Abstract
Poxviruses encode a redox system for intramolecular disulfide bond formation in cytoplasmic domains of viral proteins. Our objectives were to determine the kinetics and intracellular location of disulfide bond formation. The vaccinia virus L1 myristoylated membrane protein, used as an example, has three intramolecular disulfide bonds. Reduced and disulfide-bonded forms of L1 were distinguished by electrophoretic mobility and reactivity with monoclonal and polyclonal antibodies. Because disulfide bonds formed during 5 min pulse-labeling with radioactive amino acids, a protocol was devised in which dithiothreitol was present at this step. Disulfide bond formation was detected by 2 min after removal of reducing agent and was nearly complete in 10 min. When the penultimate glycine residue was mutated to prevent myristoylation, L1 was mistargeted to the endoplasmic reticulum and disulfide bond formation failed to occur. These data suggested that viral membrane association was required for oxidation of L1, providing specificity for the process.
Keywords: protein disulfide bonds, poxvirus morphogenesis, viral membrane
Introduction
In eukaryotic cells, stable disulfide bonds form exclusively in the relatively oxidizing environment of the endoplasmic reticulum (ER) (Frand et al., 2000; Tu et al., 2000). Poxviruses replicate in the cytoplasm of infected cells (Condit et al., 2006; Moss, 2007) and provide an exception to this rule. Poxviruses encode three cytoplasmic oxidoreductases that comprise a complete pathway for intramolecular disulfide bond formation (Senkevich et al., 2002b). E10, the first vaccinia virus (VACV) protein in this pathway, is a member of the ERV1/ALR family of flavine adenine dinucleotide-containing sulfhydryl oxidases (Senkevich et al., 2000b). A2.5, the next component, forms transient disulfide linked complexes with E10 and with the third component of the pathway, the thioredoxin G4 (Senkevich et al., 2002a; White et al., 2002). G4 also forms an intermediate with its substrate proteins. The known target proteins for this pathway are nine components of the membrane of mature virus particles that are required for cell entry and membrane fusion (Bisht et al., 2008; Brown et al., 2006; Senkevich et al., 2005). The disulfide bonds of these proteins are located within their ectodomains, which face the cytoplasm during virion assembly. The three oxidoreductases as well as each of the target membrane proteins are conserved in all members of the poxvirus family that have been analyzed. Indeed, E10 is conserved in all nucleo-cytoplasmic large DNA viruses (poxviruses, asfarviruses, iridoviruses, phycodnaviruses and mimiviruses) and G4 is conserved in all but asfarviruses (Iyer et al., 2006).
The viral oxidoreductases locate in the regions of the cytoplasm where assembly of virus particles occurs and are incorporated into virions (Senkevich et al., 2002a; Senkevich et al., 2000a; White et al., 2000). The reducing nature of the cytoplasm is unaltered by VACV infection as determined by the ratio of oxidized to reduced glutathionine (Locker and Griffiths, 1999). Glutathione, however, is not directly involved in the poxviral redox pathway because electrons are transferred through covalent protein intermediates (Senkevich et al., 2000b). The atomic structure of one of the target proteins, L1, indicates that the six cysteine side chains are relatively solvent inaccessible, probably contributing to their maintenance in the oxidized state (Su et al., 2005).
The purpose of the present study was to determine the kinetics and intracellular location of disulfide bond formation in VACV infected cells. L1, the first VACV membrane protein shown to contain intramolecular disulfide bonds, is attractive as a model substrate because it has been well characterized structurally and functionally and is conserved in all poxviruses. L1 is a myristoylated transmembrane protein of 250 amino acids (Franke et al., 1990; Ravanello and Hruby, 1994a). The structure of the 185 amino acid ectodomain has been solved to 1.5 Å and exhibits a fold consisting of bundles of α-helices packed against a pair of two-stranded β-sheets (Su et al., 2005). Analyses by mass spectrometry and X-ray crystallography have shown that the disulfides are formed between Cys-34 and Cys-57, Cys-49 and Cys-136, and Cys-116 and Cys-158 (Aldaz-Carroll et al., 2005; Su et al., 2005). Cys-34 and Cys-57 are located in loops, Cys-69 and Cys-136 in strands and Cys-116 and Cys-158 between and within helices, respectively. Although L1 had been thought to have a role in virus assembly (Ravanello and Hruby, 1994b), more recent data indicate that it is required exclusively for virus entry and membrane fusion (Bisht et al., 2008). Furthermore, L1 is a target of neutralizing antibody (Ichihashi and Oie, 1996; Wolffe et al., 1995) and is a component of experimental smallpox vaccines (Fogg et al., 2004; Hooper et al., 2000; Lustig et al., 2005).
Using pulse-chase protocols and confocal microscopy we now show that disulfide bond formation occurs within a few minutes after synthesis of L1 and that localization of L1 on viral membranes within cytoplasmic factories is required. This localization may provide specificity for oxidation of viral membrane components as opposed to other viral and cellular proteins.
