Abstract
Membrane traffic between the trans-Golgi network (TGN) and endosomes is mediated in part by the assembly of clathrin-AP-1 adaptor complex-coated vesicles. This process involves multiple accessory proteins that directly bind to the ear domain of AP-1γ via degenerate peptide motifs that conform to the consensus sequence ØG(P/D/E)(Ø/L/M) (with Ø being a large hydrophobic amino acid). Recently, γ-BAR (hereafter referred to as Gadkin for reasons explained below) has been identified as a novel AP-1 recruitment factor involved in AP-1-dependent endosomal trafficking of lysosomal enzymes. How precisely Gadkin interacts with membranes and with AP-1γ has remained unclear. Here we show that Gadkin is an S-palmitoylated peripheral membrane protein that lacks stable tertiary structure. S-Palmitoylation is required for the recruitment of Gadkin to TGN/endosomal membranes but not for binding to AP-1. Furthermore, we identify a novel subtype of AP-1-binding motif within Gadkin that specifically associates with the γ1-adaptin ear domain. Mutational inactivation of this novel type of motif, either alone or in combination with three more conventional AP-1γ binding peptides, causes Gadkin to mislocalize to the plasma membrane and interferes with its ability to render AP-1 brefeldin A-resistant, indicating its physiological importance. Our studies thus unravel the molecular basis for Gadkin-mediated AP-1 recruitment to TGN/endosomal membranes and identify a novel subtype of the AP-1-binding motif.
Keywords: Cell/Endocytosis, Cell/Exocytosis, Membrane/Trafficking, Methods/NMR, Protein/Palmitoylation, Subcellular Organelles/Endosomes
Introduction
Membrane traffic within eukaryotic cells is mediated in part by coated transport vesicles or tubules that deliver transmembrane cargo proteins and lipids to multiple intracellular destinations along the secretory and endocytic pathways (1, 2). Clathrin coat assembly (3) at the plasma membrane, the trans-Golgi network (TGN)3 /recycling endosomal boundary, and on endosomes requires mono- and heterotetrameric adaptors and a variety of accessory proteins (4). AP-1 adaptor-containing clathrin coats have been detected at the TGN, on endosomal vesicles (5–8), and on transferrin receptor (TfR)-containing tubular recycling endosomes (REs) in Madin-Darby canine kidney cells (9). AP-1 is composed of two large chains (γ1 and β1), a medium chain (μ1) required for sorting of YXXØ motif-containing transmembrane cargo, and a small chain (σ1) that fulfills a structural role in complex assembly. The ear domain of its γ1-adaptin subunit (γ-ear) serves as a platform for the recruitment of accessory and regulatory proteins, including aftiphilin, epsinR/enthoprotin/Clint (10–13), γ-synergin (14), eps15, Rabaptin-5, p56, NECAPs (15), and γ-BAR (8) (see below) to AP-1-coated membrane domains via recognition of ØG(P/D/E)(Ø/L/M) peptide motifs (with Ø being a large hydrophobic amino acid) (14, 16, 17). Most of these accessory proteins can also associate via the same peptide motifs with γ-adaptin-related monomeric GGA1–3 adaptors at the TGN.
Morphological as well as live cell-imaging studies have shown that AP-1 partitions between endosomal membranes and the TGN (6, 7), but how precisely such partitioning is accomplished has remained unclear. Membrane recruitment of AP-1 requires nucleotide exchange on the small GTPase Arf1, a process blocked by the fungal metabolite brefeldin A (BFA). Furthermore, assembly of AP-1 coats at the TGN is facilitated by phosphatidylinositol 4-kinase-mediated production of phosphoinositides (18), which may act as membrane-embedded cofactors together with Arf1-GTP and cargo proteins. Based on their molecular properties and intracellular localization, it seems likely that accessory proteins contribute to the specific targeting of AP-1 to the TGN or to endosomal membranes (4, 14, 16, 17). In support of this view, it has been shown that γ-BAR, an AP-1-binding accessory protein localized predominantly to perinuclear endosomes, can stabilize AP-1, but not the closely related GGA proteins at membranes even in the presence of BFA (8). The precise mechanism underlying this activity of γ-BAR has not been revealed. We have recently shown that γ-BAR directly interacts with the light chains of kinesin KIF5, thereby contributing to the peripheral movement of TGN-derived endosomal vesicles (19). Because γ-BAR does not harbor a curvature-sensing BAR (BIN/amphiphysin/Rvs167) domain, we propose to refer to this protein as Gadkin, for γ1-adaptin and kinesin interactor, to avoid possible confusion with BAR domain proteins.
Here we have addressed the mechanism by which Gadkin associates with AP-1 and with TGN/endosomal membranes. We show that Gadkin is a peripheral S-palmitoylated membrane protein and that this modification is absolutely required for its recruitment to TGN/endosomal membranes in living cells. Further biochemical and biophysical studies led to the discovery of a novel subtype of AP-1-binding motif within the largely unstructured Gadkin that specifically associates with the γ1-adaptin ear domain. Mutational inactivation of this novel type of motif either alone or in combination with three more conventional AP-1γ-binding peptides causes Gadkin to mislocalize to the cell surface and interferes with its ability to render AP-1 BFA-resistant. Our studies thus unravel the molecular basis for Gadkin-mediated AP-1 recruitment to TGN/endosomal membranes and identify a novel subtype of AP-1-binding motif.
EXPERIMENTAL PROCEDURES
Plasmids, Mutagenesis, and Antibodies
The full-length Gadkin coding sequence or truncated versions thereof were cloned into pEGFP-N (Clontech) for mammalian expression of C-terminally eGFP-tagged proteins. Vectors for the mammalian expression of N-terminally FLAG-tagged proteins were custom-made based on the pcDNA3 backbone (Invitrogen). For bacterial expression of N-terminally GST-tagged proteins we used the pGEX 4T-1 vector (Amersham Biosciences), for expression of His6-tagged proteins we used the pET28a(+) vector. Point mutants were generated using the QuikChange II Kit (Stratagene). The presence of the mutations was verified by double strand DNA sequencing. The following antibodies were used for detection of proteins in immunoblots, in immunofluorescence, and for immunoprecipitations: monoclonal antibodies against γ-adaptin (BD Biosciences), TfR (clone H68.4, Zymed Laboratories Inc.), clathrin heavy chain (clone TD.1), FLAG tag (M2, Sigma), glyceraldehyde-3-phosphate dehydrogenase (clone 71.1, Sigma), actin (clone AC-15, Sigma), GFP (clone 1E4, Stressgen), His tag (Novagen), and COPI (kind gift from F. T. Wieland, Heidelberg, Germany). Polyclonal antibodies against Gadkin were raised in rabbits by injecting a purified His-tagged Gadkin (amino acids 52–302). The serum was affinity-purified on cross-linked GST-tagged Gadkin (amino acids 52–302) and tested for specificity. Fluorescent dye-conjugated secondary antibodies (Alexa488, Alexa568, and Alexa594) were purchased from Molecular Probes. HRP-coupled secondary antibodies were from Dianova.
Cell Culture and Transfections
HeLa, COS7, and HEK293 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with fetal calf serum and antibiotics. Cell lines were transfected with plasmid DNA using Lipofectamine2000 (Invitrogen).
Immunofluorescence Microscopy
Cells seeded on glass coverslips were fixed with 4% paraformaldehyde, 4% sucrose in PBS, pH 7.4, for 20 min. After washing 3 times in PBS and permeabilizing for 20 min in blocking solution (30% goat serum, 0.3% Triton X-100 in PBS), primary antibody was applied in the same buffer for 1 h at room temperature. PBS-washed coverslips were incubated with secondary antibodies in blocking solution for 1 h at room temperature. Afterward coverslips were washed again three times in PBS, mounted onto glass slides using Immumount solution (Thermo Electron) supplemented with 1 μg/ml DAPI. Image acquisition was performed with a motorized Axiovert 200M inverted microscope (Carl Zeiss) equipped with a Stallion System (Intelligent Imaging Innovations). Data analysis was performed with Slidebook Software (Intelligent Imaging Innovations).
BFA Treatment
HeLa cells seeded on glass coverslips were incubated with 5 μg/ml BFA for 15 min at 37 °C in serum-containing medium prior to paraformaldehyde fixation and processing for indirect immunofluorescence microscopy.
