Abstract
Purpose
The aim of this study was to evaluate volumetric proton magnetic resonance spectroscopic imaging (MRSI) for localization of epileptogenic foci in neocortical epilepsy.
Methods
Twenty-five subjects reporting seizures considered to be of neocortical origin were recruited to take part in a 3-Tesla MR study that included high-resolution structural MRI and a whole-brain MRSI acquisition. Using a fully-automated MRSI processing protocol, maps for signal-intensity normalized N-Acetylaspartate (NAA), creatine, and choline were created, together with the relative volume fraction of grey-matter, white-matter, and CSF within each MRSI voxel. Analyses were performed using visual observation of the metabolite and metabolite ratio maps; voxel-based calculation of differences in these metabolite maps relative to normal controls; comparison of average grey- and white-matter metabolite values over each lobar volume; and examination of relative left-right asymmetry factors by brain region.
Results
Data from fourteen subjects were suitable for inclusion in the analysis. Eight subjects had MRI-visible pathologies that were associated with decreases in NAA/Creatine, which extended beyond the volume indicated by the MRI. Five subjects demonstrated no significant metabolic alterations using any of the analysis methods, and one subject had no findings on MRI or MRSI.
Conclusions
This proof of principle study supports previous evidence that alterations of MR-detected brain metabolites can be detected in tissue areas affected by neocortical seizure activity, while additionally demonstrating advantages of the volumetric MRSI approach.
Keywords: Neocortical Epilepsy, MRI, MR Spectroscopy, MRSI
Introduction
Surgical intervention for intractable epilepsy has become an established treatment option, although one that presents a considerable challenge for presurgical evaluation and localization. The prognosis of a favorable outcome following surgical removal of epileptogenic foci is greatest when several localizing studies indicate the same brain region, with established diagnostic methods including ictal video/EEG telemetry, MRI, PET, and SPECT (Kuzniecky and Devinsky, 2007; Hartman and Lesser, 2007). However, there remain several limitations of these techniques and many subjects do not present with conclusive localizing findings, particularly for neocortical epilepsy (NE), which lacks a common pathological cause as for temporal lobe epilepsies. Additional difficulties for NE include causes that can be subtle and widespread (Krsek et al., 2007), and that detection of any abnormalities within the complex structures of the neocortex by neuroimaging is problematic.
It has been shown that epileptic seizures are associated with changes in energy metabolism and that the seizure focus is associated with long-term changes of metabolite concentrations (Pan et al., 2008). In this regard, the potential for MR spectroscopy has been widely investigated, and several studies have indicated considerable potential for lateralization and localization of the epileptogenic focus. In vivo MR spectroscopy studies in humans have used detection of 31P and 1H MRS signals (Laxer, 1997), and these techniques enable detection of a number of metabolites that provide complimentary information to that provided by structural imaging methods. The most commonly applied methods use detection of proton metabolites, with the primary findings being decreased N-acetylaspartate, indicative of neuronal loss ormitochondrial dysfunction (Moffett et al., 2007), and an increased signal from choline-containing compounds, indicating alterations in cell membrane turnover. The most widely used application of MRS has been lateralization of temporal lobe epilepsy, for which diagnostic studies can be conveniently implemented using single-voxel MRS acquisitions from the hippocampi. These single-voxel MRS acquisition techniques are, however, of limited value for detection of NE, where prior localizing information that could direct placement of the volume of interest for a MRS measurement is typically unavailable. The preferred MRS technique is therefore one that can sample a wide extent of the cerebrum, and therefore best implemented using MR spectroscopic imaging (MRSI) techniques that sample a larger volume of tissue.
There exist several challenges to the implementation of MRSI methods, including long acquisition times when using conventional phase-encoding; loss of spectral information from magnetic-susceptibility induced spectral line broadening; image distortions extending from the large subcutaneous lipid signals or unsuppressed water; and limitations on signal quality and spatial resolution due to poor detection sensitivity. For these reasons, the previous applications of MRSI for studies of NE have used a variety of approaches to limit the volume of the brain from which data is sampled, including sampling over a single rectangular PRESS-selected volume in the center of the cerebrum (Lundbom et al., 2001; Garcia et al., ; Krsek et al., 2007; Leite et al., 2007; Stanley et al., 1998; Li et al., 2000); single-slice with outer volume suppression of subcutaneous lipids (Guye et al., 2005); and slice selection with inversion-recovery nulling of lipid signals (Kuzniecky et al., 1997; Mueller et al., 2004; Mueller et al., 2004; Mueller et al., 2005). Despite the fact that these reports have provided consistent evidence of MRS-detectable metabolic changes associated with NE, clear limitations remain that impede the broader application of these techniques as a diagnostic tool. Firstly, the acquisition, processing, and analysis of MRSI data require specialized personnel, and remains limited to sites that have the necessary expertise. Secondly, MRS signal detection remains sensitivity limited and problematic for reliable measurements in some regions of the brain. Finally, all prior studies have acquired data from only a fraction of the cerebrum, with notably limited sampling of frontal and temporal regions, with the result that a failure to detect metabolic alterations with NE must invariably allow for this methodological limitation.
Since little or no prior information may be available on the location of the epileptogenic focus with NE, it is highly desirable if whole-brain MRSI can be used for these diagnostic studies. In this study, a volumetric MRSI acquisition has been evaluated that has been optimized to maximize the volume of the brain from which data could be acquired. In addition, this study was implemented at 3 Tesla and used phased-array multichannel detection to enhance detection sensitivity. These methodological developments provide both increased spatial coverage and a modest sensitivity improvement over the 1.5 T MR technologies used for the previously reported studies of NE. Additional characteristics of the MRSI methods presented in this report include that the acquisition required only standard setup procedures provided from the instrument manufacturer and was implemented at this site by MR technologists, and that the MRSI data processing was fully automated. The aims of this study were therefore to evaluate this volumetric MRSI approach for localization of NE.