Results
Reduced and oxidized forms of L1 can be distinguished by electrophoretic mobility and antibody reactivity
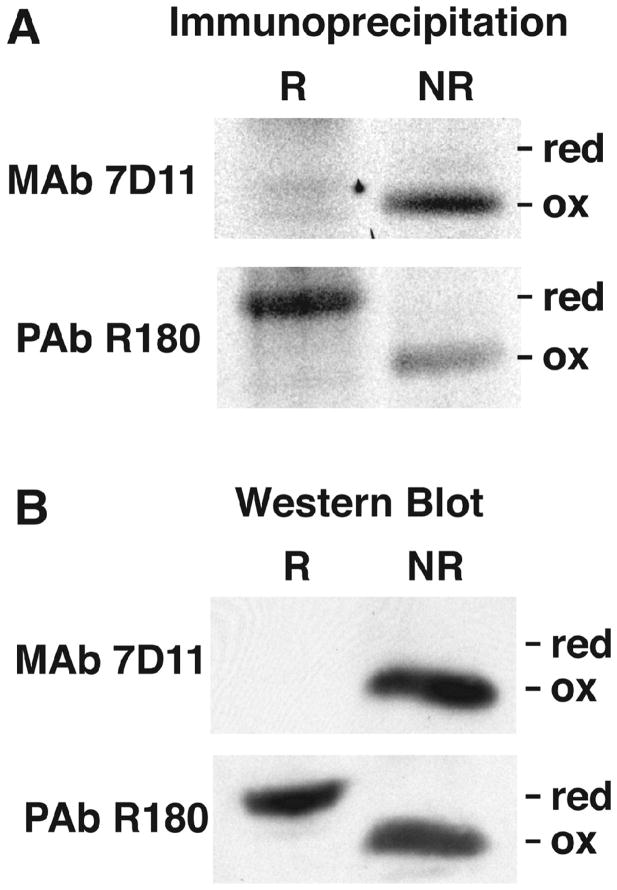
To determine the kinetics of L1 disulfide bond formation, our plan was to carry out pulse-labeling of VACV infected cells with radioactive amino acids, chase with unlabeled amino acids, and monitor the formation of intramolecular disulfide bonds. For this purpose, we needed to distinguish reduced (-SH) and oxidized (S-S) forms of L1. Previous studies (Wolffe et al., 1995) showed that the disulfide-bonded form of L1 migrates more rapidly than the reduced form by SDS-polyacrylamide gel electrophoresis (PAGE), presumably due to a more compact structure. In addition, monoclonal antibody 7D11 (abbreviated in the text as L1 MAb) specifically reacts with the disulfide-bonded form of L1 on Western blots (Wolffe et al., 1995). The polyclonal antibody R180 (abbreviated in the text as L1 PAb) prepared by injecting rabbits with a secreted recombinant form of L1 (Aldaz-Carroll et al., 2005) was expected to react with reduced and oxidized forms. In order to test the specificity of the antibodies, reduced and oxidized L1 were prepared by lysing infected cells with NP-40 in the presence of the reducing agent dithiothreitol (DTT) or in the presence of N-ethylmaleimide (NEM) to react with free cysteines and prevent disulfide interchange, respectively. The anticipated specificities of L1 MAb and L1 PAb were demonstrated by immunoprecipitation (Fig. 1A) as well as by Western blotting (Fig. 1B). The L1 PAb recognized both the reduced (~28 kDa) and oxidized (~ 25 kDa) bands, whereas L1 MAb only recognized the latter. Therefore, both electrophoretic migration and antibody reactivity distinguished reduced and oxidized forms of L1.
Fig. 1.
Specificities of L1 MAb and PAb for reduced and oxidized L1. (A) Immunoaffinity purification. BS-C-1 cells were infected with VACV and labeled from from 2 to 16 h with [35S]methionine/cysteine mixture. Lysates were prepared in the presence of DTT (R) or NEM (NR) and incubated with L1 MAb 7D11 or L1 PAb R180 complexed to protein G agarose. The proteins were eluted with sample buffer lacking reducing agent and analyzed by SDS-PAGE and autoradiography. Bands corresponding to reduced (red) and oxidized (ox) L1 are indicated. (B) Western blotting. Lysates were prepared as described in panel A and analyzed by SDS-PAGE and Western blotting with MAb 7D11 and PAb R180. Proteins were detected by chemiluminescence.