2-Bromopalmitate Treatment
HeLa cells were preincubated with 50 μm 2-bromopalmitate (dissolved in EtOH) or the same volume of EtOH for 2 h before transient transfection. Afterward the 2-bromopalmitate treatment was continued for 20 h before fixation and processing for indirect immunofluorescence microscopy.
Cell Fractionation
Cells were homogenized in 20 mm HEPES, pH 7.4, 2 mm MgCl2, 100 mm NaCl supplemented with 1 mm phenylmethylsulfonyl fluoride and mammalian protease inhibitor mixture (Sigma) by passing 15 times through a 27-gauge needle. The homogenate was centrifuged at 700 × g for 3 min to remove debris. The supernatant was centrifuged at 180,000 × g. The soluble fraction was solubilized with Triton X-100 and centrifuged again at 180,000 × g. The membrane pellet was rinsed twice with buffer, resuspended in buffer containing 1% Triton X-100, incubated for 15 min on ice for solubilization, and centrifuged at 20,000 × g for 15 min. The supernatants of the soluble and membrane protein fractions were analyzed by SDS-PAGE.
Affinity Chromatography and Immunoprecipitation Experiments
GST fusion proteins were expressed in Escherichia coli and purified from benzonase-treated bacterial lysates (to remove possible nucleic acid contaminants) using GST-bindTM resin (Novagen) according to standard protocols. Adult rat brain tissue was homogenized in 320 mm sucrose, 4 mm HEPES, pH 7.4, supplemented with 1 mm phenylmethylsulfonyl fluoride and mammalian protease inhibitor mixture (Sigma), and the postnuclear supernatant was extracted with 1% Triton X-100. The buffer concentration in the extract was adjusted to 20 mm HEPES, pH 7.4, 2 mm MgCl2, and 80–100 mm KCl or NaCl. Cleared protein extracts were prepared by ultracentrifugation at 180,000 × g. Protein extracts from cultured mammalian cells were prepared in lysis buffer containing 20 mm HEPES, pH 7.4, 2 mm MgCl2, 100 mm NaCl, and 1% Triton X-100 supplemented with 1 mm phenylmethylsulfonyl fluoride and mammalian protease inhibitor mixture (Sigma). Cell extracts were cleared by ultracentrifugation. For affinity purifications, GST fusion proteins were incubated with rat brain extracts with a protein concentration of 4–6 mg/ml for 2 h at 4 °C. Following extensive washes bound proteins were eluted in sample buffer. For immunoprecipitation experiments, antibodies were immobilized on Protein A/G-agarose beads (Santa Cruz Biotechnology) and incubated with either rat brain extracts as above or with cleared cell extracts (total protein concentration: 0.5–1 mg/ml) for 4 h at 4 °C under gentle agitation. Beads were washed extensively and eluted with sample buffer. Samples were analyzed by SDS-PAGE and immunoblotting. For direct binding assays immobilized GST fusion proteins (25 μg) were incubated for 1 h at 4 °C with 25 μg of recombinant His6-tagged Gadkin proteins and washed extensively. Complexes were analyzed by SDS-PAGE with Coomassie Blue staining.
Detection of Protein Palmitoylation Using the Biotin-switch Method
Protein palmitoylation was analyzed according to a previous study (20) using the biotin-switch method. FLAG-Gadkin-(1–140) was immunoprecipitated from transfected COS7 cells (see above) using anti-FLAG antibodies. Beads were extensively washed in 50 mm Tris buffer, pH 7.4, containing 150 mm NaCl, 2 mm MgCl2, and 0.1% Triton X-100 and treated with 25 mm N-ethylmaleimide (diluted from a freshly prepared 1 m stock solution in EtOH) in buffer for 30 min at room temperature to quench free sulfhydryl (SH)-groups. After washing, beads were treated with 1 m hydroxylamine, pH 7.4 (freshly prepared), to liberate cysteine SH-groups from palmitate moieties or 1 m Tris, pH 7.4, as control for 1 h at room temperature. Beads were washed again, and liberated cysteine SH-groups were biotinylated using 1-biotinamido-4-(4′-[maleimidoethylcyclohexane]carboxamido)butane (Pierce). Beads were extensively washed and eluted in sample buffer. Samples were analyzed by SDS-PAGE and immunoblotting. Biotinylated proteins were detected using streptavidin-HRP.
Protein Expression and Purification for NMR Studies
His6-tagged γ-ear in pET28a(+) was expressed in E. coli BL21(DE3) strain. Cultures were grown at 37 °C in M9 medium (2× salt) supplemented with 15N ammonium chloride to produce uniformly 15N-labeled proteins. Following a 4-h induction with 1 mm isopropyl-l-d-thiogalactose at 37 °C, His6-γ-ear was purified with HisTrap and Superdex 75 columns using the Akta-Prime system (Amersham Biosciences). The NMR samples contained 0.5 mm γ-ear protein in buffer (90% H2O/10% D2O, 50 mm sodium phosphate, pH 7.5, 150 mm NaCl). His6-tagged Gadkin in pET28a(+), transformed in E. coli BL21(DE3) strain was expressed in 2×YT medium (16 g/liter tryptone, 10 g/liter yeast extract, 5 g/liter NaCl) and purified using the same procedures as for γ-ear protein. Gadkin was added to γ-ear protein to obtain ratios 6:1, 3:1, 1.5:1, and 1:1.
NMR Spectroscopy
NMR experiments were performed at 300 K on Bruker DMX750 and DRX600 spectrometers in standard configuration with triple resonance cryogenic probes equipped with self-shielded single axis gradient coils. Spectra were processed using XWinNMR 2.6 (Bruker BioSpin GmbH, Rheinstetten, Germany) and analyzed using CCPNmrAnalysis (21). Following the acquisition of a 1H-15N HSQC spectrum of [15N]-γ-ear (0.5 mm in 90% H2O/10% D2O, 50 mm sodium phosphate, pH 7.5, 150 mm NaCl) at 750 MHz, WT Gadkin (Δ51) was added stepwise, and further HSQC spectra were recorded at AP-1γ-ear to Gadkin molar ratios of 6:1, 3:1, 1.5:1, and 1:1. The assignments for γ-ear were obtained from Biological Magnetic Resonance Data Bank entry 5761 (17). Two-dimensional HSQC spectra of 2H,13C,15N-labeled Gadkin Δ51 (0.5 mm in 90% H2O/10% D2O, 50 mm sodium phosphate, pH 7.5, 150 mm NaCl) were recorded at 750 MHz. WT AP-1γ-ear was added stepwise and further HSQC spectra were recorded at Gadkin-to-AP-1γ-ear-ratios of 5:1, 2:1, 1:1, and 1:2. Three-dimensional HNCO, HNCA, HN(CO)CA, HN(CA)CO, and HNCACB spectra were recorded at this final condition at 600 MHz.
CD Spectroscopy
CD spectra of 2.4 μm His6-Gadkin Δ51 in PBS containing 10 mm dithiothreitol were recorded at increasing temperatures from 25 °C to 55 °C on a JASCO J-600 spectropolarimeter (wavelength 245–190 nm, speed 20 nm/min, and resolution 0.2 nm) using a quartz cell of path length 2 mm. Three individual spectra were accumulated. The baseline was corrected by subtracting buffer runs. Selcon3 and Contin programs were applied for calculation of secondary structure content.
Limited Proteolysis
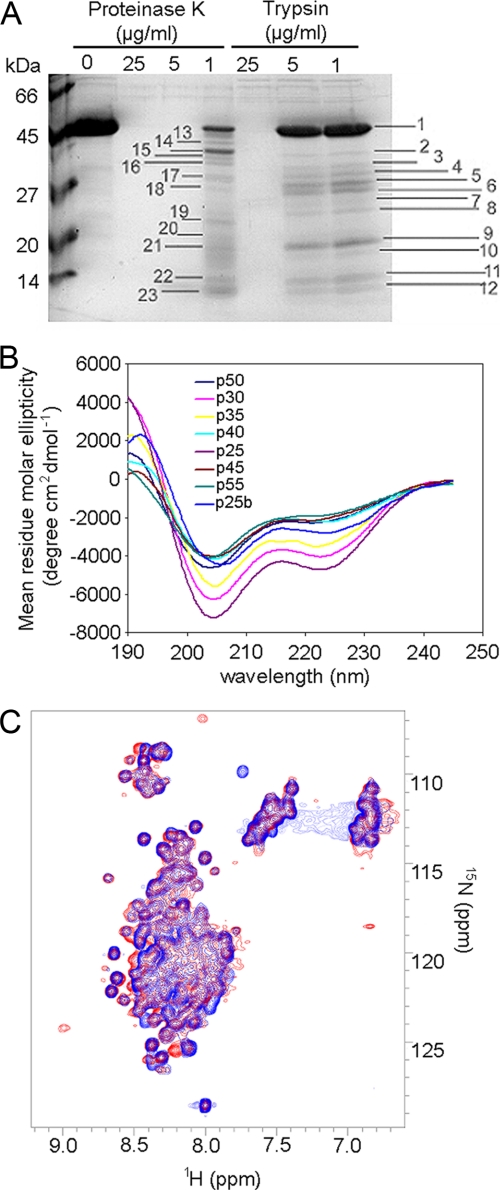
Limited proteolysis was performed with different concentrations of proteinase K and trypsin (1, 5, and 25 μg/ml) at 37 °C for 10 min. Protein bands were analyzed by in-gel digestion followed by mass fingerprinting. All fragments could be unequivocally assigned to the N-terminal portion of Gadkin (Δ51) (not shown).