Methods
Subject Selection
All subjects reporting to the University of Miami Epilepsy Center with EEG and/or clinical evidence of refractory focal epilepsy, with or without co-localizing information from structural MRI studies, were approached for participation in this study. Selection was based on seizure semiology and EEG and MRI results that indicated an epileptogenic focus of non-temporal lobe origin. All procedures were approved by the institutional human subjects research review committee, and informed consent was obtained from all participants. A total of 25 subjects were recruited, which included a second study in one subject after a 3-year time interval (i.e. a total of 26 studies). Of these studies, 3 were unsuitable for analysis due to subject motion, 5 studies were incomplete due to either the onset of a seizure or subject discomfort resulting in termination of the study, and the diagnosis for two subjects was subsequently modified and did not meet the diagnosis of focal epilepsy. The subject characteristics used for the final analysis are summarized in Table 1 together with summary of localization findings from the MRI carried out as part of this study as well as from other sources. The average age was 32, with 6 female and 9 male subjects.
Table 1.
Subject Characteristics and localization information
| Subject | Gender & Age |
Age at Onset |
Seizure Type |
Ictal EEG Localization |
MRI | Other Localization |
|---|---|---|---|---|---|---|
| 4 | F/45 | 13 | SPS/CPS | R parietal | R temporal-occipital lesion. Cortical dysplasia or infarct |
|
| 6 | M/29 | CPS | L hemisphere | R hippocampal atrophy |
MEG - lateral- frontal. |
|
| 7 | F/29 | CPS | L hemisphere | None | PET negative. |
|
| 8 | M/34 | CPS/PSG | R temporal | None | ||
| 10 | M/36 | 25 | SPS/PSG | R temporal | None | |
| 12 | M/24 | SPS/CPS | L hemisphere | L parietal and occipital atrophy |
||
| 14 | F/52 | 50 | CPS | L temporal | Nonspecific FLAIR hyperintensity in R frontal. |
|
| 16 | M/18 | 12 | CPS/PSG | Frontal | None | |
| 17 | M/57 | 2 | CPS | L temporal | L parietal lesion | |
| 18 | M/52 | CPS | L temporal | None | ||
| 22 | F/20 | 9 | CPS | L temporal-occipital | L frontal lesion; cortical dysplasia or old trauma |
|
| 23 | F/19 | >1 | CPS/PSG | Bilateral frontal, more on R |
Bifrontal T2 periventricular leukomalacia; WM loss; |
|
| 24 | M/32 | 21 | PSG | R post. temporal, parietal, occipital |
Medial R occipital, ill- defined |
|
| 25 | F/46 | 22 | CPS/PSG | None | R occipital lesion w. subtle contrast enhancement |
symptoms suggest frontal |
M: Male; F: Female; R:. Right; L: Left; CPS: complex partial seizure; SPS: simple partial seizure; PSG: partial secondary generalized.
Localization of seizure focus was based on clinical evaluation, EEG, and the clinical MRI report. Additional information from MEG or PET was available in only two subjects, for which results were either absent or inconclusive.
MR Methods
Proton MRSI studies of the brain were carried out at 3 Tesla (Siemens Trio), with five studies carried out with a quadrature bird-cage detection coil and the remainder with an eight-channel phased-array head coil. Data was acquired using a volumetric spin-echo acquisition that used two-dimensional phase encoding and echo-planar readout, frequency-selective water suppression, TE = 70 ms, and TR = 1710 ms. To suppress signal from subcutaneous lipids so as to enable measurement at the neocortex, an inversion-recovery preparation was used with TI=198 ms. After accounting for oversampling in the readout spatial and spectral dimensions the resultant image was equivalent to 50×50 voxels in-plane and 18 slices, over a field-of-view (FOV) of 280×280×180 mm, with selection of a slab of 135 mm covering the cerebrum with an angulation of −15° from the AC-PC line. The sequence parameters and associated data processing were optimized to acquire data at a higher spatial resolution than warranted from sensitivity considerations in order to maximize spatial coverage via minimization of intra-voxel magnetic-susceptibility induced spectral distortions (Ebel and Maudsley, 2003). The acquisition included a second MRSI dataset that was obtained in an interleaved manner, without water suppression, with low flip-angle excitation, and gradient-echo observation. This data provided a water reference signal with identical spatial parameters as the metabolite MRSI, and was used for several processing functions to enhance the metabolite image reconstruction and analysis. Each study also included a T1-weighted MRI (MPRAGE, Magnetization Prepared Rapid Gradient Echo) at 1 mm isotropic resolution (TR / TE / TI=2150 / 4.4 / 1100 ms, flip angle 8°), a T2-weighted (Turbo Spin Echo) MRI at 1 mm in-plane and 3 mm slice thickness (TR / TE=7000 / 104 ms), and a FLAIR (Fast Low Angle Inversion Recovery) acquisition at 1 mm in-plane, 5 mm slice thickness (TR / TE / TI=8000 / 126 / 2500 ms).
Reconstructions of the metabolite and water-reference images were carried out in a fully-automated manner using the MIDAS (Metabolite Image Data Analysis System) package (Maudsley et al., 2006; Maudsley et al., 2009). Parametric spectral analysis (Soher et al., 1998) was used to determine volumetric maps of N-acetylaspartate (NAA), total creatine (Cre), and total choline (Cho), and all parameter maps were interpolated to 64×64×32 points. The processing included correction of frequency shifts due to magnetic field inhomogeneity, and k-space extrapolation (Haupt et al., 1996) to reduce ringing artifacts from subcutaneous lipids using a mask of the scalp region derived from the T1-weighted MRI. Smoothing was applied in the spatial domain using a convolution filter applied over the brain region only, which provides some improvement in the quality in voxels at the edge of the brain relative to the more conventionally-used k-space apodization. Signal intensity normalization of the metabolite images was implemented using tissue water as an internal reference, which was derived from the water-reference SI signal.
All signal-normalized metabolite images, metabolite ratio images, and associated spectral fitting parameters, including the spectral linewidth, were then spatially transformed to a common spatial reference frame, for which the BrainWeb simulated MRI (Collins et al., 1998) was used as the target. This spatial normalization included interpolation to 2 mm isotropic voxels. For this procedure the T1-weighted MRI was used to determine the non-linear spatial transformation parameters to the reference MRI, which used mutual-information and B-spline deformation with 10 mm knot spacing (Studholme et al., 1997; Rousseau et al., 2005). All metabolite images were then spatially transformed using parameters that combined the rigid transformation between the water-reference SI and the T1-weighted MRI, and the non-linear transformation between the T1 and MNI space. The spatial reference MRI was associated with a brain atlas that mapped each of the left and right frontal, temporal, parietal, and occipital lobes, and the cerebellum.
