P., Formstecher P., Kroemer G., Lewensohn R., Zhivotovsky B. (2002) Oncogene 21, 65–77 [DOI] [PubMed] [Google Scholar]
- 27.Ghoshal K., Majumder S., Datta J., Motiwala T., Bai S., Sharma S. M., Frankel W., Jacob S. T. (2004) J. Biol. Chem. 279, 6783–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt K., Zinn R. L., Ohm J. E., McGarvey K. M., Kang S. H., Watkins D. N., Herman J. G., Baylin S. B. (2006) PLoS genetics 2, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takigawa M., Tajima K., Pan H. O., Enomoto M., Kinoshita A., Suzuki F., Takano Y., Mori Y. (1989) Cancer Res. 49, 3996–4002 [PubMed] [Google Scholar]
- 30.Chagin A. S., Chrysis D., Takigawa M., Ritzen E. M., Sävendahl L. (2006) J. Endocrinol. 188, 193–203 [DOI] [PubMed] [Google Scholar]
- 31.Antonetti D. A., Kimball S. R., Horetsky R. L., Jefferson L. S. (1993) J. Biol. Chem. 268, 25277–25284 [PubMed] [Google Scholar]
- 32.Papamarcaki T., Tsolas O. (1994) FEBS letters 345, 71–75 [DOI] [PubMed] [Google Scholar]
- 33.Janghorbani M., Van Dam R. M., Willett W. C., Hu F. B. (2007) Am. J. Epidemiol. 166, 495–505 [DOI] [PubMed] [Google Scholar]
- 34.Santana R. B., Xu L., Chase H. B., Amar S., Graves D. T., Trackman P. C. (2003) Diabetes 52, 1502–1510 [DOI] [PubMed] [Google Scholar]
- 35.Grosso S., Volta V., Vietri M., Gorrini C., Marchisio P. C., Biffo S. (2008) Biochem. Biophys. Res. Commun. 376, 65–69 [DOI] [PubMed] [Google Scholar]