Kinetics of L1 disulfide bond formation
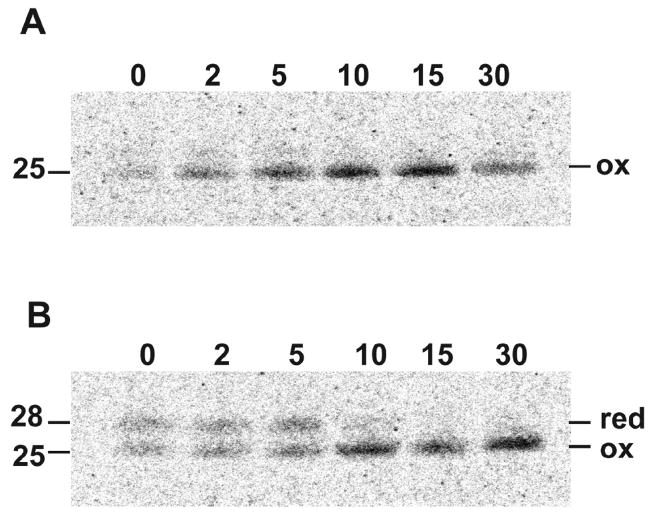
Cells that had been infected with VACV for 16 h were incubated in medium depleted of methionine and cysteine for 15 min to reduce the intracellular pools of these amino acids and then pulse-labeled with [35S]methionine and [35S]cysteine for 5 min. Following labeling, the cells were washed and the incubation at 37°C continued with unlabeled amino acids. At varying times, the cells were washed with ice-cold phosphate buffered saline, lysed in the presence of NEM and incubated with L1 MAb or L1 PAb attached to protein G beads. The bound proteins were then resolved by SDS-PAGE and the radioactivity determined by autoradiography. At 0 time, e.g. immediately after the 5 min labeling period, similar amounts of reduced and oxidized L1 were captured by L1 PAb (Fig. 2B). By 10 min, nearly all of the L1 was in the more rapidly migrating oxidized state (Fig. 2B). The 25-kDa band was the major species captured by L1 MAb and the amount increased with time (Fig. 2A). However, there was a faint 28-kDa band recognized by L1 MAb, which can be better seen in subsequent gels. The rapid kinetics indicated that folding and disulfide bond formation occurred extremely rapidly after synthesis of L1.
Fig. 2.
Kinetics of disulfide bond formation. BS-C-1 cells were infected with VACV for 16 h, incubated with methionine- and cysteine-free medium for 15 min, and then pulsed with 100 μCi of [35S]methionine/cysteine-labeling mix for 5 min. Following the pulse, cells were incubated in medium containing excess methionine and cysteine for 2 to 30 min as indicated, lysed, affinity purified with L1 MAb (A) or L1 PAB (B) and analyzed by SDS-PAGE and autoradiography. Bands corresponding to reduced (red) and oxidized (ox) L1 are indicated on the right. Numbers on the left indicate the masses of the L1 proteins in kDa determined by co-electrophoresis of marker proteins.
Kinetics of L1 disulfide bond formation following removal of DTT
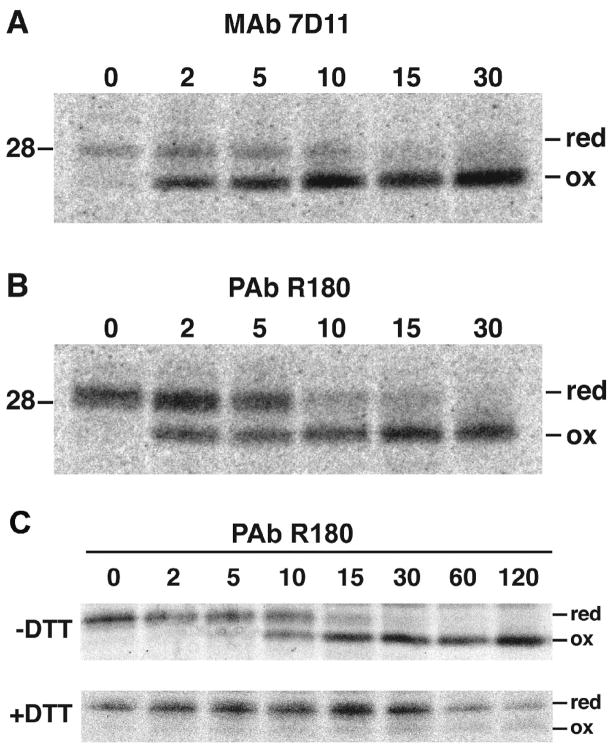
To refine our analysis, we adapted a method used by Braakman and Helenius (Braakman et al., 1992) to study disulfide bond formation in the ER. They demonstrated that addition of DTT to the medium of cells prevented disulfide bond formation of a newly synthesized protein. However, when the medium was changed, oxidation occurred rapidly and the correctly folded protein was transported through the secretory pathway. Although we did not find a reference for the use of DTT to prevent the formation of cytoplasmic disulfide bonds, we decided to adapt the procedure. The protocol used in Fig. 2 was modified by continuing the amino acid depletion step for an additional 5 min in the presence of DTT, which was maintained during the 5 min labeling period. The cells were washed and DTT removed for the chase. At 0 time, L1 PAb captured the reduced form of L1 but only a trace of oxidized L1 (Fig. 3B). Within 2 min, however, the amount of oxidized L1 increased and the conversion of reduced to oxidized L1 was nearly complete by 10 min (Fig. 3B). Similar kinetics of disulfide bond formation was determined with L1 MAb (Fig. 3A). Thus, by keeping the L1 in the reduced state during the labeling period, the kinetics of disulfide bond formation was precisely measured.