The T1-weighted MRI was also used for tissue segmentation to map grey-matter white-matter, and CSF, which was carried out using the FSL/FAST program (Smith et al., 2004; Zhang et al., 2001). The results of tissue segmentation were then resampled and smoothed so as to coincide with the MRSI FOV and spatial response function, and these images were also transformed to the standardized spatial reference frame. MRI and spectroscopic results were reviewed using the SID display module of the MIDAS package, as well as the MRIcro program (www.mricro.com).
Data Analysis
Three types of analysis were carried out:
-
Imaged-based visual analysis:
A direct visual evaluation of the metabolite images was carried out for the NAA, Cre, Cho, NAA/Cre, and Cho/Cre, and NAA/Cho images, together with the coregistered MRIs. Based on prior studies, the most sensitive marker for NE related metabolic abnormalities was anticipated to be reduced NAA; however, due to the variability of both the individual metabolite and the metabolite ratio images, changes were only considered notable if seen on both the NAA and the NAA/Cre images.
The image review also included evaluation of z-score maps for NAA and NAA/Cre that indicate the difference in the individual subject metabolite images relative to normal control values at all voxel locations (Maudsley et al., 2008). While this type of image does not generally show information that cannot also be inferred from the underlying metabolite maps, they considerably simplify interpretation for investigators that are not familiar with the normal appearance of the metabolite images, as well as providing quantitative information on the differences from control values. In brief, this procedure first calculates the mean and the standard deviation, σ, at all locations for each metabolite value using data obtained in an identical manner from an age-matched normal control group. This reference information is calculated separately for grey-matter and white-matter tissue using a regression analysis based on voxels selected from within 10 mm of each image voxel location. Using the reference mean and standard deviation images, and the actual tissue distributions from the individual NE subject under analysis, an image representing the difference from normal values was then created and scaled by the standard deviation over the normal control group to indicate the significance of the result.
The reference metabolite information required for calculation of the z-score images was obtained from an existing database (Maudsley et al., 2009) containing data acquired using the identical protocol from 43 male and 51 female subjects, aged from 18 to 66 years old. Reference images were created using subgroups of subjects selected for different age groups, namely 18 to 25; 22 to 34; 27 to 44; and 35 to 70. The overlap between these age ranges was included to provide sufficient numbers of subjects for each analysis, and the reference data selected for the analysis of each epilepsy subject was that which had the minimum difference between the subject age and the middle of the control group age range. The z-score maps were reviewed visually using a color image scale, and areas with z-scores for NAA or NAA/Cre of less than −1.96 were considered to represent significant decreases of these metabolite values. This image analysis approach is similar to that used previously (Mueller et al., 2004; Mueller et al., 2005), although performed using normative information derived from a smaller brain region in the proximity of the voxel under evaluation, as opposed to lobar-scale regions. By having detailed normative information throughout the brain volume, localized differences in normal brain metabolite concentrations are accounted for.
-
Asymmetry of regional average metabolite values:
Previous studies have indicated that although metabolic alterations cannot always be detected in specific spatial regions, diffuse changes can be observed that extend beyond the epileptogenic focus. To test for these cases, data was averaged over the lobar-scale brain regions defined in the brain atlas, and results compared with the corresponding contralateral region. Asymmetry factors were calculated for each metabolite and metabolite ratio as (Left-Right)*2 / (Left+Right). Regions of significant asymmetry were identified by looking for values greater than 3 times the standard deviation of the corresponding asymmetry values derived from the whole control group. The threshold values for assigning significant asymmetry were set as NAA=0.11; Cre=0.14; Cho=0.19; and NAA / Cre=0.11.
-
Tissue regression analysis by brain region:
Average values for all metabolites within each of the lobar brain regions were calculated for grey-matter (GM) and white-matter (WM) using a tissue regression analysis, using methods similar to that described elsewhere (Mueller et al., 2004; Mueller et al., 2005). Z-scores for each metabolite measure and tissue-type were then obtained by calculating the difference relative to the corresponding mean values from the age-matched control group, divided by the standard deviation for that metabolite parameter and region for the normal subject group. The age ranges used for the control group was matched to each subject age, using the same age groups as previously described. Z-scores of >1.96 or <−1.96 were considered significant.
Results
Fifteen studies (from 14 subjects) were included in the analysis. Of these, MRI indicated clearly identifiable lesions in four subjects, and five subjects had non-specific findings including evidence of atrophy and ill-defined signal enhancement on FLAIR images. There were no MRI findings in five subjects (33%). Summary observations for each of the MRSI analysis methods are given in Table 2.
Table 2.
Summary of MRS findings and general concordance between imaging findings and other sources of localization
| Subject | MRS Visual Localization |
MRS Regression Localization |
MRS Asymmetry |
MRI concordance |
MRS concordance |
|---|---|---|---|---|---|
| 4 | None | R temporal GM |
None | Y | N |
| 6 | R frontal ↓NAA/Cre, but not NAA. |
L frontal GM | None | N | N |
|
6 Study 2 |
R frontal ↓NAA/Cre. |
None | None | N | N |
| 7 | None | Multiple | None | n/a | N |
| 8 | R fronto- temporal ↓NAA/Cre. |
Multiple | None | n/a | Y |
| 10 | None | Multiple, due to high Cre. |
None | n/a | N |
| 12 | L parietal- occipital ↓NAA/Cre. |
L frontal, temporal, parietal, occipital |
L temporal, parietal, occipital |
Y | Y |
| 14 | None | L & R frontal and R temporal |
None | n/a | n/a |
| 16 | R frontal ↓NAA/Cre (possible). |
R frontal GM, R temporal WM, R parietal GM |
None | n/a | poss. |
| 17 | L temporal, parietal ↓NAA/Cre. |
L frontal, parietal, occipital |
L frontal, temporal, parietal |
Y | Y |
| 18 | None | None | None | n/a | N |
| 22 | L frontal ↓NAA/Cre. |
L frontal, R occipital |
None | Y | Y |
| 23 | R parietal ↓NAA/Cre. |
Multiple (high Cre) |
R temporal | Y | N |
| 24 | ↑Cho & ↓NAA in R anterior occipital. |
R temporal, R & L occipital |
None | Y | Y |
| 25 | ↓NAA R frontal; ↑Cho ↓NAA in R occipital . |
None | none | N | poss. |
R:. Right; L: Left.