Analytical Ultracentrifugation
Analytical ultracentrifugation experiments were performed in a Beckman Coulter XL-ITM analytical ultracentrifuge using the interference optics of the instrument. All experiments were performed in a buffer containing 20 mm K2PO4, pH 7.4, 150 mm NaCl, and 10 mm dithiothreitol at 20 °C. Conversion of the signal in fringe units to molar quantities used a value for the specific refractive index increment of 3.29 fringes·mg−1·ml−1. Data were acquired every 30 s until the major fraction of material had sedimented. Sedimentation traces were analyzed with the c(S) method implemented in Sedfit8.9 (National Institutes of Health). In the case of pure components, data were also analyzed by using a discrete-species model to obtain s, D, and Mw directly. Auxiliary parameters and hydrodynamic values were calculated and corrected to standard conditions using Sednterp. Preparations of pure proteins were examined at different concentrations, spanning at least one order of magnitude on the concentration scale. In the case of mixtures of AP-1γ-ear with Gadkin (Δ51) and the respective mutants, the concentration of Gadkin (Δ51) was held constant at ∼30 μm, and increasing amounts of γ-ear were added so that molar ratios (AP-1γ-ear/Gadkin (Δ51)) ranged between 0.25 and 4. One cell was filled with each of the pure components at ∼30 μm, respectively. The resulting c(S) distributions were integrated to obtain the loading concentration of protein and the average s value. The concentration of AP-1γ-ear was obtained from the concentration of total protein by subtracting the concentration of Gadkin (Δ51) that was independently determined in the same experiment. The concentration of pure AP-1γ-ear served as a control for the expected concentration of AP-1γ-ear. Average s values were plotted as a function of [AP-1γ-ear] and fitted to a model of two independent binding sites using Sedphat4.06 (NIH).
Peptide Synthetic Peptide Array on Nitrocellulose (SPOT) Synthesis
SPOT syntheses were performed on amino-functionalized Whatman 50 cellulose membranes (Whatman, Maidstone, UK) using a semi-automatic SPOT synthesizer (INTAVIS AG, Köln, Germany). The pepscan arrays were synthesized on an ester-type cellulose membrane, amino-modified by two β-alanine units (22). The substitutional and length analyses were prepared on a cellulose-(3-amino-2-hydroxypropyl)-ether (CAPE) membrane for a better signal-to-noise ratio. SPOT synthesis was predominantly performed as described in the standard SPOT synthesis protocol (23). For design and arrangement of the substitutional analysis, length analysis, and the pepscan array our in-house software (LISA) was applied. The peptide (His)6 and the peptide LASDLIVPRR were used as control spots at the upper left, upper right, and lower right corners of the synthesized arrays.
AP-1γ-ear Binding Studies of Cellulose-bound Peptides
All incubations and washing steps were carried out under gentle shaking. After washing the membrane for 10 min with EtOH and three times for 10 min with Tris-buffered saline (TBS, 50 mm Tris(hydroxymethyl)aminomethane, 137 mm NaCl, 2.7 mm KCl, adjusted to pH 8 with HCl), the membrane-bound peptide arrays were blocked for 3 h with blocking buffer (10% blocking reagent (CRB, Norwich, Great Britain) and 1% sucrose in 1:10 TBS buffer). The Gadkin-deduced cellulose-bound pepscan array was incubated with the His6 tag-labeled AP-1γ-ear (30 μg/ml), dissolved in the same blocking buffer at 4 °C for 14 h. After washing three times for 10 min with TBS, the HRP-labeled anti-His6 IgG antibody (Roche 1965085) was added (1:500). Subsequently, the membrane was washed three times for 10 min with TBS. Analysis of peptide-bound AP-1γ-ear was carried out using a chemiluminescence substrate and the Lumi-ImagerTM instrument (Roche Diagnostics, Basel, Switzerland). The substitutional and length analyses were probed for AP-1γ-ear binding with a GST-tagged moiety. Peptide-bound γ-ear was detected by adding first the anti-GST IgG antibody (G1160, Sigma) at a concentration of 1 μg/ml in blocking buffer (2 h) and subsequently after washing (3 times as described above) by adding the HRP-labeled anti-mouse IgG antibody (1 μg/ml in blocking buffer). As described above chemiluminescence was used for visualization.
RESULTS
Association of Gadkin with Membranes Is Independent of Its Ability to Bind to the AP-1 Complex
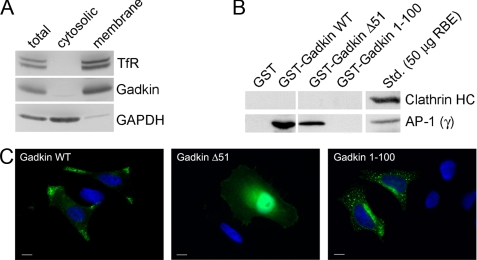
Gadkin has been identified as an AP-1-interacting peripheral membrane protein. Indeed, when HeLa cells were subjected to subcellular fractionation, endogenous Gadkin was associated with the membrane pellet, similar to the integral membrane protein TfR, whereas the soluble enzyme glyceraldehyde-3-phosphate dehydrogenase was present in the cytosolic fraction (Fig. 1A). Previous work has suggested that membrane targeting of Gadkin is mediated by the AP-1 complex (8). We tested this model by analyzing truncation mutants of Gadkin either lacking its N-terminal 51 residues (Gadkin Δ51) or its C-terminal 202 amino acids (Gadkin-(1–100)). GST-fused WT Gadkin or Gadkin Δ51 but not Gadkin-(1–100) effectively associated with AP-1 from Triton X-100-solubilized rat brain extracts. None of the fusion proteins tested bound to clathrin (Fig. 1B). To investigate the localization of these truncation mutants in living cells we transiently expressed Gadkin mutants tagged at their C-terminal end with eGFP in HeLa cells. Overexpressed WT Gadkin localized to peripheral puncta (Fig. 1C) corresponding to Tf- and AP-1-positive TGN-derived endosomal vesicles (EVs) that accumulate in the periphery due to the tight association of Gadkin with the plus-end-directed microtubule motor protein kinesin KIF5 (19). A similar dispersion of AP-1- and Tf-positive endosomes was also observed upon overexpression of c-myc- or FLAG-tagged Gadkin (data not shown). Surprisingly, we observed that Gadkin Δ51 displayed a cytoplasmic distribution in HeLa cells (Fig. 1C) despite retaining the ability to associate with AP-1 (compare Fig. 1B), whereas Gadkin-(1–100) was concentrated in the perinuclear area and on punctate, presumably endosomal structures (Fig. 1C) but failed to stably bind to AP-1 (Fig. 1C). These data therefore suggested that Gadkin might associate with membranes independently of its interaction with the AP-1 complex. Based on the truncation mutants analyzed so far, membrane binding of Gadkin involves its 100 N-terminal residues.
FIGURE 1.
The association of Gadkin with membranes is independent of its binding to AP-1. A, Gadkin is a membrane-associated protein. Total, cytosolic, and membrane fractions were prepared from HeLa cells. 50 μg of protein extracts was used for SDS-PAGE and immunoblotting using antibodies against Gadkin, the transmembrane protein TfR, and the soluble enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH). B and C, membrane association of Gadkin does not depend on its AP-1 binding ability. B, 20 μg of GST (control), GST-tagged Gadkin WT, or its truncation mutants Δ51 and 1–100 (bound to glutathione beads) were incubated with 1.5 mg of Triton X-100-solubilized rat brain extract. Bound proteins were analyzed by SDS-PAGE and immunoblotting using antibodies against AP-1 γ-adaptin or clathrin heavy chain (HC). Std, standard. C, subcellular distribution of Gadkin-eGFP truncation mutants. The localization of eGFP-Gadkin (full-length WT, Δ51, or amino acids 1–100) transiently expressed in HeLa cells was analyzed by fluorescence microscopy (green). DAPI-stained nuclei are shown in blue. Scale bar, 10 μm.