Fig. 3.
Kinetics of disulfide bond formation following DTT removal. The pulse-chase protocol was similar to that described in the legend to Fig. 2 except that the 15 min pre-incubation in methionine- and cysteine-free medium was followed by an additional 5 min incubation in the same medium containing 5 mM DTT and then pulsed with 100 μCi of [35S]methionine/cysteine-labeling mix for 5 min in the continued presence of DTT. After the chase of 2 to 30 min in the absence of DTT, the L1 protein was affinity purified with L1 MAb (A) or L1 PAB (B) and analyzed by SDS-PAGE and autoradiography. The protocol for the experiment in panel C was similar except that 5 mM DTT (+ DTT) was maintained during the chase in one sample. Abbreviations are the same as in the legend to Fig. 2.
Comparison of the ~28-kDa bands in the 0 to 5 min samples of protein captured by L1 MAb (Fig. 3A) and L1 PAb (Fig. 3B) indicated a subtle difference. The L1 MAb-associated band was relatively sharp and migrated at 28-kDa, whereas the L1 PAb-associated band was diffuse with the more intense part above 28-kDa. We suspect that partially oxidized forms of L1 migrated slightly faster than the fully reduced form and were recognized and captured by L1 MAb. The more diffuse 28- to 29-kDa band formed by proteins captured by L1 PAb would consist of reduced and partially oxidized L1.
In a variation of the above experiment, DTT was retained during the chase period; the majority of the L1 remained in the slow migrating reduced form with only a trace of the oxidized 25-kDa L1 (Fig. 3C). The amount of reduced L1 decreased between 30 and 60 min suggesting that it might be unstable under these conditions.
Viral membrane association is required for formation of disulfide bonds
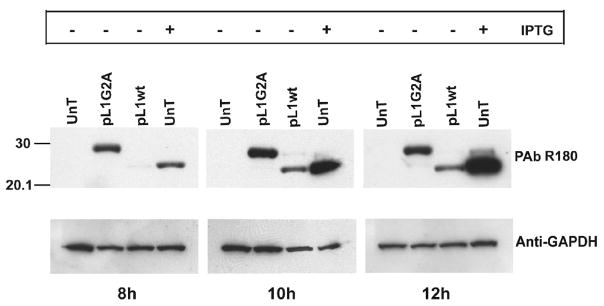
L1 has a C-terminal transmembrane domain, characteristic of proteins that associate with membranes post-translationally. Centrifugation experiments indicated that the reduced L1 present immediately after pulse-labeling was largely in the soluble fraction, whereas the oxidized L1 at 15 min was mostly in the membrane-containing pellet (data not shown). This result suggested that intramolecular disulfide-bond formation might be occurring at the viral membrane. A previous study had suggested that the N-terminal myristate moiety is necessary for incorporation of L1 into virions (Ravanello and Hruby, 1994b). To test whether myristoylation is required for oxidation of L1, we carried out an infection-transfection experiment. To prevent expression of virally encoded L1, cells were infected in the absence of inducer with vL1Ri, a recombinant VACV that has L1R regulated by the Escherichia coli lac operator (Bisht et al., 2008). The cells were then transfected with plasmids encoding wild type (wt) L1 (pL1wt) or L1 with the penultimate N-terminal amino acid changed from glycine to alanine (pL1G2A) to prevent myristoylation. As controls, cells were also infected with vL1Ri in the absence and presence of isopropyl-β-D-thiogalactopyranoside (IPTG) but were not transfected. At 8, 10 and 12 h after infection, the cells were harvested and L1 was detected by Western blotting with L1 PAb. As expected, L1 was not detected in the untransfected cells that did not receive IPTG and the L1 was entirely in the disulfide-bonded form when IPTG was present (Fig. 4). Both the G2A mutant and wt type L1 were expressed from the transfected plasmids. Importantly, the L1 G2A mutant was entirely in the reduced state whereas the wt L1 was mostly disulfide-bonded (Fig. 4). Unexpectedly, the L1 G2A mutant protein was detected earlier and in higher amounts than wt L1.
Fig. 4.
Effect of mutation of the myristoylation site of L1 on disulfide bond formation. BS-C-1 cells were infected with vL1Ri in the presence (+) or absence (−) of IPTG and transfected 1 h later with pL1wt or pL1G2A or left untransfected (UnT). After 8, 10 and 12 h the cells were lysed and analyzed by SDS-PAGE and Western blotting with L1 PAb. The blot was reprobed with antibody to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a loading control. Numbers on the left refer to position and mass of marker protein in kDa.