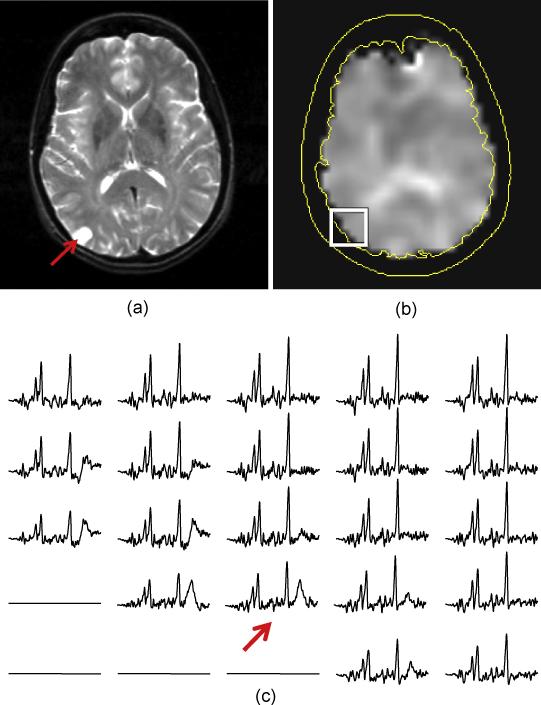
The volumetric MRSI acquisition provided consistently good spectral quality throughout approximately 70% of the cerebrum (Maudsley et al., 2009), with variable quality in orbito-frontal and basal regions. Figures 1 and 2 give examples of the ability of this MRSI acquisition to sample neocortical areas that can present difficulties for single-voxel MRS methods. In Fig. 1 are shown results for subject 4, who presented with a clearly identifiable lesion in the periphery of the right (left side of the image) occipital gyrus that was indicative of possible cortical dysplasia or infarct. The PRESS MRS measurement performed at the time of the clinical MR study of this subject was reported to be of inadequate quality due to the location and small size of the lesion. The MRSI study demonstrated excellent spectral quality right up to the cortical surface region, as indicated in the NAA/Cre image, and by the spectra selected from a region surrounding the lesion. The spectrum from the site of the lesion showed no indication of increased Cho, ruling out aggressive neoplasm; however, this study remained inconclusive for NE localization since there was no indication of decreased NAA in, or surrounding, the lesion.
Figure 1.
MRI (a) and NAA/Cre image (b) for subject 4, and example spectra (c) from the proximity of a cortical lesion in the occipital gyrus. The spectrum overlying the cortical lesion is indicated by the arrow.
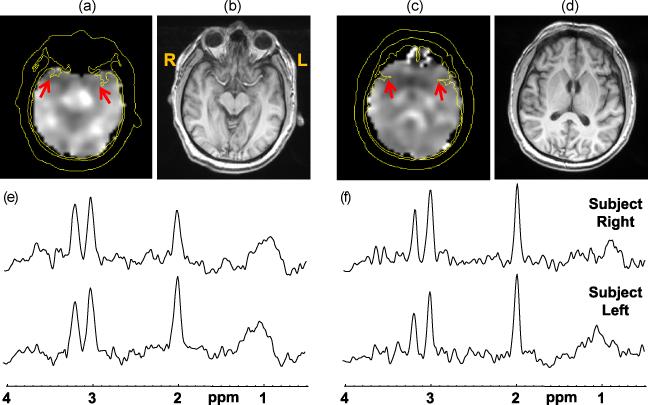
Figure 2.
Results for subject 8 at two axial slices, with the NAA/Cre images (a) and (c), and the corresponding T1-weighted MRI (b) and (d) generated by summing five 1-mm slices corresponding to the thickness of the MRSI slice. Spectra from both slices and selected from the right and left regions, as indicated by the arrows on the NAA/Cre images, are shown in (e) and (f).
In Figure 2 are shown results for subject 8, who had a negative MRI report and an EEG study that indicated localization to the right temporal lobe. The spectra selected from the temporal poles (Fig. 2e) demonstrate decreased NAA in the right side (left side of the image) relative to the contralateral side. Decreased NAA and NAA/Cre was visible in several slices, as for example shown in Fig. 2c, and extended in the superior direction over approximately a 2.5 cm. In this study the MRSI indicated that the localization was more anterior than that suggested by the EEG, and also demonstrated metabolic alterations impacting subcortical structures.