Membrane Targeting of Gadkin Requires Its S-Palmitoylation
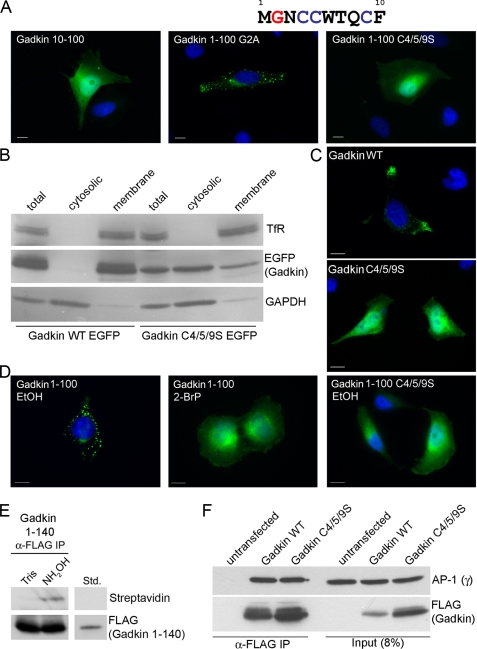
Protein association with membranes frequently is mediated by post-translational lipid modifications, including prenylation, N-myristoylation, and S-palmitoylation. Because membrane association of Gadkin does not depend on C-terminal determinants and because Gadkin lacks any obvious prenylation sites, we focused on N-myristoylation and S-palmitoylation as possible mechanisms. Gadkin contains a single glycine residue in position 2, representing a putative N-myristoylation site, as well as three cysteine residues that could potentially form acceptor sites for S-linked palmitates (Fig. 2A). To analyze the potential contribution of either type of modification to membrane targeting of Gadkin-(1–100), we created a series of mutant proteins in which either its 9 N-terminal residues had been deleted (10–100) or potential lipid modification sites had been inactivated by mutation. Gadkin-(10–100), indeed, displayed a completely cytoplasmic distribution, whereas Gadkin-(1–100, G2A) associated with peripheral membrane puncta (Fig. 2A), similar to its WT counterpart. Mutation of all three potentially palmitoylated cysteines to serines abrogated membrane targeting of Gadkin-(1–100) completely, suggesting that a cysteine-based mechanism, most likely S-palmitoylation is required for membrane association of Gadkin-(1–100). By contrast, mutation of either cysteine alone had no effect (data not shown). To analyze whether cysteines 4, 5, and 9 are also required for targeting of full-length Gadkin to membranes, we created a triple point mutant, in which these cysteines had been replaced by serines. As shown in Fig. 2B Gadkin (C4/5/9S) had largely lost its association with membranes in subcellular fractionation experiments. Moreover, transfected Gadkin (C4/5/9S)-eGFP displayed a cytoplasmic distribution in transfected HeLa cells (Fig. 2C).
FIGURE 2.
The association of Gadkin with membranes requires its S-palmitoylation. A–C, N-terminal cysteines are critical for the association of Gadkin with membranes. A, sequence of the N-terminal ten amino acids of human Gadkin. Note the presence of a single glycine at position 2 and 3 cysteines representing potential attachment sites for lipid anchors. HeLa cells were transiently transfected with plasmids encoding an N-terminal fragment of Gadkin-eGFP either lacking the first 10 amino acids (10–100) or carrying point mutations targeting glycine 2 (1–100 G2A) or cysteines 4, 5, and 9 (1–100 C4/5/9S). The subcellular localization of these Gadkin variants was analyzed by fluorescence microscopy (green). DAPI-stained nuclei are shown in blue. Scale bar, 10 μm. B and C, mutational inactivation of putative palmitoylation sites within full-length Gadkin renders the protein cytosolic. HeLa cells were transiently transfected with plasmids encoding Gadkin-eGFP WT or C4/5/9S. The subcellular distribution of the expressed proteins was analyzed by subcellular fractionation and immunoblotting (A) (as in Fig. 1A for endogenous protein) and fluorescence microscopy (B). DAPI-stained nuclei are shown in blue. Scale bar, 10 μm. D, 2-bromopalmitate treatment inhibits membrane association of Gadkin-(1–100). HeLa cells were preincubated with 50 μm 2-bromopalmitate (2-BrP, dissolved in ethanol) or the same volume of ethanol (EtOH) for 2 h, transiently transfected with plasmids encoding Gadkin-(1–100) eGFP or the C4/5/9S mutant thereof and incubated with 2-bromopalmitate or EtOH for another 20 h. The subcellular distribution of the expressed proteins was analyzed by fluorescence microscopy (green). DAPI-stained nuclei are shown in blue. Scale bar, 10 μm. E, detection of Gadkin's palmitoylation using the biotin-switch method. FLAG-Gadkin-(1–140) was immunoprecipitated from Triton X-100-solubilized extracts of transfected COS7 cells (500 μg of total protein), free sulfhydryl (SH)-groups were quenched with N-ethylmaleimide, and samples were treated with either 1 m hydroxylamine (NH2OH) to remove palmitate moieties or 1 m Tris buffer as a control. Liberated SH-groups were labeled with 300 μm 1-biotinamido-4-(4′-[maleimidoethylcyclohexane]carboxamido)butane. After elution, samples were analyzed by SDS-PAGE and immunoblotting using either an anti-FLAG tag antibody or streptavidin-HRP for detection. Std., 45 μg (9%) of cell lysate used for immunoprecipitation and biotin labeling. F, Gadkin WT or a C4/5/9S mutant bind to AP-1. COS7 cells were transiently transfected with plasmids encoding FLAG-Gadkin WT or its C4/5/9S mutant. Triton X-100-solubilized cellular extracts (350 μg of total protein) were used for co-immunoprecipitation using anti-FLAG antibodies immobilized on beads (α-FLAG IP). Bound proteins were analyzed by SDS-PAGE and immunoblotting using antibodies against FLAG and γ-adaptin (AP-1). Input, 30 μg (8%) of the cell lysates used for immunoprecipitation.
Two approaches were then taken to more directly address whether Gadkin is subject to S-palmitoylation. First, we incubated transfected HeLa cells with 2-bromopalmitate, a known inhibitor of S-palmitoylation. As expected, such treatment caused Gadkin-(1–100) to dissociate from intracellular membrane puncta, whereas treatment with the solvent alone (EtOH) had no effect (Fig. 2D). Second, palmitoylation of Gadkin-(1–140) was analyzed using the biotin-switch method (20) in which free cysteines are first protected by N-ethylmaleimide, then palmitate is dissociated from palmitoylated cysteine residues by treatment with hydroxylamine (NH2OH) liberating selectively palmitoylated cysteines that are then cross-linked to biotin and thus can be decorated with streptavidin-HRP. In agreement with the data above, immunoprecipitated FLAG-Gadkin-(1–140) was detected with streptavidin-HRP following hydroxylamine cleavage but not after treatment of samples with Tris (Fig. 2E), indicating that Gadkin-(1–140) indeed is S-palmitoylated. Finally, we assessed whether S-palmitoylation and, hence, membrane targeting is a prerequisite for AP-1 binding of Gadkin. To test this, transfected HeLa cells expressing FLAG-tagged WT or palmitoylation-defective Gadkin (C4/5/9S) were subjected to immunoprecipitation using anti-FLAG monoclonal antibodies. As seen in Fig. 2F, WT and mutant Gadkin were found to associate with endogenous AP-1 in cells with equal efficiencies. Collectively, our data indicate that Gadkin targeting to membranes requires its S-palmitoylation and occurs independently of its complex formation with AP-1. This conclusion is also in agreement with the observation that overexpressed Gadkin is capable of retaining AP-1 at membranes in the presence of BFA (8) (compare also with Fig. 7).