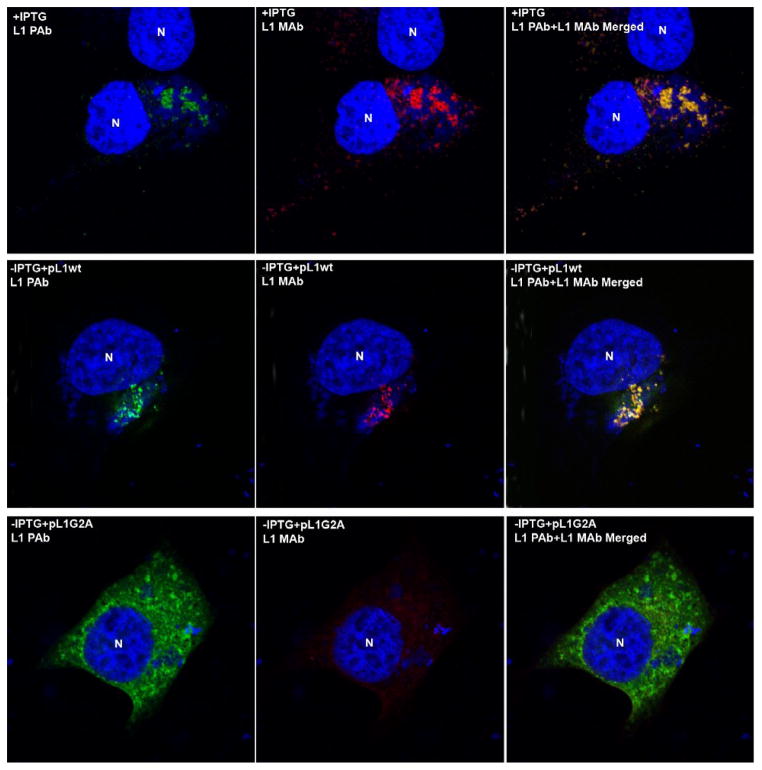
Poxvirus replication and assembly occur in cytoplasmic factories, which are typically located near the nucleus of the infected cell. Confocal microscopy was carried out to determine the intracellular locations of the wt and G2A mutant L1 proteins. Cells were infected with vL1Ri in the presence or absence of IPTG and the latter were transfected with pL1wt or pL1G2A. In the cells infected with the virus in the presence of IPTG, L1 was visualized by staining with the L1 MAb and L1 PAb. With both antibodies, L1 staining predominantly co-localized in the viral factories detected with 4′,6-diamidino-2-phenylindole (DAPI), which avidly binds double-stranded DNA (Fig. 5). The punctate L1 staining represents clusters of immature and mature virus particles (Wolffe et al., 1995). Staining for L1 was not detected when IPTG was omitted, confirming the specificity of the antibodies (data not shown). However, the images obtained when the cells were infected with vL1Ri in the absence of IPTG and transfected with pL1wt L1 were similar to those produced in the presence of IPTG (Fig. 5). In contrast, when the cells were transfected with pL1G2A, mutated L1 was only detected with the L1 PAb antibody and was dispersed throughout the cytoplasm and did not exhibit punctate staining (Fig. 5). The failure to stain the L1 G2A mutant with L1 MAb was consistent with the Western blotting experiment, which showed that the L1 G2A mutant did not form intramolecular disulfide bonds.
Fig. 5.
Intracellular localization of myristoylated and unmyristoylated L1. HeLa cells were infected with vL1Ri in the presence (top row) or absence (middle and bottom rows) of IPTG and 1 h later transfected with pL1wt (middle row) or pL1G2A (bottom row). After 16 h the cells were stained with L1 PAb and L1 MAb followed by Alexa Fluor 488 anti-rabbit antibody and Alexa Fluor 568 anti-mouse, respectively. Cells were subsequently stained with DAPI and visualized by confocal microscopy. Red, L1 MAb; green, L1 PAb; blue, DNA. N, nucleus.
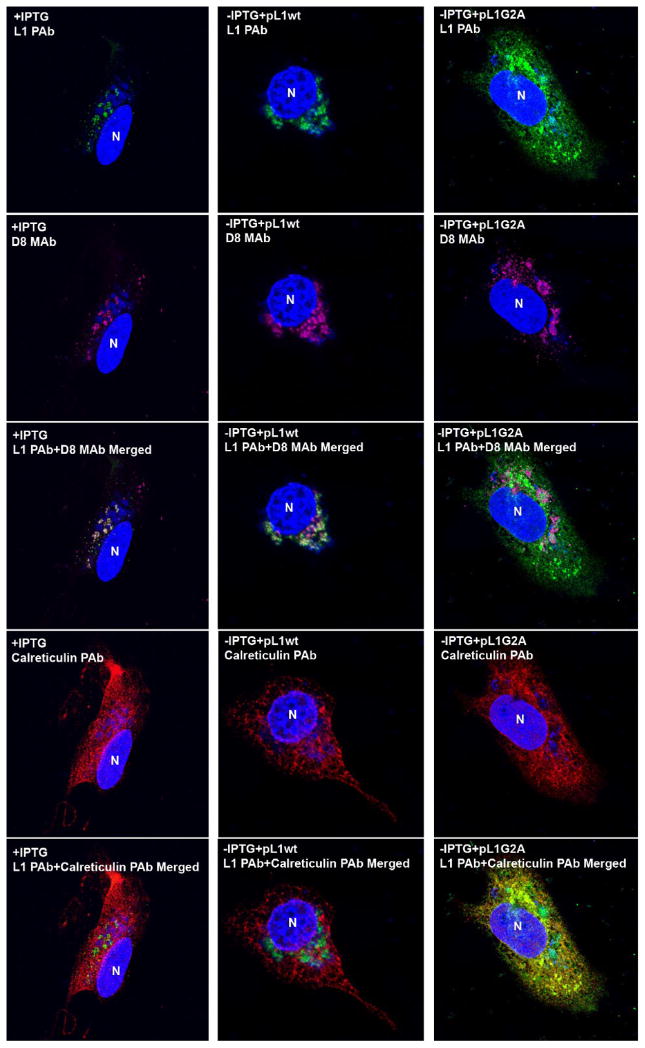
In a subsequent transfection experiment, the cells were stained with MAb to the MV membrane protein D8 and PAb to the ER resident protein calreticulin, as well as with L1 PAb. In the +IPTG samples and the −IPTG samples transfected with pL1wt, the L1 and D8 largely co-localized with each other near viral DNA and not with calreticulin (Fig. 6). In contrast, the L1 G2A mutant did not co-localize with D8 in factories but was dispersed and partly overlapped calreticulin (Fig. 6).