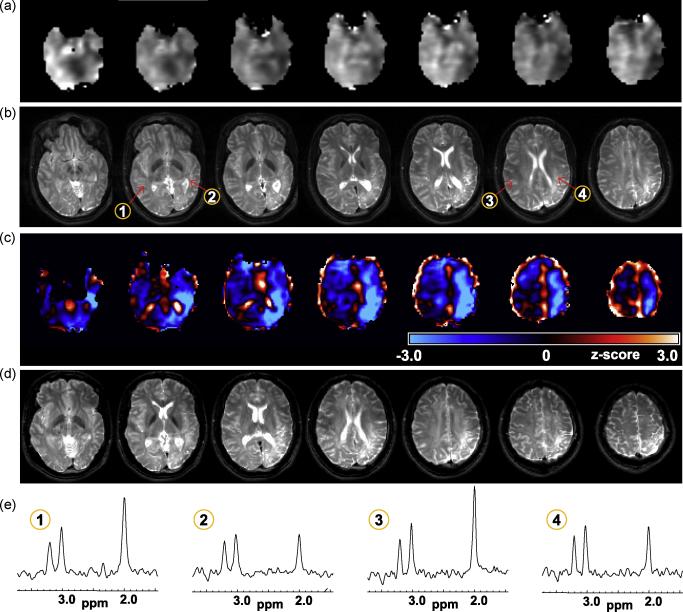
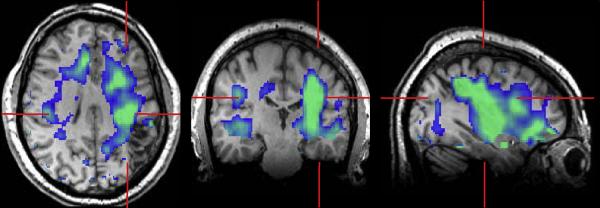
In Figures 3, 4, and 5 are shown examples for subjects that exhibited clear MRI findings. The MRI study of subject 12 indicated widened sulci and left parietal lobe atrophy, with asymmetric ventricles and diffuse signal increase on the T2-weighted MRI (Fig. 3b). Both the NAA and NAA/Cr images (Fig 3a) showed reduced signal intensity in this same region, although extending over a wider volume than would be anticipated from the MRI, which was also evident in the z-score maps (Fig. 3c). Since this z-score result is generated in the frame of the spatial reference the T2-weighted MRI is again shown in Fig. 3d following spatial registration to the reference. The z-score maps are displayed in a color scale with the light blue regions indicating significantly decreased NAA/Cre values on the order of three times the standard deviation of the normal comparison group. This image indicates widespread reductions of NAA/Cre on the left side of the brain, which is consistent with the EEG localization for this subject, and although consistent with the MRI observations the metabolic changes clearly extend over a much wider region and place greater emphasis on anterior-temporal involvement.
Figure 3.
Results for subject 12, showing (a) the NAA/Cre images at 7 axial slices and (b) the T2-weighted MRI corresponding to the center of the metabolite image slice. In (c) is shown the z-score image for NAA/Cre, which is shown in a color scale with black representing no change from normal values and running from −3σ to +3σ over the blue and red colors. The z-score image is shown in the standard spatial reference frame, and the T2 MRI is again shown in (d) following affine registration to this frame. In (e) are shown spectra from the numbered locations indicated in (b).
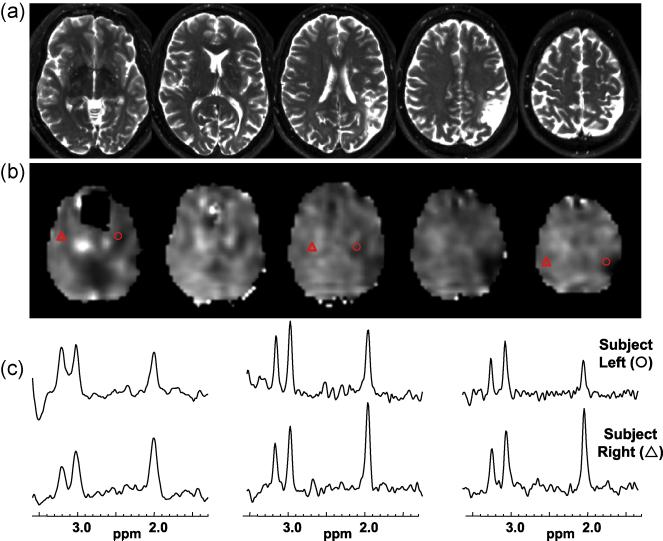
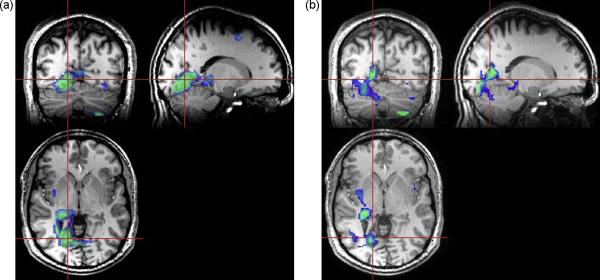
Figure 4.
Results for subject 17 that exhibits a clearly defined left parietal lesion seen on the T2-weighted MRI (a) and by decreased NAA/Cre (b). Spectra selected from contralateral anatomical locations indicated by the triangle (subject right) and circle (subject left) symbols are shown in (c) for the left, middle, and rightmost slices shown.
Figure 5.
The z-score map corresponding to the NAA/Cre image shown in Fig. 4b, shown as a color overlay thresholded to highlight voxels with NAA/Cre reduced by an amount between −1σ and −3σ, and superimposed on the T1-weighted MRI.
An additional observation from Fig. 3c is that the z-score calculation includes some voxels having anomalous values, as indicated by the white regions, that primarily occur at the edges of the brain and in ventricular regions and can be readily distinguished from the changes seen within white-matter regions. These are believed to result from increased errors in the spectral fitting where significant partial volume signal reduction occurs. Also shown in Fig. 3e are sample spectra, illustrating the consistent spectral quality obtained in this brain region and the widespread nature of the reduced NAA signal in the voxels selected from the left side of the brain.
The results of the regional tissue regression analysis for subject 12 confirmed the visual observations, with significant decreases of WM NAA/Cre for all regions within the left hemisphere, as well as increased left parietal WM and GM choline, and increased Cre in temporal and parietal lobe WM.
In Figs. 4 and 5 are shown results for subject 17, a 57 year old male that had refractory complex partial seizures since the age of two, who presented with a large left parietal lesion and EEG localization to the left temporal region. Significant loss of NAA and NAA/Cr can be seen in the lesion (Fig. 4b), but in addition these metabolic alterations can be seen to extend into white-matter regions of the temporal lobe and insula cortex, as demonstrated by comparison of spectra from contralateral locations shown in Fig. 4c. In Fig. 5 is shown the result of the z-score analysis for NAA/Cre, which demonstrates reduced values in the proximity of the lesion and extending towards the anterior temporal region. This result also illustrates the advantage of having volumetric metabolite information available. The regression analysis for this subject indicated widespread metabolic alterations with decreased NAA/Cre throughout the cerebrum, reaching greatest significance in left parietal WM and temporal GM.