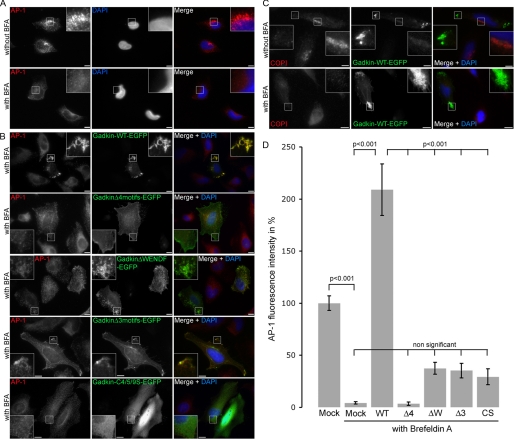
FIGURE 7.
The ability of Gadkin to prevent BFA-mediated dissociation of AP-1 from TGN/endosomal membranes depends on its S-palmitoylation and on direct binding to γ-ear. Gadkin mutants lacking the ability to bind to AP-1γ-ear (Δ4 motifs) or to undergo S-palmitoylation are unable to protect AP-1 from BFA-mediated membrane dissociation. A, HeLa cells were incubated without (top) or with (bottom) BFA and processed for indirect immunofluorescence microscopy using antibodies against AP-1. DAPI-stained nuclei are shown in blue. Scale bar, 10 μm. B, HeLa cells overexpressing Gadkin-eGFP WT or the indicated mutants were treated with BFA and processed for indirect immunofluorescence microscopy using antibodies against AP-1. DAPI-stained nuclei are shown in blue. Scale bar, 10 μm. C, HeLa cells overexpressing Gadkin-eGFP WT were treated with BFA and processed for indirect immunofluorescence microscopy using antibodies against COPI. DAPI-stained nuclei are shown in blue. Scale bar, 10 μm. D, quantification of the degree to which Gadkin WT or mutants (Δ4motifs, ΔWENDF, Δ3motifs, and C4/5/9S) protect AP-1 from BFA-induced membrane dissociation. AP-1 fluorescence intensity per cell was determined using Slidebook software and normalized to the intensity found in untransfected non-BFA treated cells. 36–76 cells were analyzed per sample. Data were pooled from two (for palmitoylation defective mutant) or four (all other mutants) independent experiments and given as mean values ± S.E. An analysis of variance multicomparison test, followed by a Tukey post-test, was used to assess statistical significance.
Gadkin Associates with a Conserved Site within the AP-1γ-ear Used Also by Other Accessory Proteins
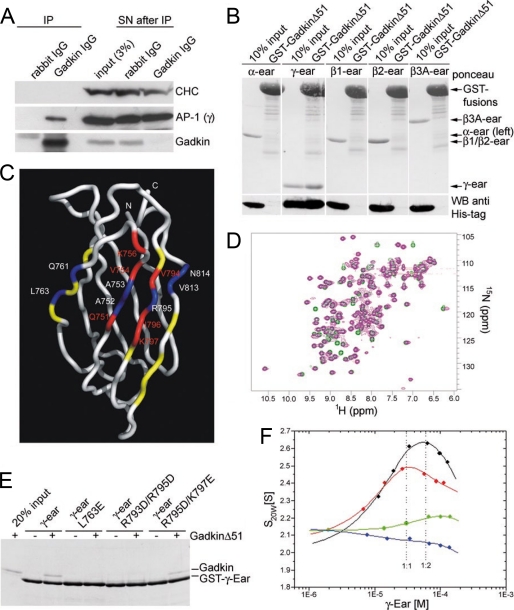
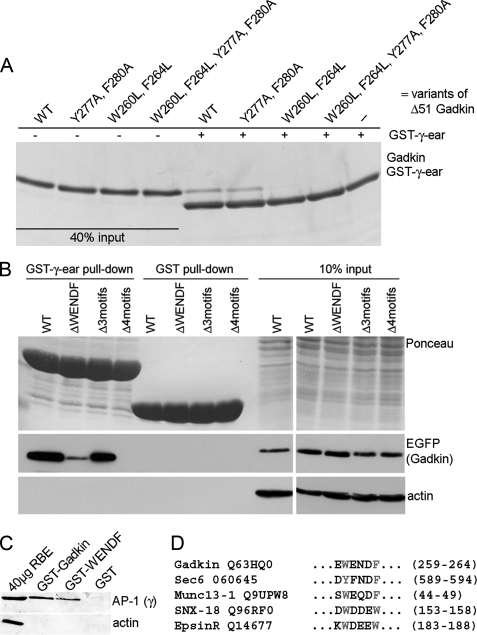
Although S-palmitoylation of Gadkin is clearly required for membrane targeting, its binding to AP-1 might still control the subcellular distribution and physiological function of Gadkin. To address this we studied the association of Gadkin with AP-1 in biochemical and biophysical detail. As expected from previous work, Gadkin and AP-1 efficiently co-immunoprecipitated from rat brain detergent extracts, suggesting that both proteins form a complex in vivo (Fig. 3A). Furthermore, in vitro binding experiments using the purified ear domains of AP-3βA, AP-2α, AP-2β, AP-1β, and AP-1γ indicated that Gadkin specifically associates with AP-1γ but not with other AP appendages (Fig. 3B). Because deletion of its N-terminal 51 residues did not impair the ability of recombinant Gadkin to bind to AP-1 (compare with Fig. 1), but greatly improved protein expression and solubility, we used this Gadkin (Δ51) for further experiments. First, we carried out analytical ultracentrifugation experiments using purified recombinant Gadkin (Δ51) and His6-AP-1γ-ear. Both γ-ear and Gadkin (Δ51) were monomeric in solution. If γ-ear and Gadkin (Δ51) were mixed, the c(S) distributions indicated the formation of a new species with an s value different from both γ-ear and Gadkin (Δ51) (supplemental Fig. 3). Integration of these distributions yielded an isotherm indicating the presence of two γ-ear binding sites within Gadkin of high (42 nm) and low affinity (high μm KD) (Table 1, Fig. 3F). To identify the cognate recognition site for Gadkin within the AP-1γ-ear, we used NMR spectroscopy. A conserved surface area on the γ-ear domain was identified by NMR chemical shift mapping. It involves amino acids Leu763, Arg793, Arg795, and Lys797 (Fig. 3, C and D, and supplemental Figs. 1 and 2) as the main binding site for Gadkin. The same site is also used by other AP-1-associated accessory proteins, including γ-synergin, aftiphilin, and enthoprotin/epsinR. This conclusion was further confirmed by site-directed mutagenesis and direct binding experiments using purified components (Fig. 3E). L763E or R793D/R795D mutants of AP-1 γ-ear had lost the ability to associate with Gadkin (Δ51), and an R795D/K797E mutant displayed reduced binding.
FIGURE 3.
Gadkin binds to a conserved surface on the ear domain of AP-1γ via multiple distinct sites. A, endogenous Gadkin and AP-1 form a complex in vivo. Anti-Gadkin antibodies or nonspecific rabbit IgGs (control) were coupled to protein A/G-agarose beads and incubated with Triton X-100 protein extracts prepared from P1 rat brains (2 mg of total protein). Bound proteins were analyzed by immunoblotting using antibodies against clathrin heavy chain, AP-1 γ-adaptin, and Gadkin. (SN: supernatant corresponding to 3% of the lysate after immunoprecipitation.) B, Gadkin specifically binds to the AP-1γ-ear but not to related ear domains. 20 μg of GST-tagged Gadkin immobilized on beads was incubated with 40 μg of His6-tagged α-, γ-, β1-, β2-, or β3A-adaptin ear domains. Samples were subjected to SDS-PAGE and immunoblotting. Membranes were stained with Ponceau S and probed with His6 tag-specific antibodies. C, NMR chemical shift mapping of the Gadkin binding surface within the AP-1γ-ear. Magnitudes of amide chemical shift changes are plotted throughout the length of the γ-ear. The backbone trace of the γ-ear is colored according to the size of the amide chemical shift changes upon binding of Gadkin. Residues colored in blue shifted, then disappeared, red ones displayed a strong shift, yellow residues a medium shift. Effects were primarily seen for highlighted residues in β-sheets 4, 5, 7, and 8. D, comparison of 1H-15N HSQC spectra of the 15N-labeled AP-1γ-ear domain in the absence (green) or presence (purple) of Gadkin (Δ51, 1:1 ratio). Changes in chemical shifts indicate changes in conformation induced by complex formation (see supplemental Figs. 1 and 2 for an assigned map and a tabulation of the chemical shift changes). E, binding of purified His6-tagged Gadkin (Δ51) to GST-γ-ear WT or the indicated mutants thereof. Immobilized GST fusion proteins (25 μg) were incubated with 25 μg of recombinant His6-Gadkin (Δ51) and washed extensively. 1/10 of each sample was analyzed by SDS-PAGE and Coomassie Blue staining. 20% (0.5 μg) of the total His6-tagged Gadkin (Δ51) used in the assay were analyzed as a standard. F, isotherms for Gadkin (Δ51) WT (black) or mutant proteins (red: Y277A/F280A; green: W260L/F264L; blue: W260L/F264L/Y277A/F280A) together with AP-1γ-ear. Dotted lines represent molar ratios of 1:1 and 1:2, respectively, based on a concentration of Gadkin (Δ51) of 30 μm. Solid lines represent fits to a model of two independent binding sites.