Fig. 6.
Co-localization of unmyristoylated L1 with ER resident protein. HeLa cells infected with vL1Ri in the presence (1st column) or absence (2nd and 3rd columns) of IPTG were transfected with pL1wt (2nd column) or pL1G2A (3rd column). Cells were stained with L1 PAb (top row), anti-D8 MAb (2nd row), and anti-calreticulin chicken PAb (4th row) followed by Alexa Fluor 488 anti-rabbit, Alexa Fluor 568 anti-mouse and Alexa Fluor 647 anti-chicken antibody and analyzed by confocal microscopy. DNA was stained with DAPI. Pink, anti-D8 MAb; green, L1 PAb; red, anti-calreticulin PAb; blue, DNA. N, nucleus.
Discussion
The poxvirus redox system is comprised of three proteins: E10, A2.5 and G4, which transfer electrons through covalent protein intermediates (Senkevich et al., 2002b). G4 interacts directly with the target protein and has been shown to form a covalent intermediate with L1. The encoding of a cytoplasmic redox system by poxviruses has raised questions regarding the specificity for viral versus cellular proteins. Since at least eight VACV proteins in addition to L1 utilize the redox system for intramolecular disulfide bond formation, sequence specificity seems unlikely. A common feature of these nine viral proteins is their possession of a transmembrane domain and localization in the mature virion membrane. Using the L1 protein as a model substrate, we showed that intramolecular disulfide bond formation occurs within minutes of protein synthesis and that association with the viral membrane is necessary.
Under standard pulse-chase conditions, L1 was already 50% disulfide bonded at the end of the 5 min labeling period. However, adding DTT to the medium just before and during the labeling period prevented oxidation. By removing DTT for the chase, it was possible to monitor the kinetics of disulfide bond formation precisely. Disulfide-bonded L1 was detected within 2 min after removal of DTT and oxidation was nearly complete between 5 and 10 min. Using a similar DTT wash out strategy, Braakman and Helenius (Braakman et al., 1992) determined that the time needed for half of influenza hemagglutinin to reach the fully oxidized form in the ER was 3 min. This time is similar to that occurring with the poxvirus cytoplasmic redox system.
N-myristoylation is a covalent modification of the penultimate glycine residue with a 14-carbon saturated fatty acid that usually occurs co-translationally upon removal of the N-terminal methionine and provides weak protein membrane and protein-protein interactions (Farazi et al., 2001). A previous study had suggested that myristoylation of the penultimate glycine was necessary for association of L1 with virions (Ravanello and Hruby, 1994b). Here we demonstrated by confocal microscopy that this mutation prevented L1 from localizing in the factory area of the cell. Instead, the mutated L1 localized with ER membranes throughout the cytoplasm. Furthermore, the unmyristoylated L1 did not contain intramolecular disulfide bonds. We do not believe that myristoylation is necessary for the correct folding of L1 since correct intramolecular disulfide bonds form when L1 is routed through the ER by engineering a signal peptide at the N-terminus, which also prevents myristoylation (Aldaz-Carroll et al., 2005). In addition, correct disulfide bonds form when unmyristoylated L1 is expressed in Escherichia coli and then refolded in vitro (Su et al., 2005). Therefore, the inability of the unmyristoylated protein to form disulfide bonds in the cytoplasm of infected cells is likely due to its failure to associate with viral membranes, where the viral oxidoreductases reside. The topology of L1 would prevent the oxidoreductases in the ER lumen from participating in disulfide bond formation. How myristoylation at the N-terminus modulates the trafficking of L1 to the viral membrane is not understood. This is particularly puzzling since F9, a disulfide bonded MV membrane protein related in sequence and structure to L1 (Su et al., 2005), is not myristoylated.