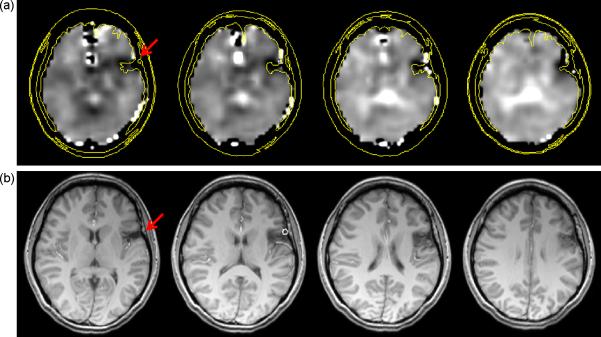
In Fig. 6 is shown the result for subject 22, which demonstrated a left frontal lesion involving the anterior insula and frontal operculum cortex, characteristic of cortical dysplasia or old trauma, and EEG localization to the left temporal-occipital region. The NAA/Cre image (Fig. 6a) is shown with an overlay indicating the brain outline and contours derived from the T1-weighted MRI. This result indicates that the metabolic findings are greater on the frontal aspect of the lesion, providing additional localization information to the MRI report. The regression analysis indicated a significant decrease of NAA/Cre in the left frontal lobe WM, but also an apparently anomalous decrease of NAA/Cre for the right occipital lobe WM.
Figure 6.
The NAA/Cre image (a) at four axial slices and the corresponding T1-weighted MRIs at 5 mm slice thickness (b) for subject 22. The arrows on the leftmost images indicate the location of a lesion and contours derived from the MRI are superimposed on the corresponding slices in (a) to illustrate the slight differences in localization between the features seen in the metabolic and structural images.
The MRI and NAA/Cre result for subject 24 is shown in Fig. 7. This subject had intractable generalized epilepsy with frequent seizures with EEG localization to the right temporal, parietal, and occipital region. The T2-weighted MRI indicated an ill-defined and mild hyperintensity in the medial right occipital lobe with no T1 contrast enhancement. A large region of decreased NAA/Cre is clearly visible in the right occipital area (Fig. 7a), which is also consistent with the area of T2 hyperintensity. However, interpretation of this MRSI result is complicated by the close proximity to the cerebellum and the temporal horn of the lateral ventricle since both of these structures are associated with significantly different concentrations of all metabolites in normal subjects (Maudsley et al., 2009). Signal changes resulting from small differences in partial volume contribution between left and right sides of the brain and the cerebellum could therefore be misinterpreted. Similarly, in the calculation of the z-score images, which depends on registration to a spatial reference, small errors in the registration can result in incorrect attribution of abnormal metabolite concentrations, although the calculation of the significance of this change is mitigated by the increased variance seen in this region for the normal subject data (an example of which can be seen in Fig. 2 of (Maudsley et al., 2009)).
Figure 7.
NAA/Cre images (a) and T1-weighted MRI (b) for subject 24. Interpretation in this portion of the occipital lobe must account for normal variations of all metabolite values in the cerebellum and periventricular regions.
The z-score maps for NAA and NAA/Cre from subject 24 are shown in Fig. 8. Both indicate significantly decreased values in the immediate region of the MRI-observed abnormality, as well as to the atrium of the right lateral ventricle, and good separation from the cerebellum. None of the asymmetry measures of this subject reached significance, although there was a trend to decreased NAA/Cr and increased Cho in the occipital lobe, presumably reflecting that the localized nature of the metabolic changes was not being detected in an analysis based on lobar-scale regions. Further evidence of the metabolic changes is presented in Fig. 9, which shows spectra integrated over 9 voxels selected from the MRI-observed abnormality and a contralateral brain region. Results of this analysis indicate a 23% reduction of NAA/Cre, 24% increase of Cho/Cre, and a 69% increase of the Cho/NAA ratio in the abnormality relative to the contralateral region, lending support to a cortical dysplasia (Mueller et al., 2005; Simister et al., 2009), although not excluding a low-grade neoplasm.
Figure 8.
Z-score map for (a) NAA, and (b) NAA/Cre for the same subject as shown in Fig. 7a. The z-score is shown as a color overlay on the T1-weighted MRI and windowed for values greater than 2σ and limited to a maximum value of 3σ. Both z-score results show good localization to the occipital-temporal region.
Figure 9.
Spectra summed over 9 voxels selected from a region of mild hyperintensity seen on the T2-weighted MRI (a) and a contralateral region (c), as indicated by the arrows on the T2 MRI (b), from subject 24.
A second subject, #25, had very similar findings to those described in the previous example with a right occipital lesion visible on the T2-weighted MRI and increased Cho and Cho/Cre and decreased NAA and NAA/Cre in this region. However, the MRSI findings for subject 25 (not shown) remained inconclusive since there were additional indications of decreased NAA/Cre in the right frontal operculum cortex. Although EEG localization was inconclusive in this subject, the seizure symptomatology implicated frontal involvement, lending support to a possible association of the frontal-lobe MRSI findings with the site of seizure activity.
Visual review of the MRSI studies indicated no notable features considered significant for five subjects (35%), and of those, the regional analyses provided no additional indication of lateralization. In one subject (#23) each of the MRS analysis approaches implicated different regions, all of which were inconsistent with the EEG localization to the frontal lobe. Possible influencing factors include that although the spectral quality for this study was good, the result also showed evidence of increased lipid signal within the brain and possible motion artifacts, and that the subject had a history of meningitis as a child and developmental delay, the effects of which on the MRS findings is unknown.
Discussion
The results of this study support previous reports that have indicated the potential of proton MRS to detect altered metabolism associated with neocortical epilepsy, while additionally demonstrating the advantages of the volumetric MRSI approach and implementation using three-Tesla instrumentation. Similar to previous reports, the findings of metabolic alterations remain variable across subjects, with approximately 50% of subjects exhibiting either no detectable metabolic changes or inconclusive findings, and widespread metabolic changes being seen in other subjects that clearly extend beyond areas of MRI-visible pathologies. From a patient evaluation perspective the MRSI measurement provided additional localization information, and in two cases, provided some evidence of localization in the absence of any MRI-observable features.
The analysis approach that was found to be the most effective relied on detection of reduced signal intensity in the NAA/Cre images, with validation by inspection of the individual NAA and Cre images. The z-score maps were found to be a valuable adjunct to this visual analysis, enabling easier interpretation of the metabolic changes. Future developments will provide these images following inverse spatial transform, thereby coregistering them to the original subject MRI.