TABLE 1.
Analytical ultracentrifugation
| Protein | sAB | KD1 | sABB | KD2 |
|---|---|---|---|---|
| S | μm | S | μm | |
| Gadkin (Δ51) WT | 2.71 | 0.042 | 5.57 | 162 |
| Gadkin (Δ51) Y277A, F280A | 2.85 | 3.04 | 117 | 15215 |
| Gadkin (Δ51) W260L, F264L | 2.63 | 104 | 3.14 | 107 |
| Gadkin (Δ51) W260L, F264L, Y277A, F280A | 2.71 (fixed) | NDa | 2.05 | ND |
a ND, not determined.
Identification of a Novel Subtype of AP-1γ-ear Binding Motif within Gadkin
The ability to associate with the ear domains of heterotetrameric clathrin adaptor complexes such as AP-1 is encoded within short linear peptide motifs, often found within unstructured regions of accessory proteins. In the case of Gadkin several putative AP-1-binding motifs had been proposed that were distributed almost over the entire protein sequence. To study the structure of Gadkin we made use of limited proteolysis, CD, and NMR spectroscopy. Treatment of purified Gadkin (Δ51) with either trypsin or proteinase K resulted in near complete breakdown of the protein (Fig. 4A). CD measurements (carried out at different temperatures; Fig. 4B and Table 2) indicated that Gadkin mainly adopts random coil conformations and only contains ∼10–20% helical structure. NMR spectroscopy (Fig. 4C) shows that the protein predominantly gives rise to random coil chemical shifts and peaks with narrow line shapes characteristic for proteins lacking stable tertiary structure. These features largely remain upon addition of AP-1γ-ear (Fig. 4C), supporting the hypothesis that the function of Gadkin is independent of stable tertiary structure. Aggregation of Gadkin can be excluded on the basis of analytical ultracentrifugation experiments, which confirm that the protein remains monodisperse at the concentration used for NMR spectroscopy (data not shown). Because we failed to detect stable secondary or tertiary structure within Gadkin following addition of AP-1γ-ear (Fig. 4C), it is likely that its AP-1 binding ability might be encoded within short linear peptide segments.
FIGURE 4.
Gadkin appears to lack stable tertiary structure. A, limited proteolysis performed with different concentrations of proteinase K and trypsin (1, 5, and 25 μg/ml) showed a pattern typical for a protein lacking stable tertiary structure. Fragments of different length were observed. Bands marked with numbers were in-gel-digested and analyzed by mass fingerprinting. All fragments could be assigned to the N-terminal portion of Gadkin (Δ51). B, CD spectra of 2.4 μm His6-tagged Gadkin (Δ51) in PBS, 10 mm dithiothreitol, taken at increasing temperature from 25 °C to 55 °C, shows the loss of structure elements with temperature increase to 40 °C. The process is irreversible; by cooling down to 25 °C (p25b) Gadkin (Δ51) remains partially unstructured. C, comparison of 1H,15N HSQC spectra of the 2H-, 15N-, and 13C-labeled Gadkin (Δ51) in the absence (blue) or presence (red) of γ-ear (1:2 ratio). Gadkin (Δ51) does not appear to adopt stable secondary structure upon addition of AP-1γ-ear.
TABLE 2.
Gadkin (Δ51) secondary structure based on CD
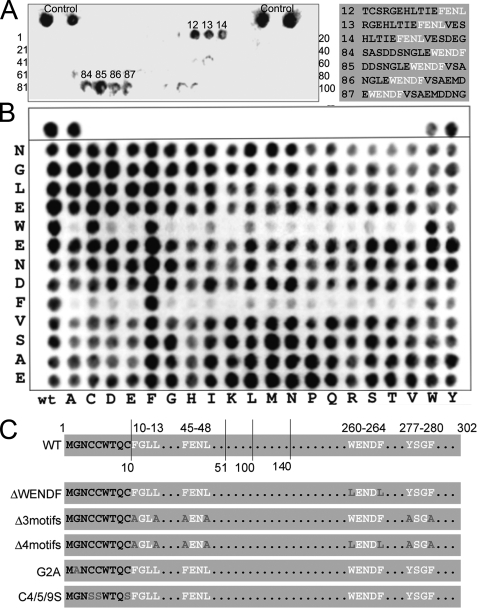
To identify such short AP-1 binding determinants within Gadkin we thus turned to cellulose-bound peptide SPOT (pepscan) arrays. To this aim the complete amino acid sequence of human Gadkin was covered by an array of cellulose-bound 15-mer peptides overlapping in sequence by three amino acids each. This matrix was incubated with GST-tagged AP-1γ-ear, washed extensively, and bound protein was detected by HRP-labeled anti-GST antibodies. Pepscan overlay assays identified two main interacting peptides (#12–14 and #84–87) containing the sequences 45FENL48 (residues critical for binding to AP-1 are highlighted in bold and italics, see also below) and 260WENDF264 (Fig. 5A). Whereas 45FENL48 resembles (although it does not strictly match) the AP-1γ-ear binding consensus ØG(P/D/E)(Ø/L/M) (16), 260WENDF264 does not. In addition to 45FENL48 Neubrandt et al. (8) had proposed two further peptides with the core sequences 10FGLL13 and 277YSGF280 as putative AP-1-binding motifs that did not yield positive signals in our assay. These motifs might moderately contribute to the overall AP-1 binding affinity of Gadkin, although they are clearly not essential (compare Fig. 5C; see also further below).
FIGURE 5.
Overlay assays using cellulose-bound peptide matrices identify a novel subtype of AP-1γ-ear-binding motif within Gadkin. A, pepscan analysis to identify AP-1γ-ear-binding peptides using overlay assays. The complete amino acid sequence of human Gadkin was covered by a cellulose-bound array of 15-mer peptides overlapping in sequence by three residues each. The matrix was incubated with GST-AP-1γ-ear (30 μg/ml) for 14 h at 4 °C. Following washes bound AP-1γ-ear was analyzed by HRP-labeled anti-GST antibodies and chemiluminescence detection. Positive control peptide spots are included at the upper left and upper right corners of the synthesized array. The sequences of identified AP-1γ-ear-binding peptides are given on the right. B, overlay AP-1γ-ear binding assay based on a pepscan substitution matrix synthesized on a modified cellulose membrane. The sequence of the peptides analyzed is indicated on the left, and substitutions are indicated at the top. The peptides His6 and LASDLIVPRR were used as control spots at the upper left and upper right corners of the synthesized arrays. Analysis of peptide-bound AP-1γ-ear was carried out using a chemiluminescence substrate and a Lumi-ImagerTM instrument. C, schematic diagram of the primary sequence of WT Gadkin and mutants used in this study. AP-1-binding motifs are printed in white, mutations are indicated in gray. The vertical lines denote the position of the different truncation mutants used.
Because the sequence WENDF does not conform to the known AP-1γ-ear binding consensus, we carried out substitution analysis using pepscan arrays. This analysis identified Trp and Phe as being critical for the interaction with AP-1γ (Fig. 5B), suggesting that 260WENDF264 might associate with corresponding hydrophobic grooves within AP-1γ-ear via a two-pin plug. Thus, WENDF and related sequences appear to constitute a novel subtype of AP-1γ-ear-binding motif. To test the importance of the WENDF motif for Gadkin (Δ51) binding to γ-ear, we created point mutants, in which Trp260 and Phe264 had been mutated to leucines and analyzed these by affinity chromatography (Fig. 6A) and analytical ultracentrifugation (Fig. 3F and Table 1). Mutant His6-Gadkin (Δ51) failed to bind to GST-γ-ear immobilized on beads (Fig. 6A) and displayed a weak, but still measurable affinity with a KD of ∼100 μm for AP-1γ-ear in analytical ultracentrifugation experiments (Fig. 3F and Table 1). Residual binding was completely abrogated by mutational inactivation of 277YSGF280 (Y277A/F280A) in a W260L/F264L mutant background (Fig. 3F and Table 1). Mutating Y277A/F280A alone eliminated the second low affinity γ-ear binding site within Gadkin (Δ51) (Fig. 3F and Table 1). This analysis thus identifies 260WENDF264 as the high and 277YSGF280 as the low affinity AP-1γ-ear binding site within Gadkin (Δ51). 45FENL48 and 10FGLL13 may provide additional AP-1-binding determinants within the N-terminal domain of Gadkin.