In conclusion, the localization of proteins on the viral membrane appears to provide specificity for disulfide bond formation. The rapidity of disulfide bond formation indicates that oxidation must occur during or very soon after insertion of L1 into the viral membrane.
Materials and methods
Cells and viruses
BS-C-1 cells were propagated in minimum essential medium with Earle’s salts supplemented with 10% fetal bovine serum, 100 units of penicillin and 100 μg of streptomycin per ml (Quality Biologicals, Gaithersburg, MD). The Western Reserve strain of VACV (ATTC VR1354) was used as the wt virus; the L1 inducible mutant vL1Ri was previously described (Bisht et al., 2008).
Plasmids
The L1 open reading frame with 36 bp upstream of the transcription start site containing the natural promoter was amplified by PCR from viral DNA and cloned in the Zero Blunt II TOPO vector (Invitrogen, Carlsbad, CA) to construct the plasmid pL1wt. Glycine, the penultimate amino acid of the L1 open reading frame, was mutated to alanine using the Quick-change site directed mutagenesis kit (Stratagene, La Jolla, CA) and cloned in the Zero Blunt II TOPO vector to produce pL1G2A.
Western blot analysis and immunoaffinity purification
Cells were lysed in 0.2% NP-40, 10 mM Tris pH 7.4, 10 mM CaCl2 and 10 mM NaCl, containing micrococcal nuclease, complete protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, IN) and 100 mM DTT or 20 mM NEM at 4°C for 10 min. Lysates were resolved by SDS-PAGE and were electrophoretically transferred onto a nitrocellulose membrane and blocked in phosphate buffered saline, pH 7.5 containing 5% dry milk. The membranes were incubated for 1 h with a 1:1000 dilution of primary antibody, washed three times with 0.05% Tween-20, and subsequently incubation with a 1:1000 dilution of horse radish peroxidase-conjugated goat anti-mouse antibody or 1:2000 dilution of horse radish peroxidase-conjugated donkey anti-rabbit antibody. Bound antibody was detected upon incubation with SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology Inc., Rockford, IL). For immunoaffinity purification, EZview Red Protein G Affinity gel (Sigma, St. Louis, MO) was used to capture antibody-protein complexes.
Kinetics of disulfide-bond formation
BS-C-1 cells were infected with VACV at a multiplicity of 10 plaque forming units per cell in a 6-well dish. After 16 h, cells were incubated with methionine- and cysteine-free medium for 15 min. Subsequently, cells were incubated with methionine- and cysteine-free medium containing 5 mM DTT for 5 min and then pulsed with 100 μCi of [35S]methionine/cysteine-labeling mix for 5 min in the continued presence of DTT. Following the pulse, cells were incubated in medium containing excess methionine and cysteine for various times (either in the presence or absence of DTT) and then scraped into cold phosphate buffered saline containing 20 mM NEM to terminate the chase period. Cells were lysed in 1% NP-40 containing NEM and complete protease inhibitor cocktail (Roche Molecular Biochemicals). Clarified cell lysates were immunopurified as described above and analyzed by SDS-PAGE and autoradiography.
Confocal microscopy
HeLa cells were infected at a multiplicity of 10 plaque forming units per cell in the presence or absence of 50 μM IPTG. Cells in the absence of IPTG were transfected with either pL1wt or pL1G2A. After 16 h the cells were fixed with 4% paraformaldehyde, quenched with 2% glycine and permeabilized with 0.1% Triton X-100. The cells were blocked with 10% fetal bovine serum, and further incubated with MAb 7D11, PAb R180, anti-D8 MAb (MAb AB1.1 kindly provided by G.L. Smith, London, UK) or anti-calreticulin chicken PAb (Abcam, Cambridge, UK). After extensive washes the cells were incubated with Alexa Fluor 568 anti-mouse IgG, Alexa Fluor 488 anti-rabbit IgG and Alexa Fluor 647 anti-chicken IgG at a dilution 1:200 and mounted on coverslips using Prolong Gold antifade reagent with DAPI (Invitrogen). Images were collected with a Leica laser scanning confocal microscope.