In no study was any significant change of Cho, or Cho/Cre, directly visualized in the metabolic images, although changes could be detected in individual quantitative analyses following signal integration over specific brain regions, as demonstrated in Fig. 9. This finding is consistent with the report of Stanley et al (Stanley et al., 1998), whereas both increased and decreased Cho were reported by Mueller at al. (Mueller et al., 2005) for dysplasia and heterotopia, respectively. While the lack of significant Cho changes in this study could simply reflect the subject selection and pathologies present, an additional consideration must include the lower reliability for the Cho measurement relative to that of Cre and NAA, as discussed below.
There are several observations from this study related to both the methodologies used and the clinical value of the measurement. With regard to the quality of the MRS measurement, the volumetric MRSI acquisition method was found to provide good coverage of the cerebrum, although there clearly remain regions of increased magnetic field homogeneity where data cannot be obtained. Sampling of the orbito-frontal and temporal pole regions, which can be problematic for any MRS measurement method, can be obtained in some cases, as shown in Fig. 2; although, this was not a consistent finding. Although the MRSI method used in this study sampled a wider region of the brain than any previous study of this patient group, the incomplete spatial sampling still remains a potential reason for the lack of findings in some subjects.
Related to this same issue of susceptibility-induced field inhomogeneity is the effect of strong magnetic field shifts in localized brain regions, resulting in incomplete water suppression and partial suppression of the Cho resonance. Despite the effective use of postprocessing methods to remove residual water signal, a common consequence of inadequate water suppression was found to be a rapidly varying baseline in the spectral region close to the Cho resonance. Due to the fact that the spectral model assumes a baseline that is more slowly varying than the metabolite resonances, the presence of large and rapidly varying residual water signals can result in less reliable estimates for the Cho peak. Although the MIDAS processing includes assessments of spectral quality as well as the accuracy of the spectral quantitation, and voxels of inadequate quality were excluded from the region-based analysis, it still remained necessary for a trained investigator to inspect all regions of interest. A more widespread application of these methods will benefit from development of improved acquisition and quality control procedures.
As indicated from previous reports, this study found that in some subjects the metabolic abnormalities were present over wide regions, which were associated with, but not necessarily matching, MRI-observed pathology. This finding is consistent with those of other MRSI studies and nuclear medicine studies (Goffin et al., 2008). It is therefore somewhat surprising that the regional-based analyses did not provide additional information over direct visual observation, and did not provide any indications of lobar-scale localization in the absence of visual localization. Several alternative approaches to the regional analysis are possible, including taking smaller regions and detection of outlying values relative to a local statistical analysis (Mueller et al., 2004), in contrast to the comparison against normal values as used in this study. An additional observation is that the lobar-scale regression analysis appeared to be consistent for the WM result, relative to visual analysis and EEG localization, although not for the GM values. Possible reasons for this may include errors in tissue segmentation, particularly with regard to CSF quantitation, and decreased spectral quality at the periphery of the imaged region that primarily affected quantitation in GM regions.
Limitations of this study include the relatively small number of subjects and that confirming information on the localization of the epileptogenic focus, for example from surgery, was not available. Additionally, information from MEG or PET studies was of limited value.
Conclusion
This study supports previous observation of reduced NAA and NAA/Cre ratio, indicative of metabolic alteration associated with tissues involved in neocortical epilepsies, and the value of this observation for epilepsy localization. The volumetric MRSI approach provides greater sampling efficiency than has previously been reported for this application, while implementation on three Tesla MRI instruments provides sufficient sensitivity to obtain better spatial resolutions in comparison to previous studies. The acquisition and processing of the volumetric MRSI method can be conveniently implemented in a routine clinical environment; however, there remains the potential for misinterpretation of common MRSI artifacts, indicating that experienced technical personnel would still be required for clinical diagnostic studies. Nevertheless, these results support the need for continued development and evaluation of the MRSI method for localization of neocortical epilepsy.
Acknowledgements
This work was supported by NIH grant R01NS041946 with additional data from control subjects acquired under R01EB00822 and R01NS055107. We thank Dr. Jose Gonzales, of the Epilepsy Foundation of South Florida, and Marinellie Vega for assistance with subject recruitment. We also thank Dr. Ken Laxer for early support in initiating this project and the developers of the MIDAS processing software implemented under R01EB00822.