FIGURE 6.
Evaluation of the impact of the newly identified WENDF motif on the Gadkin/AP-1γ-ear interaction in vitro. A, binding of purified His6-tagged Gadkin WT (Δ51) and its indicated mutants to GST-γ-ear. Immobilized GST fusion proteins (25 μg) were incubated with 25 μg of recombinant His6-tagged Gadkin (Δ51) and washed extensively. One-tenth of each sample was analyzed by SDS-PAGE followed by Coomassie Blue staining. 40% (1 μg) of His6-tagged Gadkin (Δ51) was loaded onto a gel as standard. B, association of AP-1γ-ear with Gadkin AP-1-binding motifs mutants. 25 μg of GST-γ-ear or GST were incubated with 0.5 mg (2 μg/μl) of Triton X-100-solubilized lysate of HEK293 cells expressing Gadkin-eGFP WT or the indicated mutants. Samples were analyzed by SDS-PAGE and immunoblotting for eGFP and actin. C, GST-Gadkin (Δ51) or GST-NGLEWENDFVSAE was assayed in pulldown experiments for their ability to associate with AP-1γ from rat brain extract. Samples were analyzed by immunoblotting for AP-1γ or actin. 40 μg of the input material was loaded as the standard. D, results of an EXPASY Prosite search for proteins containing putative AP-1γ-binding motifs (WY)XX(DE)(WF): Sec6 (O60645 in SWISSPROT), Munc13-1 (Q9UPW8), Sorting nexin-18 (Q96RF0), and EpsinR (Q14677).
A Novel Subtype of AP-1γ-ear Binding Motif within Gadkin Regulates Its AP-1 Binding Ability
To assess the contribution of the newly identified AP-1γ-ear-binding motifs within Gadkin to complex formation with AP-1, we transfected HEK293 cells with expression plasmids encoding Gadkin-eGFP (WT) or various mutants thereof. These included variants, in which either 260WENDF264 (ΔWENDF), the remaining three conventional γ-ear-binding motifs (Δ3 motifs), or all four motifs in combination (Δ4 motifs) had been inactivated by mutation of conserved hydrophobic residues (Fig. 5C). Detergent lysates of these cell extracts were then passed over a GST-γ-ear affinity matrix, and the amount of bound Gadkin was assessed by immunoblotting. As expected from our biochemical and biophysical analysis the ΔWENDF mutant showed a greatly diminished ability to associate with the AP-1γ-ear, whereas binding of a Gadkin mutant lacking the three conventional ØXXØ motifs (Δ3 motifs) was nearly unperturbed. Mutation of all four motifs in combination (Δ4 motifs) completely eliminated AP-1γ-ear binding (Fig. 6B). Nearly identical results were obtained if complex formation between Gadkin-eGFP and endogenous AP-1 was analyzed by co-immunoprecipitation (data not shown). These data confirm that a non-conventional AP-1γ-ear-binding motif within Gadkin is a major determinant for its association with AP-1.
We then asked whether a single WENDF motif might suffice to bind to AP-1 in vitro. Indeed, a WENDF peptide fused to GST was capable of affinity-purifying native endogenous AP-1 from rat brain extracts, whereas actin was absent from this sample (Fig. 6C). Putative AP-1-binding motifs similar to the WENDF sequence identified in Gadkin were found in a number of other proteins with a putative role in membrane traffic (Fig. 6D). These include the known AP-1 interaction partner epsinR, SNX-18, a PX-BAR domain-containing protein acting as a membrane tubulator in AP-1-dependent endosomal traffic (24), the exocytosis regulator Munc13-1 (25), and Sec6, a component of the mammalian exocyst complex, which extensively co-localizes with the AP-1B isoform (5). We speculate that all of these proteins might associate with AP-1 at least in part via this novel subtype of γ-ear-binding motif.
The Ability of Gadkin to Stabilize AP-1 at Membranes Depends on an Intact AP-1γ-ear-binding WENDF Motif and S-Palmitoylation of Gadkin
Previous work has suggested that Gadkin is involved in the control of membrane association of the AP-1 adaptor complex. More specifically, overexpression of Gadkin has been shown to prevent the BFA-induced dissociation of AP-1 from TGN/endosomal membranes (8). We used this assay system to analyze the contribution of the various AP-1γ-ear-binding motifs within Gadkin identified here and to study the importance of the S-palmitoylation of Gadkin. As expected, brief incubation of HeLa cells with BFA led to a rapid and near complete dispersion of endogenous AP-1 into the cytoplasm (Fig. 7, A and D). Overexpression of WT Gadkin prevented AP-1 dissociation from membranes and instead caused the accumulation of AP-1-positive puncta, presumably endosomes, in the cell periphery where both proteins co-localized (Fig. 7, B and D). By contrast, Gadkin-eGFP was unable to protect the related COPI complex from BFA-induced cytosolic dispersion (Fig. 7C), in agreement with its specific association with AP-1. Mutational inactivation of either the WENDF motif or of the three ØXXØ-type γ-ear-binding motifs (compare Fig. 5C) strongly diminished the ability of Gadkin to stabilize AP-1 at membranes in the presence of BFA (Fig. 7, B and D). A mutant, in which all four motifs had been inactivated (Δ4 motifs) was completely unable to protect AP-1 from BFA-induced cytosolic dispersion (Fig. 6, B and D). Lack of AP-1 binding of Gadkin mutants correlated with their redistribution to the plasma membrane (Fig. 6B). S-Palmitoylation-defective C4/5/9S mutant Gadkin localized to the cytoplasm (Fig. 7A) and also failed to prevent BFA-induced dissociation of AP-1 from membranes. We conclude that stable membrane recruitment of Gadkin depends on its S-palmitoylation. Moreover, AP-1 association is not required for membrane targeting of Gadkin per se, but regulates its localization at TGN/endosomal membranes.
DISCUSSION
The precise mechanism by which the clathrin adaptor complex AP-1 is targeted to the TGN and to endosomes has remained largely enigmatic, but accessory proteins likely contribute to it. In this work we show that the AP-1-binding accessory protein Gadkin is anchored to TGN/endosomal membranes via S-palmitoylation involving three N-terminal cysteine residues. Biochemical and biophysical analyses identify several AP-1-binding sequences within Gadkin that associate with a site on the γ-ear domain also used by other accessory proteins, including epsinR, NECAP1, rabaptin-5, and γ-synergin. These sequences include a novel subtype of γ-ear-binding motif with the consensus (WY)XX(DE)(WF). Mutational inactivation of this motif dramatically decreases the AP-1 binding affinity of Gadkin from 42 nm to >100 μm and causes a concomitant reduction in the ability of Gadkin to prevent the BFA-induced dissociation of AP-1 from TGN/endosomal membranes. Collectively, our data uncover the molecular basis for Gadkin-mediated AP-1 recruitment to TGN/endosomal membranes and identify a novel subtype of AP-1-binding motif.
Gadkin to our knowledge represents the first AP-1 binding accessory protein shown to undergo S-palmitoylation, thereby being stably anchored at membranes. This feature can also explain why overexpressed Gadkin unlike other AP-1-associated accessory proteins is capable of rendering AP-1 BFA-resistant. However, S-palmitoylation by itself is insufficient to correctly target Gadkin to the TGN/endosomal membranes. AP-1 binding-defective mutant Gadkin accumulates at the plasma membrane, suggesting that association with AP-1 is required for its recruitment to the TGN and/or to EVs. These results also fit well with the observation that a minor fraction of native endogenous Gadkin is found at plasmalemmal structures devoid of AP-1 (8). Our data thus suggest that AP-1 binding and S-palmitoylation cooperate in targeting Gadkin to the TGN and/or to EVs in living cells.