Acknowledgments
We thank Norman Cooper and Catherine Cotter of NIAID for providing cell cultures. The research was supported by the Division of Intramural Research, NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldaz-Carroll L, Whitbeck JC, Ponce de Leon M, Lou H, Pannell LK, Lebowitz J, Fogg C, White C, Moss B, Cohen GH, Eisenberg RJ. Physical and immunological characterization of a recombinant secreted form of the membrane protein encoded by the vaccinia virus L1R gene. Virology. 2005;341:59–71. doi: 10.1016/j.virol.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bisht H, Weisberg AS, Moss B. Vaccinia Virus L1 protein is required for cell entry and membrane fusion. J Virol. 2008;82:8687–8694. doi: 10.1128/JVI.00852-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Helenius J, Helenius A. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. Embo J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E, Senkevich TG, Moss B. Vaccinia virus F9 virion membrane protein is required for entry but not virus assembly, in contrast to the related l1 protein. J Virol. 2006;80:9455–9464. doi: 10.1128/JVI.01149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit RC, Moussatche N, Traktman P. In a nutshell: structure and assembly of the vaccinia virion. Adv Virus Res. 2006;66:31–124. doi: 10.1016/S0065-3527(06)66002-8. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- Fogg C, Lustig S, Whitbeck JC, Eisenberg RJ, Cohen GH, Moss B. Protective immunity to vaccinia virus induced by vaccination with multiple recombinant outer membrane proteins of intracellular and extracellular virions. J Virol. 2004;78:10230–10237. doi: 10.1128/JVI.78.19.10230-10237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand AR, Cuozzo JW, Kaiser CA. Pathways for protein disulphide bond formation. Trends Cell Biol. 2000;10:203–210. doi: 10.1016/s0962-8924(00)01745-1. [DOI] [PubMed] [Google Scholar]
- Franke CA, Wilson EM, Hruby MD. Use of a cell-free system to identify the vaccinia virus L1R gene product as the major late myristylated virion protein M25. J Virol. 1990;64:5988–5996. doi: 10.1128/jvi.64.12.5988-5996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper JW, Custer DM, Schmaljohn CS, Schmaljohn AL. DNA vaccination with vaccinia virus L1R and A33R genes protects mice against a lethal poxvirus challenge. Virology. 2000;266:329–339. doi: 10.1006/viro.1999.0096. [DOI] [PubMed] [Google Scholar]
- Ichihashi Y, Oie M. Neutralizing epitopes on penetration protein of vaccinia virus. Virology. 1996;220:491–494. doi: 10.1006/viro.1996.0337. [DOI] [PubMed] [Google Scholar]
- Iyer LA, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Locker JK, Griffiths G. An unconventional role for cytoplasmic disulfide bonds in vaccinia virus proteins. J Cell Biol. 1999;144:267–279. doi: 10.1083/jcb.144.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig S, Fogg C, Whitbeck JC, Eisenberg RJ, Cohen GH, Moss B. Combinations of polyclonal or monoclonal antibodies to proteins of the outer membranes of the two infectious forms of vaccinia virus protect mice against a lethal respiratory challenge. J Virol. 2005;79:13454–13462. doi: 10.1128/JVI.79.21.13454-13462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Poxviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2905–2946. [Google Scholar]
- Ravanello MP, Hruby DE. Characterization of the vaccinia virus L1R myristylprotein as a component of the intracellular virion envelope. J Gen Virol. 1994a;75:1479–1483. doi: 10.1099/0022-1317-75-6-1479. [DOI] [PubMed] [Google Scholar]
- Ravanello MP, Hruby DE. Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virus assembly. J Virol. 1994b;68:6401–6410. doi: 10.1128/jvi.68.10.6401-6410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich T, White C, Weisberg A, Granek J, Wolffe E, Koonin E, Moss B. Expression of the vaccinia virus A2.5L redox protein Is required for virion morphogenesis. Virology. 2002a;300:296–303. doi: 10.1006/viro.2002.1608. [DOI] [PubMed] [Google Scholar]
- Senkevich TG, Ojeda S, Townsley A, Nelson GE, Moss B. Poxvirus multiprotein entry-fusion complex. Proc Natl Acad Sci USA. 2005;102:18572–18577. doi: 10.1073/pnas.0509239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, Weisberg A, Moss B. Vaccinia virus E10R protein is associated with the membranes of intracellular mature virions and has a role in morphogenesis. Virology. 2000a;278:244–252. doi: 10.1006/viro.2000.0656. [DOI] [PubMed] [Google Scholar]
- Senkevich TG, White CL, Koonin EV, Moss B. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc Natl Acad Sci USA. 2000b;97:12068–12073. doi: 10.1073/pnas.210397997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, White CL, Koonin EV, Moss B. Complete pathway for protein disulfide bond formation encoded by poxviruses. Proc Natl Acad Sci USA. 2002b;99:6667–6672. doi: 10.1073/pnas.062163799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su HP, Garman SC, Allison TJ, Fogg C, Moss B, Garboczi DN. The 1.51-A structure of the poxvirus L1 protein, a target of potent neutralizing antibodies. Proc Natl Acad Sci USA. 2005;102:4240–4245. doi: 10.1073/pnas.0501103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–1574. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- White CL, Senkevich TG, Moss B. Vaccinia virus G4L glutaredoxin is an essential intermediate of a cytoplasmic disulfide bond pathway required for virion assembly. J Virol. 2002;76:467–472. doi: 10.1128/JVI.76.2.467-472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Weisberg AS, Moss B. A Glutaredoxin, encoded by the G4L gene of vaccinia virus, is essential for virion morphogenesis. J Virol. 2000;74:9175–9183. doi: 10.1128/jvi.74.19.9175-9183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe EJ, Vijaya S, Moss B. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology. 1995;211:53–63. doi: 10.1006/viro.1995.1378. [DOI] [PubMed] [Google Scholar]