References
- Collins DL, Zijdenbos AP, Kollokian V, Sled JG, Kabani NJ, Holmes CJ, Evans AC. Design and construction of a realistic digital brain phantom. IEEE Transactions on Medical Imaging. 1998;17:463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- Ebel A, Maudsley AA. Improved spectral quality for 3D MR spectroscopic imaging using a high spatial resolution acquisition strategy. Magn Reson Imaging. 2003;21:113–120. doi: 10.1016/s0730-725x(02)00645-8. [DOI] [PubMed] [Google Scholar]
- Garcia PA, Laxer KD, van der Grond J, Hugg JW, Matson GB, Weiner MW. Correlation of seizure frequency with N-acetyl-aspartate levels determined by 1H magnetic resonance spectroscopic imaging. Magnetic Resonance Imaging. 1997;15:475–478. doi: 10.1016/s0730-725x(96)00327-x. [DOI] [PubMed] [Google Scholar]
- Goffin K, Dedeurwaerdere S, Van Laere K, Van Paesschen W. Neuronuclear assessment of patients with epilepsy. Semin Nucl Med. 2008;38:227–239. doi: 10.1053/j.semnuclmed.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Guye M, Ranjeva JP, Le Fur Y, Bartolomei F, Confort-Gouny S, Regis J, Chauvel P, Cozzone PJ. 1H-MRS imaging in intractable frontal lobe epilepsies characterized by depth electrode recording. Neuroimage. 2005;26:1174–1183. doi: 10.1016/j.neuroimage.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Hartman AL, Lesser RP. Update on epilepsy and cerebral localization. Curr Neurol Neurosci Rep. 2007;7:498–507. doi: 10.1007/s11910-007-0077-4. [DOI] [PubMed] [Google Scholar]
- Haupt CI, Schuff N, Weiner MW, Maudsley AA. Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn Reson Med. 1996;35:678–687. doi: 10.1002/mrm.1910350509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krsek P, Hajek M, Dezortova M, Jiru F, Skoch A, Marusic P, Zamecnik J, Kyncl M, Tichy M, Komarek V. (1)H MR spectroscopic imaging in patients with MRI-negative extratemporal epilepsy: correlation with ictal onset zone and histopathology. Eur Radiol. 2007;17:2126–2135. doi: 10.1007/s00330-007-0594-1. [DOI] [PubMed] [Google Scholar]
- Kuzniecky R, Devinsky O. Surgery Insight: surgical management of epilepsy. Nat Clin Pract Neurol. 2007;3:673–681. doi: 10.1038/ncpneuro0663. [DOI] [PubMed] [Google Scholar]
- Kuzniecky R, Hetherington H, Pan J, Hugg J, Palmer C, Gilliam F, Faught E, Morawetz R. Proton spectroscopic imaging at 4.1 tesla in patients with malformations of cortical development and epilepsy. Neurology. 1997;48:1018–1024. doi: 10.1212/wnl.48.4.1018. [DOI] [PubMed] [Google Scholar]
- Laxer KD. Clinical applications of magnetic resonance spectroscopy. Epilepsia. 1997;38(Suppl 4):S13–17. doi: 10.1111/j.1528-1157.1997.tb04534.x. [DOI] [PubMed] [Google Scholar]
- Leite CC, Lucato LT, Sato JR, Valente KD, Otaduy MC. Multivoxel proton MR spectroscopy in malformations of cortical development. AJNR Am J Neuroradiol. 2007;28:1071–1075. doi: 10.3174/ajnr.A0511. discussion 1076–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LM, Cendes F, Andermann F, Dubeau F, Arnold DL. Spatial extent of neuronal metabolic dysfunction measured by proton MR spectroscopic imaging in patients with localization-related epilepsy. Epilepsia. 2000;41:666–674. doi: 10.1111/j.1528-1157.2000.tb00226.x. [DOI] [PubMed] [Google Scholar]
- Lundbom N, Gaily E, Vuori K, Paetau R, Liukkonen E, Rajapakse JC, Valanne L, Hakkinen AM, Granstrom ML. Proton spectroscopic imaging shows abnormalities in glial and neuronal cell pools in frontal lobe epilepsy. Epilepsia. 2001;42:1507–1514. doi: 10.1046/j.1528-1157.2001.15301.x. [DOI] [PubMed] [Google Scholar]
- Maudsley AA, Darkazanli A, Alger JR, Hall LO, Schuff N, Studholme C, Yu Y, Ebel A, Frew A, Goldgof D, Gu Y, Pagare R, Rousseau F, Sivasankaran K, Soher BJ, Weber P, Young K, Zhu X. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. doi: 10.1002/nbm.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, Bloomer C. Mapping of brain metabolite distributions by volumetric proton MR Spectroscopic Imaging (MRSI) Magn Reson Med. 2009;61:548–559. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudsley AA, Domenig C, Sheriff S. Reproducibility of serial whole-brain MR spectroscopic imaging. NMR Biomed. 2009;22:1–7. doi: 10.1002/nbm.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudsley AA, Studholme C, Govindaraju V. International Society for Magnetic Resonance in Medicine. Toronto: 2008. Tissue-dependent analysis of metabolic alterations in the brain by MR spectroscopic imaging; p. 1604. [Google Scholar]
- Moffett JR, Ross B, Arun P, Madhavarao CN, Namboodiri AMA. N-Acetylaspartate in the CNS: From neurodiagnostics to neurobiology. Progress in Neurobiology. 2007;81:89–131. doi: 10.1016/j.pneurobio.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, D. LK, Barakos JA, Cashdollar N, Flenniken DL, Vermathen P, Matson GB, Weiner MW. Identification of the epileptogenic lobe in neocortical epilepsy with proton MR spectroscopic imaging. Epilepsia. 2004;45:1580–1589. doi: 10.1111/j.0013-9580.2004.53003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Laxer KD, Barakos JA, Cashdollar N, Flenniken DL, Vermathen P, Matson GB, Weiner MW. Metabolic characteristics of cortical malformations causing epilepsy. J Neurol. 2005;252:1082–1092. doi: 10.1007/s00415-005-0819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Laxer KD, Cashdollar N, Flenniken DL, Matson GB, Weiner MW. Identification of abnormal neuronal metabolism outside the seizure focus in temporal lobe epilepsy. Epilepsia. 2004;45:355–366. doi: 10.1111/j.0013-9580.2004.27603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JW, Williamson A, Cavus I, Hetherington HP, Zaveri H, Petroff OA, Spencer DD. Neurometabolism in human epilepsy. Epilepsia. 2008;49(Suppl 3):31–41. doi: 10.1111/j.1528-1167.2008.01508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Maudsley AA, Ebel A, Darkazanli A, Weber P, Sivasankaran K, Yu Y, Studholme C. Proc Soc Photo Opt Instrum Eng. Vol. 5747. San Diego: 2005. Evaluation of sub-voxel registration accuracy between MRI and 3D MR spectroscopy of the brain; pp. 1213–1221. [Google Scholar]
- Simister RJ, McLean MA, Barker GJ, Duncan JS. Proton MR spectroscopy of metabolite concentrations in temporal lobe epilepsy and effect of temporal lobe resection. Epilepsy Res. 2009;83:168–176. doi: 10.1016/j.eplepsyres.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis III: Application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn Reson Med. 1998;40:822–831. doi: 10.1002/mrm.1910400607. [DOI] [PubMed] [Google Scholar]
- Stanley JA, Cendes F, Dubeau F, Andermann F, Arnold DL. Proton magnetic resonance spectroscopic imaging in patients with extratemporal epilepsy. Epilepsia. 1998;39:267–273. doi: 10.1111/j.1528-1157.1998.tb01371.x. [DOI] [PubMed] [Google Scholar]
- Studholme C, Hill DL, Hawkes DJ. Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimization of voxel similarity measures. Medical Physics. 1997;24:25–35. doi: 10.1118/1.598130. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]