One of the most interesting outcomes of this study is the identification of a novel subtype of AP-1γ-binding motif with the consensus sequence (WY)XX(DE)(WF). Our biochemical and biophysical data suggest that this motif associates with a conserved site on the γ-ear domain of AP-1 with high nanomolar affinity, thereby potentially providing Gadkin with privileged access to AP-1. Other sequences resembling conventional γ-ear-binding motifs, such as the 45FENL48 sequence within the N-terminal domain of Gadkin, also contribute to complex formation with AP-1. However, even these motifs within Gadkin do not conform strictly to the known γ-ear binding consensus sequence ØG(P/D/E)(Ø/L/M) found in most other AP-1-associated accessory proteins, including γ-synergin, aftiphilin, eps15, epsinR, or rabaptin-5 (16). The use of AP-1-binding motifs significantly deviating from the known consensus thus appears to distinguish Gadkin from these other factors and fits well with the observation that Gadkin selectively associates with the γ-ear domain of AP-1 but not with that of the related GGA proteins (8).4 Selectivity for AP-1 versus GGA binding within accessory proteins may thus be encoded at least in part by variations from the consensus sequence as well as by the use of non-conventional subtypes of γ-ear-binding motifs.
In agreement with this proposal, motifs conforming to the (WY)XX(DE)(WF) consensus can be found within the known AP-1-binding proteins epsinR (10, 12) (alternatively termed enthoprotin (13) or Clint (11)) and SNX18 (24), the exocytosis regulator Munc13-1 (25), and the exocyst component Sec6. For epsinR and SNX18, complex formation with AP-1 has been demonstrated, whereas Munc13-1 and Sec6 have not yet been shown to bind to AP-1 in vitro. However, a putative direct interaction between Sec6 and AP-1 is likely, based on the fact that exocyst selectively associates with AP-1B-positive endosomal membranes and that expression of AP-1B facilitates exocyst recruitment during basolateral cargo sorting in epithelial cells (5). Future studies will need to address this point in more detail.
Little is known so far regarding the precise physiological function of the Gadkin-AP-1 complex characterized here in biochemical detail. Overexpression and knockdown studies have implicated Gadkin in mannose 6-phosphate receptor- dependent sorting of lysosomal enzymes (8), a pathway known to involve an endosomal pool of AP-1 (7). Whether or not Gadkin is also involved in other postulated functions of AP-1 such as the formation of Weibel-Palade bodies (26), maturation of secretory granules (27), or basolateral sorting of endosomally delivered cargo in polarized cells (5), remains to be investigated. The precise mapping of the AP-1 binding determinants described here should pave the way for a more thorough investigation of these possibilities. We have recently shown that Gadkin directly binds to the light chains of kinesin KIF5 via a site non-overlapping with any of the AP-1 binding determinants identified in the present work (19). This positions Gadkin at the interface of AP-1-mediated sorting of transmembrane cargo and transport of TGN-derived EVs along the microtubule-based cytoskeleton. Such a function is very compatible with the observed high affinity interaction of Gadkin with AP-1 via the motifs identified here and with its S-palmitoylation-dependent stable association with membranes. Future studies capitalizing on our results will need to address the physiological role of the Gadkin-AP-1 complex in kinesin-driven transport of TGN/endosomal membranes in more detail in vivo.
Supplementary Material
This work was supported by Deutsche Forschungsgemeinschaft (DFG Grants HA2686/1-3 and 1-4 to V. H. and T. M.), SFB 449/TP A11 (to V. H.) and Z1 (to R. V.), and DFG Exc 257-Neurocure.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
T. Maritzen, M. R. Schmidt, V. Kukhtina, V. A. Higman, H. Strauss, R. Volkmer, H. Oschkinat, C. G. Dotti, and V. Haucke, unpublished data.
- TGN
- trans-Golgi network
- AP-1
- clathrin adaptor complex 1
- BFA
- brefeldin A
- Gadkin
- γ1-adaptin and kinesin interactor
- EV
- endosomal vesicle
- HRP
- horseradish peroxidase
- RE
- recycling endosome
- Tf
- transferrin
- TfR
- transferrin receptor
- WT
- wild type
- BAR
- BIN/ amphiphysin/Rvs167
- eGFP
- enhanced green fluorescent protein
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline
- DAPI
- 4′,6-diamidino-2-phenylindole
- TBS
- Tris-buffered saline
- SH
- sulfhydryl.
REFERENCES
- 1.Bonifacino J. S., Traub L. M. (2003) Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 2.Mellman I. (1996) Annu. Rev. Cell Dev. Biol. 12, 575–625 [DOI] [PubMed] [Google Scholar]
- 3.Kirchhausen T. (2000) Annu. Rev. Biochem. 69, 699–727 [DOI] [PubMed] [Google Scholar]
- 4.Traub L. M. (2005) Biochim. Biophys. Acta 1744, 415–437 [DOI] [PubMed] [Google Scholar]
- 5.Fölsch H., Pypaert M., Maday S., Pelletier L., Mellman I. (2003) J. Cell Biol. 163, 351–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinners I., Tooze S. A. (2003) J. Cell Sci. 116, 763–771 [DOI] [PubMed] [Google Scholar]
- 7.Meyer C., Zizioli D., Lausmann S., Eskelinen E. L., Hamann J., Saftig P., von Figura K., Schu P. (2000) EMBO J. 19, 2193–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neubrand V. E., Will R. D., Möbius W., Poustka A., Wiemann S., Schu P., Dotti C. G., Pepperkok R., Simpson J. C. (2005) EMBO J. 24, 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Futter C. E., Gibson A., Allchin E. H., Maxwell S., Ruddock L. J., Odorizzi G., Domingo D., Trowbridge I. S., Hopkins C. R. (1998) J. Cell Biol. 141, 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirst J., Motley A., Harasaki K., Peak Chew S. Y., Robinson M. S. (2003) Mol. Biol. Cell 14, 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalthoff C., Groos S., Kohl R., Mahrhold S., Ungewickell E. J. (2002) Mol. Biol. Cell 13, 4060–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills I. G., Praefcke G. J., Vallis Y., Peter B. J., Olesen L. E., Gallop J. L., Butler P. J., Evans P. R., McMahon H. T. (2003) J. Cell Biol. 160, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wasiak S., Legendre-Guillemin V., Puertollano R., Blondeau F., Girard M., de Heuvel E., Boismenu D., Bell A. W., Bonifacino J. S., McPherson P. S. (2002) J. Cell Biol. 158, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirst J., Borner G. H., Harbour M., Robinson M. S. (2005) Mol. Biol. Cell 16, 2554–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritter B., Philie J., Girard M., Tung E. C., Blondeau F., McPherson P. S. (2003) EMBO Rep. 4, 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattera R., Ritter B., Sidhu S. S., McPherson P. S., Bonifacino J. S. (2004) J. Biol. Chem. 279, 8018–8028 [DOI] [PubMed] [Google Scholar]
- 17.Wasiak S., Denisov A. Y., Han Z., Leventis P. A., de Heuvel E., Boulianne G. L., Kay B. K., Gehring K., McPherson P. S. (2003) FEBS Lett. 555, 437–442 [DOI] [PubMed] [Google Scholar]
- 18.Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., Yin H. L. (2003) Cell 114, 299–310 [DOI] [PubMed] [Google Scholar]
- 19.Schmidt M. R., Maritzen T., Kukhtina V., Higman V. A., Doglio L., Barak N. N., Strauss H., Oschkinat H., Dotti C. G., Haucke V. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15344–15349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou H., Subramanian K., LaGrassa T. J., Markgraf D., Dietrich L. E., Urban J., Decker N., Ungermann C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17366–17371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 22.Wenschuh H., Volkmer-Engert R., Schmidt M., Schulz M., Schneider-Mergener J., Reineke U. (2000) Biopolymers 55, 188–206 [DOI] [PubMed] [Google Scholar]
- 23.Landgraf C., Panni S., Montecchi-Palazzi L., Castagnoli L., Schneider-Mergener J., Volkmer-Engert R., Cesareni G. (2004) PLoS Biol 2, E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Håberg K., Lundmark R., Carlsson S. R. (2008) J. Cell Sci. 121, 1495–1505 [DOI] [PubMed] [Google Scholar]
- 25.Varoqueaux F., Sons M. S., Plomp J. J., Brose N. (2005) Mol. Cell. Biol. 25, 5973–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lui-Roberts W. W., Collinson L. M., Hewlett L. J., Michaux G., Cutler D. F. (2005) J. Cell Biol. 170, 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dittie A. S., Hajibagheri N., Tooze S. A. (1996) J. Cell Biol. 132, 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.